Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Self-assembly of ultra-small micelles from amphiphilic lipopeptoids†
King Hang Aaron
Lau
 *a,
Valeria
Castelletto
b,
Thomas
Kendall
c,
Jan
Sefcik
d,
Ian W.
Hamley
*a,
Valeria
Castelletto
b,
Thomas
Kendall
c,
Jan
Sefcik
d,
Ian W.
Hamley
 *b,
Mehedi
Reza
e and
Janne
Ruokolainen
e
*b,
Mehedi
Reza
e and
Janne
Ruokolainen
e
aWestCHEM/Department of Pure & Applied Chemistry, University of Strathclyde, 99 George Street, Glasgow, G1 1RD, UK. E-mail: Aaron.Lau@strath.ac.uk
bDepartment of Chemistry, University of Reading, Whiteknights, Reading, RG6 6AD, UK. E-mail: I.W.Hamley@reading.ac.uk
cEPSRC Doctoral Training Centre in Continuous Manufacturing and Crystallisation, Department of Chemical and Process Engineering, University of Strathclyde, 75 Montrose Street, Glasgow, G1 1XJ, UK
dDepartment of Chemical and Process Engineering, University of Strathclyde, 75 Montrose Street, Glasgow, G1 1XJ, UK
eDepartment of Applied Physics, Aalto University School of Science, P.O. Box 15100, FI-00076, Aalto, Finland
First published on 25th January 2017
Abstract
Poly(N-substituted glycine) “peptoids” constitute a promising class of peptide-mimetic materials. We introduce the self-assembly of lipopeptoids into spherical micelles ca. 5 nm in diameter as well as larger assemblies by varying the peptoid sequence design. Our results point to design rules for the self-assembly of peptoid nanostructures, enabling the creation of stable, ultra-small peptidomimetic nanospheres.
Peptidic systems that self-assemble into nanostructures have captured great attention due to their resemblance to proteins and other biological structures. In particular, a large literature describes peptide amphiphiles, specifically lipo-peptides, that show significant potential in biomedicine.1 Nanofibrils or worm-like micelles are the most commonly reported structural motif.1c,2 They are typically observed to arise from the self-assembly of peptide headgroups with (extended) beta-sheet structures. Spherical micelles, formed if hydrogen bonding between peptide chains is de-emphasized,3 have been less commonly reported.2b Naturally occurring lipopeptides that form micelles highlight the biomedical potential of such nanostructures.4
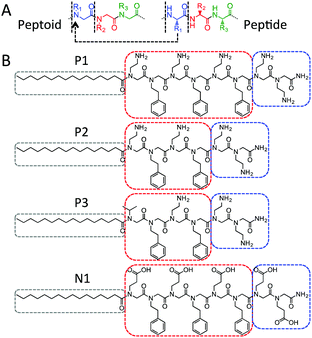
Peptoids are structural isomers of peptides with sidechain attachment shifted to the amide nitrogens (Fig. 1A).5 As a class of novel peptide-mimetic polymers, they are emerging as a versatile platform for materials applications and they possess several attractive features for biomedical applications due to the sidechain shift.5,6 First, proteolysis is essentially inhibited,5,7 which enables applications requiring resistance against biodegradation (e.g. antimicrobial peptidomimics,8 and long-term biointerfaces and biomaterials).6a,9 Second, sequence-specific peptoids may be conveniently synthesized by a “submonomer” solid phase protocol10 and bioactive sequences may be discovered from combinatorial library searches.5,11 Moreover, peptoids are conformationally flexible since they can adopt both cis- and trans-backbone conformations and they lack intra- and inter-backbone hydrogen bonding.5 A rich family of secondary structures – helices, ribbons and sheets – may be induced by focusing on sidechain selection and design,5 and these have been exploited for protein-mimetic structures,12 cell membrane penetrating helices,8a,13 and scaffolds for displaying biorecognition sequences.14
However, peptoid self-assembly into nanostructures has received relatively less attention, despite the fact that polypeptoids obtained from living polymerization could self-assemble similar to conventional block-copolymers (BCP),15 and sequence-specific peptoids have been used as models for studying conventional BCP self-assembly.16 Among the limited peptoid-specific structures reported, nanosheets appear to be a prominent morphology. An alternating hydrophilic–hydrophobic motif has been shown to form nanosheets extending tens of microns laterally.14 Hybrid oligo(peptoid–peptide) diblock amphiphiles have been shown to form hydrogels composed of nanosheets or nanofibers depending on the appended peptide sequence,17 and strips of nanosheets may curl into nano- or micro-tubules.18 Interestingly, sequence-specific control of a basic structural form such as self-assembled micelles has not been reported.
Inspired by peptide self-assembly, we coupled palmitoyl lipid “tails” to amphiphilic peptoid sequences to direct the self-assembly of the resulting lipopeptoids (Fig. 1B). The narrow cross-section of the palmitoyl chain ensures that the peptoids form a relatively large head-group. A pair of ionizable residues was also introduced at the C-terminus to enhance head-group solubility. The combined geometric and solubility head-to-tail asymmetries were expected to promote the self-assembly of small well-defined micelles with a high surface curvature.19
The main peptoid sequence has an alternating XY amphiphilic motif, where X is an ionizable residue and Y is the aromatic N-phenylethyl glycine (Npe). Npe residues have been reported to contribute to the assembly of peptoid nanosheets through hydrophobic and π–π interactions,14 and we speculated that these interactions would also stabilize any self-assembled lipopeptoid structure. P1/P2/P3 have positively (P) charged Nae (N-aminoethyl glycine) residues. The number of XY motifs is decreased from 3 in P1 to 2 in P2, to probe whether shortening the peptoid relative to the hydrophobic palmitoyl tail influences the self-assembly. P3 is the same length as P2 but the last Nae residue is replaced by Nval (the analogue of valine) to further probe the effect of increased hydrophobicity. N1 has glutamic acid analogues (Nglu) in place of Nae to create a negatively (N) charged counterpart to P1 for verifying the self-assembly of the underlying amphiphilic design.
The peptoids were synthesized using standard submonomer solid phase synthesis. In contrast to lipopeptides (a.k.a. peptide amphiphiles) with alternating amphiphilic motifs that often lead to nanofibers composed of extended beta-sheet structures,1c,20 we found that the lipopeptoids assembled into well-defined and very stable micelles.
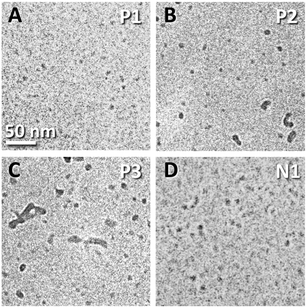
Fig. 2 shows typical cryo-TEM images of the nanostructures formed from lipopeptoids dissolved at a concentration of 1 wt% in water. The analogous sequences P1 and N1 in particular are seen to form uniform spherical nanostructures 4–6 nm in diameter (Fig. 2A and D). Similar “nanospheres” were also observed in the P2 and P3 samples, but these co-existed with larger structures varying in size and shape, especially for P3 (Fig. 2B and C).
The nanometer size, uniformity, and spherical shape of the P1 and N1 nanostructures are indicative of the formation of spherical micelles. The observation of discrete P2 and P3 structures with at least one dimension in the 10 nm range is also consistent with micellar structures. In fact, micelle formation was anticipated by the dissolution behaviour of the lipopeptoids and RP-HPLC measurements.
HPLC shows that P1 was the most hydrophilic. However, it eluted from a C18 column only at a high 57.5% acetonitrile (ACN) in water (Fig. S1, ESI†). P2 eluted at slightly higher 58.5% ACN while P3 eluted at 72% ACN. N1 was expected to appear the most hydrophobic because Nglu would not be ionized under the standard acidic HPLC conditions used (0.1% TFA – trifluoroacetic acid), and it eluted at the highest 76% ACN. Sequences eluting at such high organic phase contents would ordinarily not be soluble in water. Nonetheless, adding water to lyophilized P1, P2 and P3 immediately resulted in clear solutions even at concentrations as high as 100 mg mL−1 (see ESI†). N1 was also readily dissolved in water with the addition of a base (NaOH; to neutralize the TFA co-lyophilized after HPLC purification and to aid Nglu ionization). Such apparent solubility is consistent with the self-assembly of lipopeptoids into micelles with ionic peptoid shells that were able to sequester the hydrophobic palmitoyl cores.
Pyrene fluorescence assays provide further evidence of micelle self-assembly. Focusing on P1/P2/P3, Fig. 3 shows the critical aggregation concentration (CAC) of lipopeptoid self-assembly,3a which occurred at 0.1 wt% for P1 and P2 (i.e. 0.8 mM and 1 mM, respectively) and at ∼0.04 wt% for P3 (i.e. ∼0.4 mM). The CAC is lowest for P3, which corresponds to its later elution in HPLC. The slightly lower CAC for P2 compared to P1 indicates it is more hydrophobic, which is also consistent with P2's slightly later HPLC elution. This result also points to the relative importance of the hydrophobic palmitoyl tail (P2 has fewer peptoid residues than P1) compared to the proportion of solubilizing peptoid residues (P2 has a higher 4![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2 Nae
2 Nae![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Npe ratio than P1's 5
Npe ratio than P1's 5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3). The increasing aggregation propensity implied by the decreasing CAC from P1 to P3 could also explain the increasing appearance of larger structures across the series (Fig. 2).
3). The increasing aggregation propensity implied by the decreasing CAC from P1 to P3 could also explain the increasing appearance of larger structures across the series (Fig. 2).
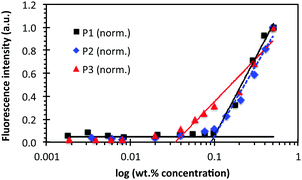 | ||
| Fig. 3 Pyrene fluorescence intensity plotted against log concentration of P1, P2 and P3. The intensities were normalized to the highest values recorded at 0.5 wt% lipopeptoid. The pyrene concentration was 1.3 × 10−5 wt% and fluorescence was measured at 373–375 nm. The breaks in the plots identify the CAC. See Fig. S3 (ESI†) for representative full spectra of the fluorimetry measurements. | ||
Small angle X-ray scattering (SAXS) further characterized the physical dimensions of the lipopeptoid assemblies. Samples above the CACs at 1, 5 and 10 wt% were measured, and all exhibited similar results. The 1 wt% dataset, corresponding to the cryo-TEM samples, is shown in Fig. 4. A simple spherical core and shell model (with shell thickness R1 and core radius R0) was used to analyze the SAXS data. Excellent fits were obtained and the structural and scattering parameters obtained are shown in Table 1.
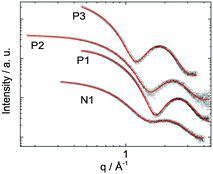 | ||
| Fig. 4 SAXS form factor data of the lipopeptoids P1, P2, P3 and N1 dissolved at 1 wt% in water (symbols) and fitting by SASfit according to a spherical core–shell model (red traces). The fitted parameters and structural dimensions are described in Table 1. | ||
The average micelle diameter Dtotal was 5.4 nm and 4.2 nm for the P1 and N1 micelles, respectively. These dimensions are in excellent agreement with the TEM results, given the resolution of the unstained cryo-TEM images and the uncertainty in the SAXS fitting. Dtotal of P2 micelles (i.e. Dtotal(P2)) is 5.2 nm and matches the fraction of spherical structures seen in cryo-TEM (Fig. 2B). The fact that the Dtotal(P2) is not higher, as might be expected from the larger structures seen in TEM, may be due to reduced contrast for this sample. Similar to P2, SAXS measured only a slightly larger Dtotal(P3) = 7.0 nm, corresponding to the fraction of smaller micelles seen in TEM (Fig. 2C). Nonetheless, the polydispersities obtained increased from 0.44 (P1) to 0.57 (P2) and then to 0.71 (P3), which is consistent with the increased morphological inhomogeneity seen in TEM images.
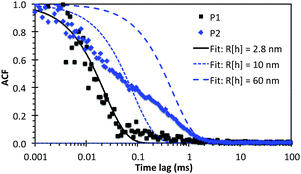
Dynamic light scattering (DLS) measurements focused on P1 and P2 to better understand the SAXS and cryo-TEM data. Although DLS is significantly more sensitive for larger particles, we have been successful in using the technique to characterize molecular clusters with hydrodynamic radii (Rh) as small as 1 nm.21Fig. 5 shows the measured normalised intensity autocorrelation functions (ACF) together with the theoretical ACFs calculated for monodisperse populations of particles with various Rh. The P1 data was best fitted using Rh = 2.8 nm (i.e. Dtotal = 5.6 nm), which is in excellent agreement with the diameters measured by both SAXS and cryo-TEM. For P2, the initial ACF decay at short τ < 0.02 ms overlaps with the P1 data, indicating the presence of micelles with diameter ∼5 nm. However, a broad shoulder extends to 2 ms, corresponding to a relatively small polydisperse population of particles with Rh ranging from 10 to 60 nm. The presence of a small population of such larger micelles (detected due to their more intense scattering) may stand out prominently in TEM images (Fig. 2C), but may not be detected by SAXS (Table 1).
The consistent micelle diameters measured by cryo-TEM, SAXS and DLS, using samples prepared separately and at multiple concentrations above the CAC, give confidence to the measured dimensions and indicate reproducible self-assembly. Indeed, P1 and P2 samples re-measured by DLS after storage for 11 weeks at 4 °C gave very similar results (average Rh = 3 nm; see Fig. S5 and Table S3, ESI†), which demonstrated that the lipopeptoid designs gave highly stable micelles.
Interestingly, the observed nanostructures are smaller than expected from the dimensions of the lipopeptoids (e.g. P1 and N1 have contour lengths = 4.8 nm).‡ A simplistic picture of spherical micelles with fully extended sequences would result in a diameter twice the contour length, i.e. around 10 nm for P1. Also, SAXS indicated the same ∼1 nm peptoid shell thickness R1 for all designs (Table 1) despite their different sequence lengths. Since it has been shown that peptoid backbones are inherently flexible, with persistence lengths ranging from 0.5 to 1 nm,22 we speculate that the peptoid sequences in the micelles are “bent” over to form relatively thin shell layers. The shell thickness would then be dictated by the same peptoid chain cross-section for all sequences (Fig. 1). Indeed, such a configuration could facilitate the ordering of the hydrophilic Nae/Nglu residues towards the water interface and the hydrophobic Npe residues towards the palmitoyl core. Thus lipopeptoid micelles with “ultra-small” diameters less than indicated by the peptoid sequence lengths could be formed.
The formation of micellar structures from all the lipopeptoids, as intended, suggests that the designed geometric asymmetry and head-to-tail hydrophobicity contrast are useful for controlling peptoid self-assembly. In comparison, previously reported lipopeptoids with lower degrees of asymmetry (using “fatter” double-tailed phospholipids and more hydrophobic residue motifs) formed spherical aggregates 80–600 nm in diameter.23 In addition, the fact that backbone hydrogen bonding is suppressed in peptoids might also enhance the importance (and conceptual simplicity) of basic geometric and hydrophobicity considerations in peptoid self-assembly. Indeed, as a counterpoint, computer simulation models of peptide self-assembly suggest that weakened hydrogen bonding favors the formation of spherical peptide micelles over beta-sheets and cylindrical morphologies.3b
In conclusion, we designed lipopeptoids with high geometric and hydrophobicity head-to-tail asymmetries by coupling palmitoyl tails to amphiphilic peptoid sequences, in order to direct peptoid self-assembly into micelles. Unprecedented ultra-small, uniform and stable spherical micelles with ca. 5 nm diameters and 0.1 wt% CAC were obtained by appropriate peptoid sequence design. The micelles resemble many globular proteins in size and could be explored as protein mimics. The small size could be due to the inherent conformational flexibility of the peptoid backbone, and could facilitate the transport of these micelles across tissues and blood vessels for drug delivery and biosensing applications. Increases in the sequence hydrophobicity were observed to lead generally to increased aggregation and polydispersity. Comparison with previous reports suggests that the basic considerations of the overall molecular shape and the distribution of hydrophobicity content could play prominent roles in peptoid self-assembly.
KHAL thanks Mikyoung Goo for her outstanding assistance with peptoid synthesis, and the Strathclyde Academic Investment Scheme and Tenovus Scotland (S15/29) for financial support. IWH thanks EPSRC for the award of a Platform grant (EP/L L020599/1). We are grateful to the ESRF for the award of beamtime (ref. MX-1769) and to Gabriele Gachin for support during the beamtime. TK thanks EPSRC and the Doctoral Training Centre in Continuous Manufacturing and Crystallisation (EP/K503289/1) for funding. Research data available at: http://dx.doi.org/10.15129/15d4068d-b268-45fd-bd6a-22b0cbae2ede.
References
- (a) M. J. Webber, E. A. Appel, E. W. Meijer and R. Langer, Nat. Mater., 2016, 15, 13–26 CrossRef CAS PubMed; (b) J. Boekhoven and S. I. Stupp, Adv. Mater., 2014, 26, 1642–1659 CrossRef CAS PubMed; (c) F. Versluis, H. R. Marsden and A. Kros, Chem. Soc. Rev., 2010, 39, 3434–3444 RSC.
- (a) T. Aida, E. W. Meijer and S. I. Stupp, Science, 2012, 335, 813–817 CrossRef CAS PubMed; (b) J. F. Miravet, B. Escuder, M. D. Segarra-Maset, M. Tena-Solsona, I. W. Hamley, A. Dehsorkhi and V. Castelletto, Soft Matter, 2013, 9, 3558–3564 RSC.
- (a) A. Dehsorkhi, V. Castelletto and I. W. Hamley, J. Pept. Sci., 2014, 20, 453–467 CrossRef CAS PubMed; (b) Y. S. Velichko, S. I. Stupp and M. O. de la Cruz, J. Phys. Chem. B, 2008, 112, 2326–2334 CrossRef CAS PubMed.
- I. W. Hamley, Chem. Commun., 2015, 51, 8574–8583 RSC.
- A. S. Knight, E. Y. Zhou, M. B. Francis and R. N. Zuckermann, Adv. Mater., 2015, 27, 5665–5691 CrossRef CAS PubMed.
- (a) K. H. A. Lau, Biomater. Sci., 2014, 2, 627–633 RSC; (b) C. Secker, S. M. Brosnan, R. Luxenhofer and H. Schlaad, Macromol. Biosci., 2015, 15, 881–891 CrossRef CAS PubMed.
- S. M. Miller, R. J. Simon, S. Ng, R. N. Zuckermann, J. M. Kerr and W. H. Moos, Drug Dev. Res., 1995, 35, 20–32 CrossRef CAS.
- (a) A. M. Czyzewski, H. Jenssen, C. D. Fjell, M. Waldbrook, N. P. Chongsiriwatana, E. Yuen, R. E. Hancock and A. E. Barron, PLoS One, 2016, 11, e0135961 Search PubMed; (b) M. L. Huang, S. B. Y. Shin, M. A. Benson, V. J. Torres and K. Kirshenbaum, ChemMedChem, 2012, 7, 114–122 CrossRef CAS PubMed.
- K. H. A. Lau, T. S. Sileika, S. H. Park, A. M. L. Sousa, P. Burch, I. Szleifer and P. B. Messersmith, Adv. Mater. Interfaces, 2015, 2, 1400225 CrossRef PubMed.
- R. N. Zuckermann, J. M. Kerr, S. B. H. Kent and W. H. Moos, J. Am. Chem. Soc., 1992, 114, 10646–10647 CrossRef CAS.
- Y. Gao and T. Kodadek, Chem. Biol., 2013, 20, 360–369 CrossRef CAS PubMed.
- B. C. Lee, T. K. Chu, K. A. Dill and R. N. Zuckermann, J. Am. Chem. Soc., 2008, 130, 8847–8855 CrossRef CAS PubMed.
- A. M. Czyzewski and A. E. Barron, AIChE J., 2008, 54, 2–8 CrossRef CAS.
- E. J. Robertson, A. Battigelli, C. Proulx, R. V. Mannige, T. K. Haxton, L. Yun, S. Whitelam and R. N. Zuckermann, Acc. Chem. Res., 2016, 49, 379–389 CrossRef CAS PubMed.
- C. U. Lee, T. P. Smart, L. Guo, T. H. Epps and D. H. Zhang, Macromolecules, 2011, 44, 9574–9585 CrossRef CAS PubMed.
- A. M. Rosales, R. A. Segalman and R. N. Zuckermann, Soft Matter, 2013, 9, 8400–8414 RSC.
- Z. D. Wu, M. Tan, X. M. Chen, Z. M. Yang and L. Wang, Nanoscale, 2012, 4, 3644–3646 RSC.
- (a) R. C. Elgersma, G. E. Mulder, J. A. W. Kruijtzer, G. Posthuma, D. T. S. Rijkers and R. M. J. Liskamp, Bioorg. Med. Chem. Lett., 2007, 17, 1837–1842 CrossRef CAS PubMed; (b) H. K. Murnen, A. M. Rosales, J. N. Jaworski, R. A. Segalman and R. N. Zuckermann, J. Am. Chem. Soc., 2010, 132, 16112–16119 CrossRef CAS PubMed.
- J. N. Israelachvili, Intermolecular and Surface Forces, Academic Press, San Diego, 3rd edn, 2011, pp. 535–576 Search PubMed.
- E. De Santis and M. G. Ryadnov, Chem. Soc. Rev., 2015, 44, 8288–8300 RSC.
- A. Jawor-Baczynska, J. Sefcik and B. D. Moore, Cryst. Growth Des., 2013, 13, 470–478 CAS.
- A. M. Rosales, H. K. Murnen, S. R. Kline, R. N. Zuckermann and R. A. Segalman, Soft Matter, 2012, 8, 3673–3680 RSC.
- (a) C.-Y. Huang, T. Uno, J. E. Murphy, S. Lee, J. D. Hamer, J. A. Escobedo, F. E. Cohen, R. Radhakrishnan, V. Dwarki and R. N. Zuckermann, Chem. Biol., 1998, 5, 345–354 CrossRef CAS PubMed; (b) Y. U. Konca, K. Kirshenbaum and R. N. Zuckermann, Int. J. Nanomed., 2014, 9, 2271–2285 CrossRef PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available: Methods and materials, as well as dissolution, HPLC and DLS data. See DOI: 10.1039/c6cc09888f |
| ‡ Calculated from 0.25 nm for every pair of methylenes and 0.35 nm per peptoid residue.14 |
| This journal is © The Royal Society of Chemistry 2017 |