Feature driven classification of Raman spectra for real-time spectral brain tumour diagnosis using sound
Ryan
Stables
a,
Graeme
Clemens
bc,
Holly J.
Butler
b,
Katherine M.
Ashton
d,
Andrew
Brodbelt
d,
Timothy P.
Dawson
d,
Leanne M.
Fullwood
c,
Michael D.
Jenkinson
e and
Matthew J.
Baker
*b
aDigital Media Technology Laboratory, Millennium Point, City Centre Campus Birmingham City University, West Midlands, B47XG, UK
bWestCHEM, Department of Pure and Applied Chemistry, University of Strathclyde, Technology and Innovation Centre, 99 George Street, Glasgow, G11RD, UK. E-mail: matthew.baker@strath.ac.uk; Twitter:@ChemistryBaker; Fax: +44 (0)1415484822; Tel: +44 (0)1415484700
cCentre for Materials Science, Division of Chemistry, University of Central Lancashire, Preston, PR12HE, UK
dNeuropathology, Lancashire Teaching Hospitals NHS Trust, Royal Preston Hospital, Sharoe Green Lane North, Preston, PR29HT, UK
eThe Walton Centre for Neurology and Neurosurgery, The Walton Centre NHS Trust, Lower Lane, Liverpool, L97LJ, UK
First published on 12th October 2016
Abstract
Spectroscopic diagnostics have been shown to be an effective tool for the analysis and discrimination of disease states from human tissue. Furthermore, Raman spectroscopic probes are of particular interest as they allow for in vivo spectroscopic diagnostics, for tasks such as the identification of tumour margins during surgery. In this study, we investigate a feature-driven approach to the classification of metastatic brain cancer, glioblastoma (GB) and non-cancer from tissue samples, and we provide a real-time feedback method for endoscopic diagnostics using sound. To do this, we first evaluate the sensitivity and specificity of three classifiers (SVM, KNN and LDA), when trained with both sub-band spectral features and principal components taken directly from Raman spectra. We demonstrate that the feature extraction approach provides an increase in classification accuracy of 26.25% for SVM and 25% for KNN. We then discuss the molecular assignment of the most salient sub-bands in the dataset. The most salient sub-band features are mapped to parameters of a frequency modulation (FM) synthesizer in order to generate audio clips from each tissue sample. Based on the properties of the sub-band features, the synthesizer was able to maintain similar sound timbres within the disease classes and provide different timbres between disease classes. This was reinforced via listening tests, in which participants were able to discriminate between classes with mean classification accuracy of 71.1%. Providing intuitive feedback via sound frees the surgeons’ visual attention to remain on the patient, allowing for greater control over diagnostic and surgical tools during surgery, and thus promoting clinical translation of spectroscopic diagnostics.
1. Introduction
Malignant gliomas are among the most lethal of cancers with a 20-year average reduction in life expectancy, the highest of any cancer and just 6% of adults with a high grade glioma (glioblastoma (GB)) survive for more than 5 years’ post-diagnosis of a malignant brain tumour.1,2 Brain tumours can be grouped into two main classes, primary and metastatic tumours. Primary brain tumours such as GB originate within the central nervous system (CNS) with tumour types after the glial cells to which they show morphological similarities.3 Around 13![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 people in the UK are diagnosed with brain cancer every year, of which, 60% are metastatic tumours, which have originated from primary cancers outside the CNS.4 The major primary tumours that metastasise to the brain are lung (50%), breast (15–25%) melanoma (5–20%) and all others (5–30%).5 The identification of the organ of origin increases the efficiency of treatment and patient survival, however in approx. 15% of metastatic cases the primary location is unknown.6
000 people in the UK are diagnosed with brain cancer every year, of which, 60% are metastatic tumours, which have originated from primary cancers outside the CNS.4 The major primary tumours that metastasise to the brain are lung (50%), breast (15–25%) melanoma (5–20%) and all others (5–30%).5 The identification of the organ of origin increases the efficiency of treatment and patient survival, however in approx. 15% of metastatic cases the primary location is unknown.6
Following detection of a brain tumour by conventional imaging modalities (e.g. MRI) a biopsy and resection is a likely course of action, particularly for high grade and metastatic cancers. Complete tumour removal during surgery is a strong indicator of recurrence-free survival with increased median overall survival for those patients undergoing macroscopic resection compared to tumour debulking and biopsy.7 The recurrence rates for Grade II and II meningiomas are 13.8% with complete resection and 46.7% for incomplete resection.8 The current process is unable to accurately identify tumour margins and definitive histopathological diagnosis is generally too time consuming for responsive action during surgery. It should also be noted that it is often not possible to remove all of a tumour since they frequently involve eloquent areas of the brain and the surgery itself would leave the patient with an unacceptable neurological deficit e.g. hemiparesis or dysphasia. There is a pressing need for a tool that can identify the origin of a tumour (e.g. primary vs. metastasis) and enable the identification of tumour margins (e.g. brain tumour vs. normal) dynamically during surgery.
Vibrational spectroscopic techniques, such as Raman and FTIR, have been shown to be sensitive to the hallmarks of cancer.9–15 They are non-destructive, simple to operate and require minimal sample preparation. Inelastically (Raman) scattered light from molecules under irradiation is wavelength-shifted with respect to the incident light by molecular vibrations. The Raman spectrum is complementary to that of IR where incident light is absorbed at the resonant frequency of a particular bond or group. Different biomolecules exhibit distinct responses to varying wavelengths of light; the resulting spectrum can be thought of as a ‘fingerprint’ or ‘signature’ of the sample. Spectroscopic analysis allows the objective classification of biological material on a molecular level.12 Several groups have investigated the use of Raman spectroscopy to discriminate brain tissue. In particular, Gajjar et al. reported the ability of Raman spectroscopy to differentiate between brain tumour and healthy brain tissue.16,17 They observed successful discrimination between different brain tumour types in their study. They reported that cancerous brain tissue could be discriminated from healthy brain tissue on low-E substrates based on spectral peaks at: 997 cm−1 (phospholipids and glucose-I-phosphate), 1077 cm−1 and 1446 cm−1 (lipids and proteins), 1241 cm−1 (amide III), 1460 cm−1 (cytosine) and 1654 cm−1 (amide I). We have previously published research investigating the sample preparation and use of Raman spectroscopy for spectral histopathology of metastatic brain cancer and primary sites of origin.17,18 This study showed the use of multivariate analysis and visual inspection of Raman spectra from brain tumour tissue samples. Using the ratio of 620 cm−1 (C–C twisting mode of phenylalanine) to 782 cm−1 (cytosine uracil ring breathing mode of nucleotide) versus 721 cm−1 (symmetric choline C–N stretch/adenine) to 620 cm−1, disease discrimination was achievable at sensitivities and specificities of 85.71–100% and 94.44–100% respectively.
Raman spectroscopy for clinical diagnostics has seen concerted development of Raman probes to identify and delineate tumour margins during surgery. Proof-of-concept studies were initially conducted upon fixed tissue, predominately Formalin Fixed Paraffin Processed (FFPP), that are routinely used for histopathological analysis; however, due to the biochemical alterations that occur as a consequence of chemical fixation, largely evident in the lipid associated spectral regions, investigation in fresh tissue is spearheading clinical implementation.19 Pioneering work has shown the applicability of fibre-optic Raman probes for intra-operative analysis of the brain20 (with recent patient preliminary trials performed during brain surgery), stomach,21 oesophagus22 and lymph nodes.23 Mahadevan-Jansen24 has reported the successful discrimination of cervical precancers using in vivo Raman spectroscopy and Krafft has recently shown the in vivo detection of metastases in mouse brain.25 Optical biopsy of breast tissue has also allowed rapid discrimination of cancerous tissues and has also derived diagnostic markers for cancer prognosis.26–28 These examples highlight the potential of Raman probes in the clinic; however, clinical implementation still remains a significant challenge. Primarily, data feedback is too slow and reliant upon interpretation of complex spectra or multivariate analysis (MVA) plots, requiring additional expertise at the point of interpretation. Although rapid spectral acquisition and automated analysis is possible, a typical diagnosis of a small spectral dataset could take up to several minutes due to the complexity and quality of the Raman spectrum. These time constraints are further amplified when larger datasets are acquired, such as those obtained in spectroscopic imaging experiments. A process that enables real-time, easy to understand feedback of the molecularly specific data to the surgeon during surgery would prove extremely useful.
In a recent study we have introduced a method for subjectively discriminating between spectra by synthesising sounds from vibrational spectroscopic data for the detection of differentiated and undifferentiated stem cells.29 In this study, we demonstrate a real-time methodology for band-wise feature extraction, sound synthesis and feedback from Raman spectra to enable the real-time discrimination of metastatic brain tumours, primary brain tumours (GB) and normal tissue. By extracting features prior to diagnosis via MVA, the analytical time constraints are reduced, and a real-time feedback is possible. Furthermore, by using sonification combined with a Raman probe, surgeons are able to analyse tissue during surgery, providing a responsive diagnostic environment for patient benefit. Providing data feedback via sound frees the surgeons’ visual attention to remain focused on the surgical resection and eases translation of Raman spectroscopy as a surgical aid.
2. Materials and methods
Our primary experiment investigates the influence of band-wise spectral feature extraction on brain tumour classification, when compared against existing dimensionality reduction methods. To do this, we compare the classification accuracies of three classifiers when presented with sub-band features and principal components. The three classifiers chosen for this experiment were a K-Nearest Neighbour classifier (KNN), a Support Vector Machine (SVM) and a Linear Discriminant Analysis classifier (LDA). Each for the classifiers were trained with the same dataset partitions and cross-validated. Additionally, in order to show the salience of the selected features and the relevance of corresponding spectral bands, we analyse the magnitude spectra and run feature selection on the extracted features. To demonstrate the efficacy of the feature-driven approach to tissue sample classification, we then use data sonification to generate audio samples, thus illustrating the technique's potential for use with in vivo auditory feedback. To do this, synthesized audio clips are generated for each of the tissue samples using a frequency modulation (FM)-synthesis technique, then subjectively evaluated by non-specialist participants via listening tests.2.1 Dataset
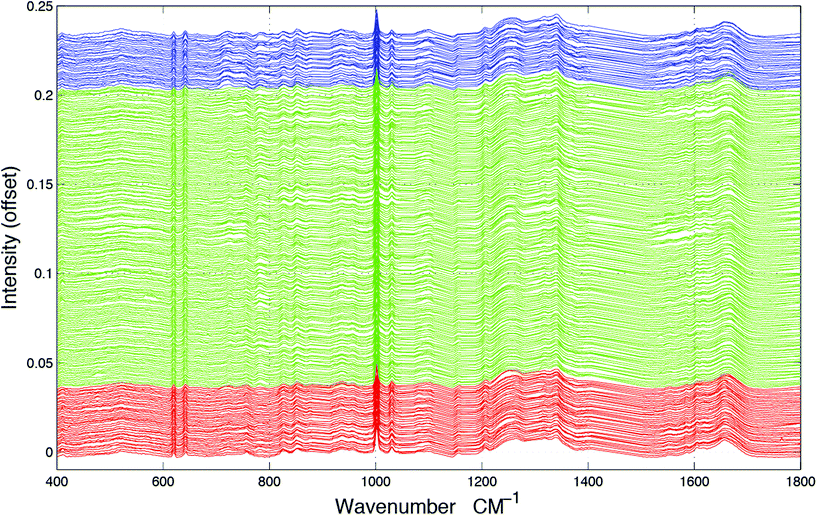
Tissue sections were obtained from FFPP tissue blocks from the Brain Tumour North West (BTNW) bio-bank and were then cut using a microtome to a thickness of 10 μm and placed onto CaF2 (Crystran, UK) substrates for spectroscopic measurements under ethical approval (BTNW/WRTB 13_01). A total of 48 tissue samples were obtained from 41 patients, 7 of whom produced normal brain samples, 5 of whom produced World Health Organisation (WHO) grade IV GB brain tumour samples and 29 of whom produced metastatic brain cancer samples. All tissue sections were placed onto CaF2 substrates were de-waxed using 3 × 5 minute baths of Histoclear, followed by 3 × 5 minute baths of ethanol. Images of the tissue can be seen in our previous study.18Raman spectroscopic measurements were carried using a Horiba Jobin-Yvon LabRAM HR800 spectrometer, with an air-cooled CLDS 785 nm laser, combined with a single edge filter (cut off 100 cm−1 and an output power of 300 mW). Spectra were acquired using a 0.75 numerical aperture ×60 objective (LUMPlanFLN, Olympus) immersion lens with the confocal hole set to 100 μm for spectral acquisition. Immersion Raman spectroscopy was carried out by submerging the tissue placed onto CaF2 substrates into deionised water during spectral collections. In total, 952 spectra were collected from the tissue set and within-class samples were averaged in groups of 4 to mitigate against noise. For the normal tissue samples, 157 samples were averaged to 39 spectra, the GB samples were averaged from 127 to 31 spectra and the metastatic samples were averaged from 668 to 167 samples, resulting in a dataset of 237 spectra, as shown in Fig. 1.
2.2 Data preprocessing
Pre-processing was carried out on the raw data using LabSpec 6 spectroscopy software suite (HORIBA scientific) and SpecToolbox: an empirically developed Matlab toolbox, made especially for the processing and analysis of vibrational spectroscopy data. The raw spectra exhibit a low frequency oscillating band, which the Raman bands sit on, often referred to as the spectral background. This is considered to be a direct result of morphology dependent scattering of the incident light and Raman lines that cause non-collimated entry into the spectrometer as stray light when using lasers above 500 nm. Although the use of an immersion lens reduced the light scattering phenomenon through liquid to tissue, producing a better refractive index matching than air to tissue, low frequency spectral backgrounds were still present in the raw data recorded using the immersion lens. To correct for this, LabSpec 6 software was used to subtract a fitted fifth order polynomial from each recorded spectrum (5th order polynomial fit and 7 points of smoothing), producing flat baselines for all data recorded, which is essential when comparing spectra using multivariate analysis. Paraffin Raman peaks situated at 882–912 cm−1, 1051–1071 cm−1, 1115–1143 cm−1, 1163–1187 cm−1, 1284–1305 cm−1 and 1407–1501 cm−1 were also removed from the spectra to account for any residual paraffin remaining in the FFPP tissue.14 Data was normalised before spectral analysis so as to ensure that there is a commonality between the spectra being compared through adjusting each peak in each spectrum to its own internal standard. In this case vector normalisation was chosen, where each wavenumber absorption variable of the spectrum is squared and summed, then divided by the square root of the total, thus scaling all spectra.2.3 Feature extraction and variable ranking
Recent studies involving the spectroscopic analysis of pathological samples adopts a linear dimensionality reduction approach to feature subset derivation as a precursor to classification using a supervised learning algorithm.18,30,31 Generally, this includes the projection of datapoints in a Raman spectrum onto a lower dimensional subspace using a technique such as PCA coupled with a kernel-based classifier such as a support vector machine (SVM). Due to the limitations of real-time analysis, we investigate alternative feature derivation techniques in an attempt to reduce the computational load, thus providing us with a system capable of reacting to changes in cell-type by providing auditory feedback to the user with minimal delay time. To do this we propose the extraction of the most discriminatory characteristics from each spectrum using common statistical techniques and domain knowledge. This involves the extraction of sub-band features from each of the spectra in our dataset, followed by an analysis of feature saliency using an information-gain-based feature selection algorithm.In order to capture the discriminatory characteristics of each spectrum, we employed sub-band decomposition, where the data in each band represents a region of spectral interest bounded by two predefined bins. The bands were selected subjectively, based on their association with a predefined molecular vibration. The global characteristics of each spectrum were calculated using broadband features taken from all available wavenumbers, with a lower bound of 400 cm−1 and an upper bound of 1800 cm−1. In order to remove redundant information, statistical descriptors were taken from each band with the intention of representing the spectral shape and energy in a low-dimensional form. The spectral energy of each band was estimated using a peak measurement and Root Mean Squared (RMS) Energy. To represent the spectral shape of each band, standard statistical moments were extracted; these include spectral centroid, skew and kurtosis. Here, the centroid (given in eqn (1)) acts as a weighted mean of the spectral band, hence representing a central point.
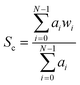 | (1) |
The skew (given in eqn (2)) defines the asymmetry of a band.
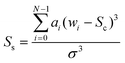 | (2) |
The kurtosis (given in eqn (3)) describes the extent to which a band exhibits a central peak.
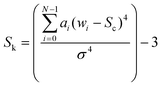 | (3) |
In each of the equations, ai denotes the ith Raman coefficient, wi represents the ith wavenumber and σ is the standard deviation of the distribution. Once extracted, the features are vector normalised to allow for transferability and comparisons are made using paired inter-band ratios. This allows us to not only analyse the behaviour of single-band characteristics over each target class, but it allows us to preserve any first order interdependencies that may exist between pairs of bands. To investigate the salience of each feature, the single band and inter-band features are concatenated to form a feature matrix and variable ranking is applied using information gain. This involves comparing the distribution of each feature, taken across all training samples for a given target class with each of the other target classes for the same feature. The result is a list of features, ranked by their saliency with the target class vector. We can then apply a threshold to remove features that fall below a desired level of salience, leaving us with a lower-dimensional representation of the spectra. Once this is done, we evaluate the performance of the highest scoring features using a non-linear classifier and compare them to existing methods of feature derivation.
2.4 Classification
In order to evaluate the performance of sub-band feature extraction, three classifiers (KNN, SVM and LDA) are trained with the first ten principal components (chosen as they retain >90% of the variance in the data), then with empirically derived features. The prediction errors are then compared including the sensitivities and specificities of each model. The first model KNN is an instance-based classifier, selected for its simplicity. The technique uses Euclidean distance to map a datapoint to a tissue group during a testing phase, based on the corresponding classes of the K-nearest datapoints. In our experiment, we optimise K to 5 by running an iterative error reduction procedure with the labelled data. Secondly we use an SVM classifier with a Radial Basis Function (RBF) kernel, a technique that has proven to be successful with spectroscopic data, when trained with spectral principal components.3,30,31 The cost (C) and gamma (γ) parameters of the RBF kernel were optimised via an iterative training procedure and empirically set to 362.03 and 0.003 respectively. The final classification method uses LDA, a form of multivariate analysis that attempts to maximise the inter-class variance based on a-priori knowledge of class distributions. LDA has been shown to be particularly effective for the classification of Raman.18,32,33 With LDA classification, the preceding dimensionality reduction stage is judged to be particularly important due to common issues with over-fitting.18,33In the proposed topology, the PCA phase is compared to sub-band feature extraction by using both types of data as a precursor to the model training phase. In the PCA-based models, the number of loadings are determined by the preservation of >90% variance in the data, resulting in 10 dimensions. In the feature extraction model, we derive an initial feature matrix of 115 variables, which is subsequently reduced to 45 after the application of variable ranking. To compare the performance of each classifier, 10-fold cross validation was used, where each test and training partition was used consistently across all three classifiers, with both the principal component and feature-based input types.
2.5 Sonification
In order to demonstrate a potential application of the feature-driven approach, we simulate the conditions of real-time auditory feedback by using a limited number of discriminatory features (defined by the variable ranking stage) to control the parameters of a FM synthesizer. By controlling the parameters in this way, we are able to provide variations in sound timbre based on the tissue samples being analysed. Due to the multi-dimensional nature of the spectral feature data, and the low number of parameters available for FM synthesis, the mapping process between spectral and auditory parameters is non-trivial. With a low number of features we are able to provide direct mappings to parameters such as carrier frequency, modulation frequency and modulation index, however the optimal number of features is generally higher, resulting in a trade-off between classification accuracy and complexity.To verify the performance of the sonification technique, subjective listening tests were implemented whereby listeners were asked to classify 30 samples based on their dissimilarity. In total, 25 listeners aged between 18–40 participated in the listening test, all of whom had normal hearing and some experience with either spectroscopic analysis or medical diagnostics. Persistent samples were presented in pairs, and were randomly selected from a dataset of 237 samples. The samples were chosen in equal quantities from one of three classes: normal, metastatic and GB. The presented pairs could be taken from different classes or from the same class, giving an indication of both inter-class and intra-class dissimilarity. Participants were asked to listen to each pair of samples and provide a similarity rating between 0 < n < 100, where 0 is very dissimilar and 100 is very similar. The results were then compared to evaluate the technique's capability of generating samples with sufficient perceptual variability.
To aid in audible discrimination, a training stage was presented to the group, in which a randomised set of labelled samples were available to the listener, with exposed class groupings. After this, the samples were removed and the participants were presented with unlabelled data, which they were asked to partition into 3 groups of potentially varying size. Participants were not made aware of class representations and were asked to assign groupings based on relative similarity as opposed to the relationship to the training data.
3. Results & discussion
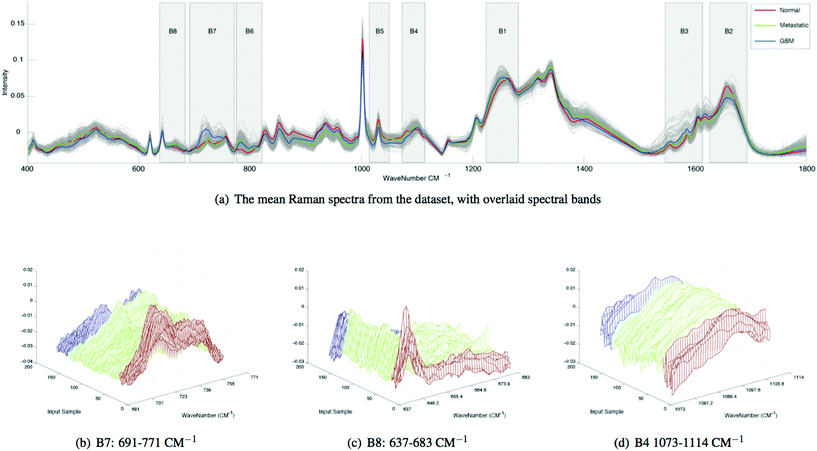
We extract the aforementioned spectral features from 8 sub-bands in each spectrum and compare the bands using ratios. The spectral sub-bands chosen for this experiment, along with their molecular representations are presented in Table 1. Here, the bands are empirically derived based on the following groupings of molecular vibrations: within B1, beta-sheet structural proteins can be observed between ∼1240–1260 cm−1, alpha-helix structural proteins can be observed between ∼1260–1280 cm−1 and P![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O asymmetric stretching vibrations due to nucleic acids can be observed at ∼1220–1250 cm−1. Within B2, alpha-helix structural proteins can be observed at ∼1650–1665 cm−1 and beta-sheet structural proteins can be observed between ∼1665–1680 cm−1. Within B3, nucleic acids and amino acids such as tryptophan can be observed between ∼1575–1580 cm−1, Amide III can be observed around ∼1550 cm−1 and other amino acids are evident at ∼1610 cm−1. Within B4, P
O asymmetric stretching vibrations due to nucleic acids can be observed at ∼1220–1250 cm−1. Within B2, alpha-helix structural proteins can be observed at ∼1650–1665 cm−1 and beta-sheet structural proteins can be observed between ∼1665–1680 cm−1. Within B3, nucleic acids and amino acids such as tryptophan can be observed between ∼1575–1580 cm−1, Amide III can be observed around ∼1550 cm−1 and other amino acids are evident at ∼1610 cm−1. Within B4, P![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O symmetric vibrations from nucleic acids or cell membrane phospholipids can be observed at ∼1080 cm−1 and nucleic acids are evident between ∼1080–1114 cm−1. Within B5 amino acids such as phenylalanine have been attributed to ∼1030 cm−1. Within B6, ring breathing vibrations from nucleic acids can be observed at ∼782 cm−1. Within B7, choline (group (H3C)N+) can be attributed to vibrations at ∼720 cm−1 and amino acids such as tryptophan are evident between ∼740–756 cm−1. Within B8, amino acids such as tyrosine and C–C twisting vibrations from nucleic acids cause vibrations around ∼642 cm−1 and nucleic adios are evident between ∼650–680 cm−1.
O symmetric vibrations from nucleic acids or cell membrane phospholipids can be observed at ∼1080 cm−1 and nucleic acids are evident between ∼1080–1114 cm−1. Within B5 amino acids such as phenylalanine have been attributed to ∼1030 cm−1. Within B6, ring breathing vibrations from nucleic acids can be observed at ∼782 cm−1. Within B7, choline (group (H3C)N+) can be attributed to vibrations at ∼720 cm−1 and amino acids such as tryptophan are evident between ∼740–756 cm−1. Within B8, amino acids such as tyrosine and C–C twisting vibrations from nucleic acids cause vibrations around ∼642 cm−1 and nucleic adios are evident between ∼650–680 cm−1.
| Band no | Molecular assignment | Region bounds (cm−1) | Contribution |
|---|---|---|---|
| B1 | Amide III (proteins) and nucleic acids (DNA and RNA) | 1224–1282 | 0.424 |
| B2 | Amide I (proteins) | 1626–1693 | 0.429 |
| B3 | Nucleic acids and amino acids | 1546–1613 | 0.413 |
| B4 | Nucleic acids and phospholipids | 1073–1114 | 0.440 |
| B5 | Amino acids (proteins) | 1014–1050 | 0.425 |
| B6 | Nucleic acids | 775–821 | 0.403 |
| B7 | Phospholipid and amino acids | 691–771 | 0.476 |
| B8 | Nucleic acids and amino acids | 637–683 | 0.461 |
3.1 Feature representation
Components in B7, specifically around 721 cm−1 tentatively assigned to symmetric stretching vibration of choline group (H3C)N+ may provide information regarding the disease status of the sample.34,35 Choline makes up the head group of phosphatidylcholine, an essential phospholipid abundantly found in eukaryotic cell membranes.15,36–39 Previous studies using magnetic resonance spectroscopy (MRS) have shown choline content to increase in GB tissue and all primary brain tumours. The increase in choline signal from GB and primary brain tissue has been thought to be attributed to increased cell density as a result of increased cellular proliferation, and over-expression of choline transporters and choline kinase enzymes40–43 It is important to note, that due to the use of FFPP tissue in this study, any lipid related peaks should be tentatively interpreted due to the significant impact chemical fixation has on these spectral regions.In addition to the intensity around 721 cm−1 in B7, some of the metastatic spectra and all GB spectra are shown to have an increase in signal intensity in the 740–760 cm−1 frequency range (also encapsulated by B7). Activity around 750 cm−1 can be associated with the essential amino acid known as tryptophan. Previous studies have already documented the role tryptophan has on primary gliomas; primary brain tumours showing high levels of tryptophan uptake using the alpha-[(11)C] Methyl-L-tryptophan (AMT) as a PET tracer combined with magnetic resonance imaging (MRI).44 Similarly, Kamson et al. suggest that the AMT-PET tracer coupled with MR imaging has the potential to differentiate between high-grade gliomas and brain metastases, something which is difficult using conventional MRI.45,46 An increase in spectral intensities from the Raman bands situated around 750 cm−1 (B7) and between 1578–80 cm−1 (B3), from spectra recorded from GB tissue, suggests agreement between our data and studies using the AMT-PET tracer to distinguish GB brain tumours, indicating increased tryptophan levels. However, a Raman band situated around 750 cm−1 may also be associated with molecular vibrations from lactic acid.
Activity around 782 cm−1 in B6 can be tentatively assigned to ring breathing vibrations from nucleotides of DNA and RNA (uracil, thymine and cytosine ring breathing vibrations).15,38 An increase in Raman scattering intensity in this region has previously been associated with a greater concentration of nucleic acids. An increase in nucleic acids present in cancerous tissue can again be correlated with an increase in cell density as a consequence of increased cellular proliferation.18,47,48 This may also explain why spectra recorded from cancerous tissue samples show increases in signal from Raman bands situated at ∼650 (B8), ∼1100 (B4), 1220–1250 (B1) and ∼1575 cm−1 (B3). Vibrations around B3 have previously been attributed to vibrations of molecules from nucleic acid molecular structures.48 A study by Wang et al., investigating cancerous bladder tissue with Raman spectroscopy, also showed intensity around the 782 cm−1 region in B6 to be significantly elevated from cancerous tissues when compared against normal tissue spectra.49
Spectral differences can also be observed at Raman bands attributed to proteins. Fig. 2 and 3 both show that cancerous tissue spectra have a different Raman band structure to the normal tissue spectra from the Amide III band (B1). Here, cancerous Raman spectra exhibit an increased intensity between 1220–1260 cm−1, shifting the overall central energy position of the band to ∼1255 cm−1, whereas normal spectra have greater spectral intensity at ∼1265–1270 cm−1, shifting the central position of the Raman band to a higher vibrational frequency. These shifts are captured using the spectral centroid feature, potentially explaining the salience of features 1 and 4 (the centroid B1/B4 and the centroid of B1 respectively), as listed in Table 2. This may suggest that both metastatic and GB brain tissue has an increase in concentration of beta-structural proteins than alpha-helix structural proteins. The increased variance depicted in metastatic and GBM tissue samples in Fig. 3, may be indicative of augmented biological variability that would be expected in a proliferating tumour, which is effectively defined as uncontrolled and therefore variable growth characteristics.
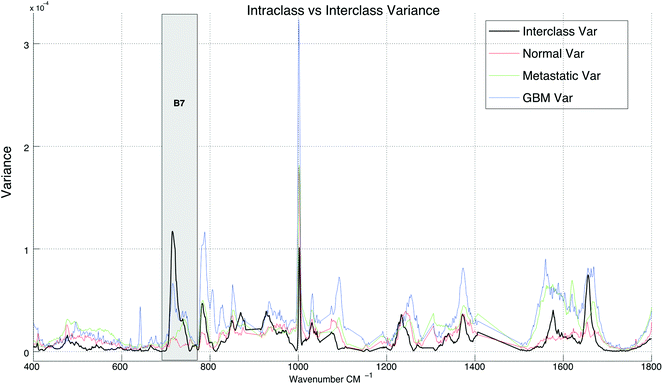 | ||
| Fig. 3 A comparison between interclass and intraclass variance exhibited by the spectra across all three tissue sample groups. | ||
| Rank | Feature | Band no(s) | Ratio | FScore | Rank | Feature | Band no(s) | Ratio | FScore | Rank | Feature | Band no(s) | Ratio | F Score |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | Centroid | B1/B4 | 1 | 0.611 | 18 | peakFreq | B2/B7 | 1 | 0.512 | 35 | peakFreq | B7/B8 | 1 | 0.456 |
| 2 | peakAmp | B2/B8 | 1 | 0.6 | 19 | peakAmp | B8 | 0 | 0.51 | 36 | peakAmp | B7/B1 | 1 | 0.455 |
| 3 | peakFreq | B4/B7 | 1 | 0.59 | 20 | peakFreq | B5/B7 | 1 | 0.494 | 37 | Centroid | B7/B8 | 1 | 0.453 |
| 4 | Centroid | B1 | 0 | 0.584 | 21 | peakFreq | B7/B3 | 1 | 0.493 | 38 | RMS | B2/B8 | 1 | 0.448 |
| 5 | peakAmp | B8/B2 | 1 | 0.579 | 22 | peakAmp | B7/B3 | 1 | 0.488 | 39 | Kurtosis | B6/B2 | 1 | 0.439 |
| 6 | peakFreq | B7/B2 | 1 | 0.567 | 23 | Centroid | B3/B4 | 1 | 0.479 | 40 | Centroid | B4/B6 | 1 | 0.437 |
| 7 | peakAmp | B5/B8 | 1 | 0.564 | 24 | peakAmp | B3/B7 | 1 | 0.477 | 41 | Kurtosis | B2/B6 | 1 | 0.433 |
| 8 | peakAmp | B2/B7 | 1 | 0.554 | 25 | Centroid | B4/B3 | 1 | 0.477 | 42 | RMS | B2/B7 | 1 | 0.428 |
| 9 | peakFreq | B7/B4 | 1 | 0.551 | 26 | peakFreq | B7/B5 | 1 | 0.474 | 43 | peakFreq | B8/B7 | 1 | 0.427 |
| 10 | peakAmp | B8/B5 | 1 | 0.549 | 27 | peakFreq | B1/B4 | 1 | 0.474 | 44 | peakFreq | B1/B3 | 1 | 0.419 |
| 11 | peakAmp | B7 | 0 | 0.547 | 28 | peakFreq | B7 | 0 | 0.474 | 45 | RMS | B6/B5 | 1 | 0.417 |
| 12 | Centroid | B4 | 0 | 0.536 | 29 | Centroid | B7 | 0 | 0.469 | 46 | peakAmp | B1/B2 | 1 | 0.414 |
| 13 | Centroid | B4/B7 | 1 | 0.535 | 30 | RMS | B7/B2 | 1 | 0.465 | 47 | RMS | B5/B6 | 1 | 0.414 |
| 14 | peakAmp | B7/B5 | 1 | 0.53 | 31 | RMS | B5/B8 | 1 | 0.464 | 48 | peakAmp | B2/B1 | 1 | 0.414 |
| 15 | Centroid | B2/B7 | 1 | 0.521 | 32 | peakFreq | B4/B1 | 1 | 0.463 | 49 | RMS | B1/B5 | 1 | 0.413 |
| 16 | Centroid | B5/B7 | 1 | 0.516 | 33 | RMS | B7/B5 | 1 | 0.46 | 50 | RMS | B8/B5 | 1 | 0.413 |
| 17 | peakFreq | B3/B7 | 1 | 0.513 | 34 | peakFreq | B6/B4 | 1 | 0.458 |
Our data also shows spectral differences around the amide I and II Raman bands (B1 and B2) when comparing diseased against normal tissue spectra. The spectral differences seen in B1 are particularly interesting, with metastatic cancerous tissue shifting to a higher frequency; the central energy position of the amide I band of metastatic cancerous tissue being ∼1670 cm−1, whereas for GB and normal tissue spectra the centroid of the amide I band is around 1655 cm−1. From Table 2, an amide I Raman band with a frequency of ∼1670 cm−1 suggests a protein structure conformational change as a result of disease, with the metastatic amide I Raman band showing a greater concentration of beta-sheet structural proteins than both GB and normal tissue, which are shown to have primarily alpha-helix structural proteins present in their tissue.
Differences in conformational protein structure have been described before when using vibrational spectroscopy to differentiate between diseased and healthy tissue.50 Gniadecka et al. also found similar spectral differences between cancerous and normal tissue spectra from the amide I band.51 A further study by Gajjar et al. also showed that an increase in Raman signal intensity between 1230–1250 cm−1 (B1) and changes to amide I secondary structure were important spectral differences when distinguishing normal and cancerous brain tumour tissue.17 Importantly, the study showed that in cancerous tissue there is a shift in the position of the amide I Raman band position, moving towards to a higher frequency position when compared against normal tissue spectra.
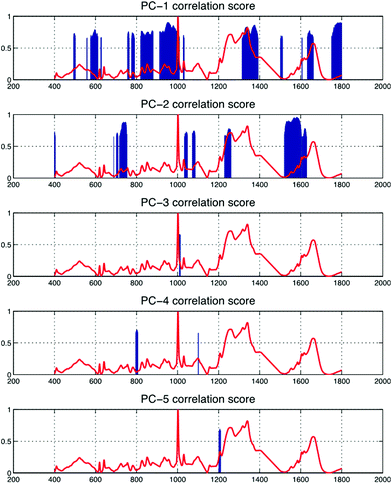
In Table 1, band-wise contribution is calculated by taking the mean feature selection score from features derived from each band (including ratios) this is on a 1 to negative 1 scale. The table shows the band with the highest mean score is B7 (consisting predominantly of phospholipids and amino acids), which has a value of 0.476 and the band with the least impact overall is B6, which consists primarily of nucleic acids. The bands with the highest contribution are illustrated in Fig. 2, in which subtle intra-class variances can be observed between the spectra from each tissue group. This is reinforced in Fig. 3, in which the interclass variance for B7 is particularly high when compared with the intraclass variance across each tissue group. This property suggests that the band could yield discriminatory features for classification due to positive class separability. In addition to this, B7 is particularly highly correlated with the 1st principal component of the magnitude spectrum (Fig. 4). This suggests that the band represents a significant portion of the variance within the data as the PCA algorithm derives new features by ranking eigenvectors of the covariance matrix. Similarly, PC 2 is positively correlated with B1 and B3, PC3 is positively correlated to B5, PC4 is positively correlated to B6 and PC5 is positively correlated to B1. From Fig. 3 it is also evident that metastatic and GBM samples exhibit an increased degree of variance within their spectra, in comparison to control samples.
Table 2 shows the 50 highest ranked features from the feature selection investigation. Here it is clear that peak location (peakFreq), peak amplitude (peakAmp) and spectral centroid (centroid) were salient descriptors, particularly when extracted from bands B1, B4 and B7. 88% of the highest 50 features were inter-band ratios, suggesting that the interaction between spectral regions is an important factor in discriminating between classes. This is significant, given that linear dimensionality reduction techniques such as PCA do not necessarily model complex interactions between spectral bins.
In order to evaluate the salience of the sub-band feature extraction approach, we compare the model's classification accuracy, including sensitivity and specificity to other common protocols such as the reduced dimensionality model presented in recent studies using an SVM with PCs taken directly from the spectral coefficients,31 or using an LDA-based classifier.52 To do this, 10-fold cross validation was applied to the dataset and the PCs were substituted for ranked feature vectors. Overall, this significantly improved the accuracy of both the KNN and the SVM-based classification techniques, and had a small negative effect on the LDA-method. For the KNN classifier, the mean accuracy increased by 25% from 66.02% to 91.02%, with a P-value of 0.00001. The feature extraction approach improved the SVM classification accuracy by 26.25% from 70.76% to 97.01%, with a p-value of 0.000001. Finally, the mean classification accuracy of the LDA classifier dropped from 96.54% to 95.38% (−1.16%), with a p-value of 0.193, thus the difference is deemed to be negligible. The results of the classification experiment are presented in Fig. 5.
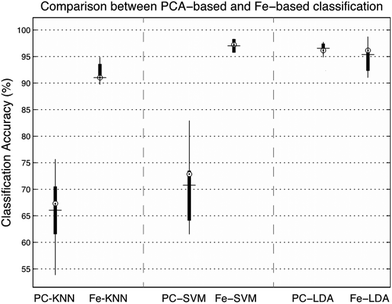 | ||
| Fig. 5 Comparison between PCA and feature-driven classification, measured across three classifiers using 10-fold cross validation, with randomly assigned training and testing test partitions. | ||
To further examine the effects of feature extraction on tissue classification, the sensitivity (rate of correctly classified positive predictions) and specificity (rate of correctly classified negative predictions) were measured across each tissue group, show in Table 3. The results show that both sensitivity and specificity are generally improved through feature extraction for the KNN and SVM based classifiers, and slightly reduced using LDA. For the KNN technique, the mean improvement is 28.21% for sensitivity and 14.07% for specificity, for SVM, the mean improvement is 57.06% for sensitivity and 32.96% for specificity and for LDA the mean reduction in sensitivity is 3.32% and the mean reduction in specificity is 1.78%. When measured across all three types of classification, the class with the highest mean sensitivity is the metastatic group with 91.36% accuracy, and the class with the highest specificity is the GB tissue group, with 96.19% accuracy.
| Method | Class 1: Normal | Class 2: Metastatic | Class 3: GBM |
|---|---|---|---|
| 2*PC-KNN | |||
| Sens | 60.34 | 92.85 | 35.67 |
| Spec | 92.55 | 58.78 | 94.8 |
| 3*Fe-KNN | |||
| Sens | 94.48 | 92.39 | 86.63 |
| Spec | 94.82 | 96.73 | 96.79 |
| 3*PC-SVM | |||
| Sens | 13.03 | 68.49 | 23.84 |
| Spec | 83.32 | 18.81 | 87.92 |
| 3*Fe-SVM | |||
| Sens | 91.33 | 94.64 | 90.59 |
| Spec | 97.19 | 93.78 | 97.96 |
| 3*PC-LDA | |||
| Sens | 92.94 | 99.82 | 84.79 |
| Spec | 98.72 | 92.32 | 99.78 |
| 3*Fe-LDA | |||
| Sens | 90.69 | 99.98 | 76.91 |
| Spec | 98.75 | 86.82 | 99.9 |
Overall, the performance of feature-based classification is promising, with a significant improvement in classification evident in KNN and SVM techniques (+25% and +26.25% increase in classification accuracy) and a negligible decrease in accuracy for the PC-LDA technique (−1.16%). This suggests that the process of extracting statistical attributes from each Raman spectrum provides a suitable alternative to reducing its dimensionality using PCA. This means that not only are we able to rapidly generate inputs to a classifier, but we are also able to expose more information regarding the underlying molecular contributions. This is due to the relative ambiguity of each principal component when compared against the spectral shape descriptors, derived using statistical moments.
3.2 Data sonification
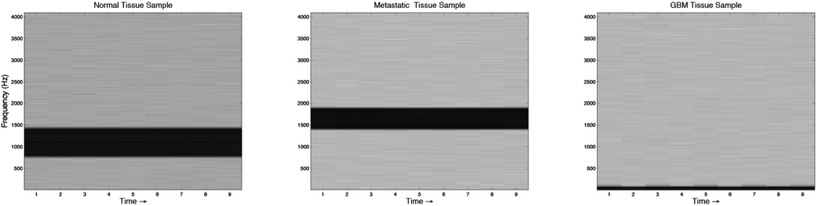
Sonification was performed by mapping the top five features from the feature extraction experiment to parameters of an FM Synthesizer. Here, the B1/B4 centroid, B2/B8 peak amplitude, B2/B7 peak frequency, B1 centroid and B8/B2 peak amplitude were used to control the f0, modulation index, modulation frequency, decay time, amplitude and sample-length parameters of the FM-synthesizer respectively. Using these features, the audio samples generated using the technique were intended to have different sound timbre between tissue groups and similar timbre within tissue groups. An example audio clip from each of the groups is shown in Fig. 6, this illustrates the sound spectra of each audio sample using a spectrogram representation.To subjectively evaluate the synthesized audio samples, participants were asked to discriminate between three sample groups, based on the timbre of each sample. The results from the test show that the mean classification accuracy was 71.1%, with a mean variance of 3.4% measured across participants (Fig. 7). The tissue type that was easiest to audibly classify was the metastatic group, which had a mean classification accuracy of 86.7%, this was followed by the GB group, which had a mean score of 72.2% and the normal group, which had a mean classification accuracy of 54.4%, this suggests the metastatic is significantly easier to discriminate, with a p-value of 0.000018.
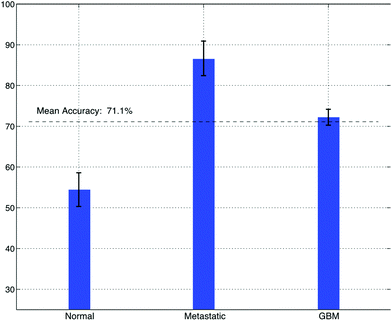 | ||
| Fig. 7 The accuracy of brain cancer identification from a test cohort of 20 people, using only sound timbre as a discriminatory aid. | ||
In order to generate audible feedback, thus providing clinicians and surgeons with multimodal feedback, potentially in real-time, the features with the highest contribution were mapped to the parameters of an FM synthesizer. To do this, the centroid of B1/B4, peak amplitude of B2/B8, peak frequency of B4/B7, centroid of B1 and peak amplitude of B8/B2 were mapped to the fundamental frequency, modulation frequency, modulation index, envelope (central peak), and loudness. Although the reduced feature subset is able to yield accuracies of (88.99% SVM) and (71.35% LDA), the subjective classification accuracy based on the sonified samples is comparatively low at 71.77%. Whilst this is not a significant reduction, it suggests there is scope for improvement to either the feature mapping process, potentially involving the integration of a higher-dimensionality feature subset to the synthesis parameter space, or a modification to the sonification methodology. The subjective nature of this approach may contribute to this level of performance, due to the effects of age, sex, ear sensitivity as well as emotional impacts that will affect the performance of each listener. A larger study interrogating a wider cohort may approach this issue and improve classification levels. This future study should also aim to address the users of this auditory feedback approach, primarily neurosurgeons, and thus the features of this demographic should be matched accordingly.
4. Conclusion
We have shown improvements in classification of Raman spectra from tissue using a feature-driven technique with a mean improvements classification accuracy of a KNN classifier by 25% and an SVM classifier by 26.25%, when used as an alternative to spectral dimensionality reduction (PCA). The technique is also shown to perform similarly to PCA, when used in conjunction with an LDA classifier, exhibiting a negligible increase in mean error (1.16%).As feature extraction allows us to observe direct relationships between the classifier inputs and the Raman bands, we are able to hypothesise that there are relationships between molecular groupings and the class assignment of samples. In particular, the variance in B7 represents changes in choline and tryptophan and intensity in B6 can be attributed to ring breathing vibrations from nucleotides of DNA and RNA. Our results also correlate with previous literature, suggesting that B3 exhibits tissue-class variation due to varying levels of tryptophan between GB and metastatic cancers.46
As an application of the feature extraction technique, we show that sonification can be used to create audio samples from a reduced subset of the extracted features using FM synthesis, thus enabling auditory feedback in near real-time providing potential opportunities for impact in tumour tissue border detection. After implementing listening tests with 25 participants, the mean subjective classification accuracy was 77.1%. The features were selected via a variable ranking stage (using information gain) and yielded a mean accuracy of 80.17% when measured across all three classifiers.
This investigation presents an alternative method of deriving classifier input-data from Raman spectra and demonstrated that it is suitable for the sonification of cancerous and noncancerous tissue samples. Our results suggest that a feature selection approach as opposed to a dimensionality reduction, both aids classification efficacy, but also reduces computational burden. In doing so, in vivo spectroscopic diagnosis in a surgical environment is increasingly achievable. By depicting that this diagnostic output can also be converted to auditory feedback in an effective manner, we illustrate a potential translational technique that may aid a clinician during endoscopic procedures.
Acknowledgements
The authors acknowledge the support from Rosemere Cancer Foundation, Brain Tumour North West, EPSRC, the Defence Science & Technology Laboratory (DSTL) and the Sydney Driscoll Neuroscience Foundation for funding.References
- N. G. Burnet, S. J. Jefferies, R. J. Benson, D. P. Hunt and F. P. Treasure, Br. J. Cancer, 2005, 92(2), 241–245 Search PubMed.
- Cancer research UK: Statistics and outlook for brain tumours. http://www.cancerresearchuk.org/about-cancer/type/brain-tumour/treatment/statistics- and-outlook- for-brain-tumours, accessed: 10–08-2015.
- J. R. Hands, K. M. Dorling, P. Abel, K. M. Ashton, A. Brodbelt, C. Davis, D. Dawson, M. D. Jenkinson, R. W. Lea, C. Walker and M. J. Baker, J. Biophotonics, 2014, 7(3–4), 189–199 CrossRef PubMed.
- Brain Research Trust: About Brain Tumours, http://www.brt.org.uk/brain-tumours, accessed: 10-08-2015.
- R. Soffieti, P. Cornu, J. Y. Delattre, R. Grant, F. Graus, W. Grisold, J. Heimans, J. Hildebrand, P. Hoskin, M. Kallijo, P. Krauseneck, C. Marosi, T. Siegal and C. Vecht, Brain metastases, in European Handbook of Neurological Managments, ed. N. E. Gilhus, M. P. Barnes and M. Brainin, Wiley-Blackwell, 2nd edn, 2011, ch. 1, pp. 437–445 Search PubMed.
- F. M. BergnercN, B. Romeike, R. Reichart, R. Kalff, C. Krafft and J. Popp, European Conference on Biomedical Optics, 2011, 80870X, Optical Society of America.
- R. Rampling, A. James and V. Papanastassiou, J. Neurol., Neurosurg. Psychiatry, 2004, 75(Suppl. 2), ii24–ii30 Search PubMed.
- K. Violaris, V. Katsarides, M. Karakyriou and V. Sakellariou, Neurosci. J., 2012, 1–4 Search PubMed.
- M. J. Baker, J. Trevisan, P. Bassan, R. Bhargava, H. J. Butler, K. M. Dorling, P. R. Fielden, S. W. Fogarty, N. J. Fullwood and K. A. Heys, et al. , Nat. Protoc., 2014, 9(8), 1771–1791 CrossRef PubMed.
- N. Stone, C. Kendall, J. Smith, P. Crow and H. Barr, Faraday Discuss., 2004, 126, 141–157 RSC.
- C. Kallaway, L. M. Almond, H. Barr, J. Wood, J. Hutchings, C. Kendall and N. Stone, Photodiagn. Photodyn. Ther., 2013, 10(3), 207–219 CrossRef PubMed.
- A. Mahadevan-Jansen and R. R. Richards-Kortum, J. Biomed. Opt., 1996, 1(1), 31–70 CrossRef PubMed.
- D. I. Ellis and R. Goodacre, Analyst, 2006, 131(8), 875–885 RSC.
- O. J. Old, L. M. Fullwood, R. Scott, G. R. Lloyd, L. M. Almond, N. A. Shepherd, N. Stone, H. Barr and C. Kendall, Anal. Methods, 2014, 6(12), 3901–3917 RSC.
- G. Clemens, J. R. Hands, K. M. Dorling and M. J. Baker, Analyst, 2014, 139(18), 4411–4444 RSC.
- S. E. Taylor, K. T. Cheung, I. I. Patel, J. Trevisan, H. F. Stringfellow, K. M. Ashton, N. J. Wood, P. J. Keating, P. L. Martin-Hirsch and F. L. Martin, Br. J. Cancer, 2011, 104(5), 790–797 CrossRef PubMed.
- K. Gajjar, L. D. Heppenstall, W. Pang, K. M. Ashton, J. Trevisan, I. I. Patel, V. Llabjani, H. F. Stringfellow, P. L. Martin-Hirsch, T. Dawson and F. Martin, Anal. Methods, 2013, 5(1), 89–102 RSC.
- L. M. Fullwood, G. Clemens, D. Griffiths, K. M. Ashton, T. P. Dawson, R. W. Lea, C. Davis, F. Bonnier, H. J. Byrne and M. J. Baker, Anal. Methods, 2014, 6, 3948–3961 RSC.
- L. M. Fullwood, D. Griffiths, K. M. Ashton, T. Dawson, R. W. Lea, C. Davis, F. Bonnier, H. J. Byrne and M. J. Baker, Analyst, 2014, 139(2), 446–454 RSC.
- M. Jermyn, K. Mok, J. Mercier, J. Desroches, J. Pichetee, K. Saint-Arnaud, L. Bernstein, M. C. Guiot, K. Petrecca and F. Leblond, Sci. Transl. Med., 2015, 7(274), 274ra19 CrossRef PubMed.
- S. Duraipandian, M. S. Bergholt, W. Zheng, K. Yu Ho, M. The, J. Guan Yeoh, J. Bok Yan So, A. Shabbir and Z. Huang, J. Biomed. Opt., 2012, 17(8), 081418–081418 CrossRef PubMed.
- C. Kendall, N. Stone, N. Shepherd, K. Geboes, B. Warren, R. Bennett and H. Barr, J. Pathol, 2003, 200(5), 602–609 CrossRef PubMed.
- J. D. Horsnell, J. A. Smith, M. Sattlecker, A. Sammon, J. Christie-Brown, C. Kendall and N. Stone, Surgeon, 2012, 10(3), 123–127 CrossRef PubMed.
- U. R. S. Utzinger, D. L. Heintzelman, A. Mahadevan-Jansen, A. Malpica, M. Follen and R. Richards-Kortum, Appl. Spectrosc., 2001, 55(8), 955–959 CrossRef.
- M. Kirsch, G. Schackert, R. Salzer and C. Krafft, Anal. Bioanal. Chem., 2010, 398(4), 1707–1713 CrossRef PubMed.
- B. Brozek-Pluska, J. Musial, R. Kordek, E. Bailo, T. Dieing and H. Abramczyk, Analyst, 2012, 137(16), 3773–3780 RSC.
- H. Abramczyk, B. Brozek-Pluska, J. Surmacki, J. Jablonska-Gajewicz and R. Kordek, Prog. Biophys. Mol. Biol., 2012, 108(1–2), 74–81 CrossRef PubMed.
- H. Abramczyk and B. Brozek-Pluska, Anal. Chim. Acta, 2015, 909, 91–100 CrossRef PubMed.
- D. Vicinanza, R. Stables, G. Clemens and M. J. Baker, International Conference on Auditory Display (ICAD14), New York, USA, June, 2014.
- N. Bergner, B. F. M. Romeike, R. Reichart, R. Kalff, C. Krafft and J. Popp, Analyst, 2013, 138, 3983–3990 RSC.
- C. Lacombe, V. Untereiner, C. Gobinet, M. Zater, G. D. Sockalingum and R. Garnotel, Analyst, 2015, 140(7), 2280–2286 RSC.
- T. J. Harvey, C. Hughes, A. D. Ward, E. Correia Faria, A. Henderson, N. W. Clarke, M. D. Brown, R. D. Snook and P. Gardner, J. Biophotonics, 2009, 2(1–2), 47–69 CrossRef PubMed.
- F. L. Martin, M. J. German, E. Wit, T. Fearn, N. Ragavan and H. M. Pollock, J. Comput. Biol., 2007, 14(9), 1176–1184 CrossRef PubMed.
- C. Krafft, L. Neudert, T. Simat and R. Salzer, Spectrochim. Acta, Part A, 2005, 61(7), 1529–1535 CrossRef PubMed.
- N. Stone, C. Kendall, N. Shepherd, P. Crow and H. Barr, J. Raman Spectrosc., 2002, 33(7), 564–573 CrossRef.
- R. E. Kast, G. K. Serhatkulu, A. Cao, A. K. Pandya, H. Dai, J. S. Thakur, V. M. Naik, R. Naik, M. D. Klein and G. W. Auner, et al. , Biopolymers, 2008, 89(3), 235–241 CrossRef PubMed.
- B. Jimenez, R. Mirnezami, J. Kinross, O. Cloarec, H. C. Keun, E. Holmes, R. D. Goldin, P. Ziprin, A. Darzi and J. K. Nicholson, J. Proteome Res., 2013, 12(2), 959–968 CrossRef PubMed.
- I. Notingher, Sensors, 2007, 7(8), 1343–1358 CrossRef.
- L. Zheng, C. M. McQuaw, M. J. Baker, N. P. Lockyer, J. C. Vickerman, A. G. Ewing and N. Winograd, Appl. Surf. Sci., 2008, 255(4), 1190–1192 CrossRef PubMed.
- D. Bertholdo, A. Watcharakorn and M. Castillo, Brain Neuroimaging Clinics of North America, 2013, 23(3), 359–380 CrossRef PubMed.
- C. H. A. Tan and E. H. A. Tan, World J. Nucl. Med., 2012, 11(1), 30 CrossRef PubMed.
- P. D. St-Coeur, M. Touaibia and M. Cuperlovic-Culf, Genomics, Proteomics Bioinf., 2013, 11(4), 199–206 CrossRef PubMed.
- R. K. Gupta, T. F. Cloughesy, U. Sinha, J. Garakian, J. Lazareff, G. Rubino, L. Rubino, D. P. Becker, H. V. Vinters and J. R. Alger, J. Neuro-Oncol., 2000, 50(3), 215–226 CrossRef CAS PubMed.
- C. Plathow and W. A. Weber, J. Nucl. Med., 2008, 49(Suppl. 2), 43S–63S CrossRef CAS PubMed.
- C. Juhasz, D. C. Chugani, O. Muzik, D. Wu, A. E. Sloan, G. Barger, C. Watson, A. K. Shah, S. Sood and E. L. Ergun, et al. , J. Cereb. Blood Flow Metab., 2006, 26(3), 345–357 CrossRef CAS PubMed.
- D. O. Kamson, S. Mittal, A. Buth, O. Muzik, W. J. Kupsky, N. L. Robinette, G. R. Barger and C. Juhasz, Mol. Imaging, 2013, 12(5), 327 Search PubMed.
- H. J. Byrne, K. M. Ostrowska, H. Nawaz, J. Dorney, A. D. Meade, F. Bonnier and F. M. Lyng, in Optical Spectroscopy and Computational Methods in Biology and Medicine, Springer, 2014, pp. 355–399 Search PubMed.
- H. J. Byrne, G. Sockalingum and N. Stone, RSC Analytical Spectroscopy Monographs No. 11, Biomedical Applications of Synchrotron Infrared Microspectroscopy, 2011 Search PubMed.
- L. Wang, Y. Liu, J. Zeng and L. Huang, Spectrosc. Spectral Anal., 2012, 32(1), 123–126 CAS.
- T. Yamada, N. Miyoshi, T. Ogawa, K. Akao, M. Fukuda, T. Ogasawara, Y. Kitagawa and K. Sano, Clin. Cancer Res., 2002, 8(6), 2010–2014 CAS.
- M. Gniadecka, P. A. Philipsen, S. Sigurdsson, S. Wessel, O. F. Nielsen, D. H. Christensen, J. Hercogova, K. Rossen, H. K. Thomsen and R. Gniadecki, et al. , J. Invest. Dermatol., 2004, 122(2), 443–449 CrossRef CAS PubMed.
- M. J. Baker, E. Gazi, M. Brown, J. H. Shanks, P. Gardner and N. W. Clarke, Br. J. Cancer, 2008, 99(11), 1859–1866 CrossRef CAS PubMed.
| This journal is © The Royal Society of Chemistry 2017 |