Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Nanostickers for cells: a model study using cell–nanoparticle hybrid aggregates†
Benjamin
Brunel‡
a,
Grégory
Beaune‡
a,
Usharani
Nagarajan
a,
Sylvie
Dufour
bc,
Françoise
Brochard-Wyart‡
*de and
Françoise M.
Winnik‡
*afg
aWPI International Center for Materials Nanoarchitectonics (MANA), National Institute for Materials Science (NIMS), 1-1 Namiki, Tsukuba, Ibaraki 305-0044, Japan. E-mail: Francoise.brochard-wyart@curie.fr; francoise.winnik@umontreal.ca
bInserm, U955, Equipe 6, Créteil, 94000, France
cUniversité Paris Est, Faculté de Médecine, Créteil 94000, France
dLaboratoire Physico Chimie Curie, Institut Curie, PSL Research University, CNRS UMR168, 75005, Paris, France
eSorbonne Université, UPMC Univ Paris 06, 75005, Paris, France
fDepartment of Chemistry and Faculty of Pharmacy, University of Montreal, CP 6128 Succursale Centre Ville, Montreal, QC H3C3J7, Canada
gDepartment of Chemistry and Faculty of Pharmacy, University of Helsinki, Helsinki, Finland
First published on 31st August 2016
Abstract
We present direct evidence that nanoparticles (NPs) can stick together cells that are inherently non-adhesive. Using cadherin-depleted S180 murine cells lines, which exhibit very low cell–cell adhesion, we show that NPs can assemble dispersed single cells into large cohesive aggregates. The dynamics of aggregation, which is controlled by diffusion and collision, can be described as a second-order kinetic law characterized by a rate of collision that depends on the size, concentration, and surface chemistry of the NPs. We model the cell–cell adhesion induced by the “nanostickers” using a three-state dynamical model, where the NPs are free, adsorbed on the cell membrane or internalized by the cells. We define a “sticking efficiency parameter” to compare NPs and look for the most efficient type of NP. We find that 20 nm carboxylated polystyrene NPs are more efficient nanostickers than 20 nm silica NPs which were reported to induce fast wound healing and to glue soft tissues. Nanostickers, by increasing the cohesion of tissues and tumors, may have important applications for tissue engineering and cancer treatment.
Nanoparticles (NPs) are frequently used in nanomedicine and medical diagnostics where they serve as delivery agents, actuators, or imaging agents.1–3 We discovered that bare nanoparticles can enhance cellular adhesion and mimic the specific homotypic interactions between membrane proteins, known as cadherins. The study was prompted by a report of Leibler et al. indicating that bare silica NPs can act as a glue for soft tissues, such as liver sections,4 and induce fast healing of deep wounds in the skin and liver of rats.5 This new property of NPs hints that NPs could act as a cell/cell adhesive, but there has been no unequivocal demonstration of this phenomenon. We decided to carry out our study with cadherin-depleted mouse sarcoma S180 line cells to evaluate their cell assembly and aggregation.6 S180 cells form weakly cohesive, small and rough aggregates, whereas transfected clones with a high level of cadherin expression form large cohesive aggregates that are very compact and smooth.7 We demonstrate here that NPs can trigger the self-assembly of dispersed single cadherin-depleted cells into cohesive aggregates. We monitor the dynamics of aggregation, which are ruled by cell diffusion and cell–cell collision, and model the cell–cell adhesion induced by the NPs, or nanostickers, using a three-state dynamical model where the NPs are free, adsorbed on the membrane, or internalized by the cells.
The study was conducted as follows. First, we monitored the formation of S180 cell aggregates as a function of time in the absence of NPs and in the presence of NPs at a given concentration Ce. This first set of measurements gave us the “sticking efficiency parameter” (G) of each type of NP. Then, we measured G versus the concentration of NPs. The dynamics of cell aggregation were modeled by analogy to the collision-induced aggregation of Brownian particles8 that leads them to stick to each other with a probability P that depends upon cell–cell adhesion. This model leads to the definition of G as the normalized sticking probability. The nanosticker-induced cell–cell adhesion was evaluated using a three-state dynamical model, where NPs are either free in the cell culture medium, adsorbed on the cell membrane, or internalized. The most efficient nanostickers were identified by varying the model parameters. The study was conducted with silica NPs (SiO2) similar to those used by Leibler et al. and four types of polystyrene NPs, either negatively charged under physiological conditions, denoted as “Carbo” or positively charged, denoted as “Amine”. Their physico-chemical properties are listed in Table S1 (ESI†). The NPs selected were non-toxic to cells within the time and concentration domains of this study, as evaluated by the CCK-8 assay (see the ESI† for details). The polystyrene particles were fluorescently labeled for the visualization of NP-treated cells by confocal fluorescence microscopy.
Aggregates of S180 cells were obtained using the hanging droplet method9 (see the ESI† for experimental details). Droplets of the cell suspensions in the cell culture medium were deposited on a Petri dish cover. The cover was placed, inverted, on top of a container containing a phosphate buffer for humidity control. As time progresses, cells fall to the bottom of the droplets. They diffuse on this 2D surface and meet other cells. In the case of adherent cells, encounters result in the formation of groups of cells that adhere to each other to form clusters and, eventually, large aggregates. To monitor the progress of aggregate formation as a function of time, we prepared 7 covers, each holding 20 droplets of cell dispersion. The inverted covers were placed over the containers, and the 20 hanging droplets were collected at various times over a period of 2 days. Each recovered droplet was placed on an untreated glass coverslip and observed by optical microscopy. Micrographs recorded at 15 min, 6 h, and 2 days after initiation of the study are presented in Fig. 1A, with cells alone, and Fig. 1B, with cells treated with Carbo20 NPs. Droplets collected at t = 15 min contain only single cells whether Carbo20 NPs were added or not. After a period of 6 h, the droplet of S180 cells alone (A) presents mostly isolated cells, a few groups of 3 or 4 cells, and some larger clusters. In the case of cells treated with Carbo20 NPs (B) we observed clusters of cells and a few isolated cells. The total number of entities per unit area, circled in red, is named N. After a 2 day incubation, S180 cells alone (A) form a few loose clusters, surrounded by isolated cells and small groups, whereas the NP-treated cells aggregate in a single spheroid, approximately 250 μm in diameter, surrounded by a few isolated cells.
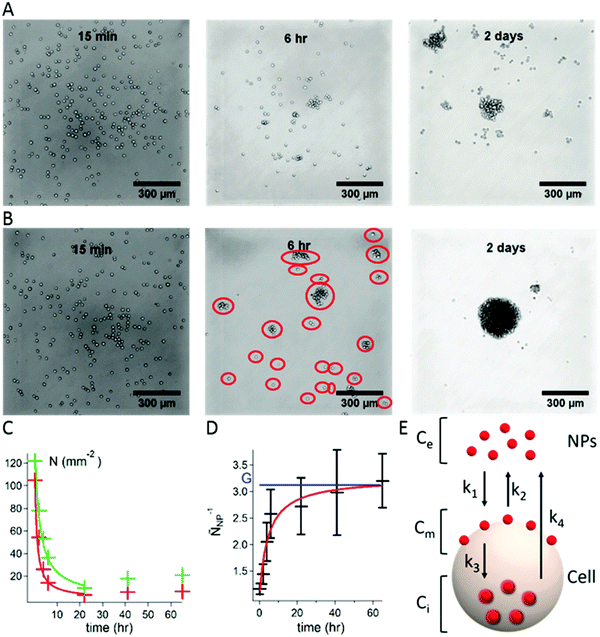 | ||
| Fig. 1 Formation of aggregates of S180 cells using the hanging droplet method. (A) Optical micrographs of droplets containing S180 cells collected 15 min, 6 h, and 2 days after initiation of the experiment. (B) Optical micrographs recorded under the same conditions in the case of a mixture of S180 cells and Carbo20 (volume fraction Φe = 5 × 10−6i.e. Ce = 4.7 × 1011 NPs mL−1). The red circles highlight isolated entities defined as groups (N). (C) Plot of the change with incubation time of the number of groups N per unit area observed in optical micrographs of droplets. Green data points and line: S180 cells alone; red data points and line: S180 cells with Carbo20 (Φe = 5 × 10−6i.e. Ce = 4.7 × 1011 NPs mL−1). The lines are fits to the data using eqn (1). (D) Plot of the change with incubation time of ÑNP−1 (Carbo20, Φe = 5 × 10−6i.e. Ce = 4.7 × 1011 NPs mL−1). The blue horizontal line indicates the value of G; the red line is the fit of the data points with eqn (2). Error bars represent the standard error over 20 drops. (E) Kinetic model describing the distribution of NPs between the bulk, the surface of the cell membrane, and the interior of the cell. | ||
The number (N) of groups per unit area was determined for a large number of micrographs collected over 70 h and plotted as a function of incubation time for S180 cells treated, or not treated, with NPs (see Fig. 1C, Carbo20 NPs). As time elapses, the number of groups (N) decreases (the groups become larger). At very long times (t > 24 h), N increases slightly as a consequence of cell division and subsequent rearrangement of cells into new aggregates.
The decrease of N with time was modeled as follows.10 Once all the cells have fallen to the bottom of the drop, they diffuse on this surface with a mean diffusion rate V, such that a single cell (with a diameter dcell) covers a surface equal to V·dt·dcell during the time interval dt. Thus, for N number of cells covering a surface area, a cell encounters an average of V·dt·dcell·N cells. We call the rate of collision K = V·dcell. If P is the probability for two cells to stick together, then K·P is the rate of successful collisions. Progressively, cells form larger groups of cells (N is the number of groups per unit area), which obey the same dynamics, assuming K·P to be independent of the size of the group. For a time period dt, each group fuses with P·K·dt·N groups. It means that among the N groups, P·K·dt·N·N/2 fusions occur. The evolution of N is governed by dN/dt = −PKN2/2. After integration, the number of groups per unit area N(t) decreases as:
| N(t) = N0/(1 + N0PKt/2) | (1) |
We can see from Fig. 1C that N decreases faster for cells treated with NPs. This is expressed in the fitting function by the difference between Pcontrol and PNP. The enhanced adhesion is quantified by the ratio of the probabilities PNP/Pcontrol = 2.8 ± 0.3.
To compare the dynamics of aggregation with and without NPs, we design the ratio:
| ÑNP−1 = Ncontrol/NNP = (1 + N0PNPKt/2)/(1 + N0PcontrolKt/2) | (2) |
The aim is to calculate Cm because the adhesive effect is due to the NPs located on the membrane. The spatial distribution of NPs is evaluated using the dynamical model shown in Fig. 1E. The NPs are located (i) in the bulk solution, with a concentration Ce assumed to be constant with time as the NPs are in excess; (ii) on the cell membrane, with a surface concentration Cm; and (iii) internalized in the cell, with a concentration Ci. The internalization of NPs and their removal are crucial for therapeutic applications. The mechanisms of endocytosis and exocytosis of NPs are well described in ref. 13.
To describe the adsorption, the endocytosis and exocytosis of NPs in the cells, we introduce four rate constants defined in Fig. 1E: k1 and k2 associated with the adsorption and desorption of the NPs on the cell membrane, k3 with the internalization by endocytosis and k4 with the removal of NPs by exocytosis. From a dimensional analysis, we can write k1 ≈ rp3τon−1, where τon is the adsorption time; k2 ≈ τoff−1, where τoff is the desorption time; k3 ≈ rp3τin−1, where τin is the internalization time; and k4 ≈ τex−1, where τex is the exit time, which is assumed to be independent of rp.
The kinetic equations determining the distributions of NPs and in particular Cm are:
| dCm/dt = k1Ce(Cms − Cm) − k2Cm − k3Cm(Cis − Ci) | (3) |
| dCi/dt = k3(A/V)Cm(Cis − Ci) − k4Ci | (4) |
The ratio A/V in eqn (4) is associated with the transfer of the NPs from the surface into the cell volume and is equal to 3/R, where R is the cell radius. The volume of the droplet is large compared to the volume of the cells and we can assume that Ce remains constant (Fig. S2, ESI†).
The internalized NPs are assumed to enter in the cell by endocytosis of the membrane decorated with NPs and to exit the cell by exocytosis are ruled by eqn (3) and (4). The stationary state leads to the NPs internalized concentration Ci and to the surface distribution of the NPs Cm.
In the stationary regime, dCm/dt = 0 and dCi/dt = 0, which allow us to derive Ci and Cm.
| k1Ce(Cms − Cm) − k2Cm − k3Cm(Cis − Ci) = 0 | (5) |
| k3(A/V)Cm(Cis − Ci) − k4Ci = 0 | (6) |
| k1Ce(Cms − Cm) − k2Cm − k4CiR/3 = 0 | (7) |
(I) Regime of internalization Ce < Ce*
In this regime, eqn (7) with Cm = 0 leads to Ci = (3k1/Rk4)CeCms. Ci increases up to Cis which defines the threshold concentration Ce*. Ce* = (Rk4/3k1)(Cis/Cms) = (rp2/l2)Cis, where rp is the radius of the nanoparticles and l is a characteristic length defined by l2 = 3k1/Rk4 (in dimension, l2 ≈ (τex/τon)rp3/R). The model leads to simple scaling laws for the threshold concentration c*, where the NPs are internalized, that involves a characteristic length l. The measurement of l leads to a derivation of the exit time τex divided by the adsorption time, τon.
(II) Regime of adsorption of the beads on the membrane Ce > Ce*
In this regime the cell is saturated with nanoparticles, and eqn (7) has to be written with Ci = Cis:
k 1 C e(Cms − Cm) − k2Cm = (R/3)k4Cis. It leads to the solution for Cm given by:
| Cm/Cms = (Ce − Ce*)/((Ce − Ce*) + CL) | (8) |
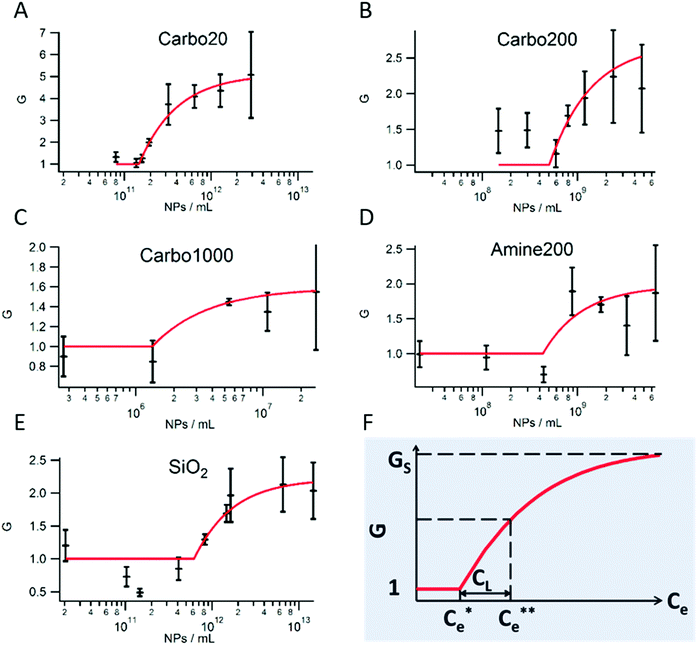
Eqn (8) shows that this regime is ruled by a Langmuir law characterized by the Langmuir concentration CL, but with a shift in the concentration (Ce − Ce*). When Ce − Ce* > CL, i.e. Ce > Ce** = (k2/k1) + 2Ce*, Cm reaches the saturation value Cms and G tends to Gs ≈ 1 + βUCms. Assuming that the NPs are in close contact at saturation (Cms = 1/(πrp2)) and that β = Ac/kBT (Ac being the contact area of colliding cells), Gs = 1 + n*U/kBT, where n* = AcCms is the number of NPs per cell–cell collision. By fitting the experimental efficiency parameter G(Ce) as shown in Fig. 2 with this model we can obtain the 3 parameters Ce*, Ce** and Gs for all types of particles. Their values are listed in Table 1.
| NP | C e* [NPs mL−1] | Φ e* | l 2/rp2 | C e** [NPs mL−1] | G S |
|---|---|---|---|---|---|
| Carbo20 | 1.5 ± 0.1 × 1011 | 1.7 × 10−6 | 1.1 ± 0.7 × 103 | 2.6 ± 0.5 × 1011 | 4.8 ± 0.7 |
| Carbo200 | 5 ± 2 × 108 | 1.5 × 10−6 | 1.3 ± 0.5 × 103 | 1.0 ± 0.8 × 109 | 2.7 ± 0.8 |
| Carbo1000 | 1.4 ± 0.5 × 106 | 9.4 × 10−7 | 2.1 ± 0.8 × 103 | 3 ± 4 × 106 | 1.6 ± 0.4 |
| Amine200 | 4.3 ± 0.6 × 108 | 1.8 × 10−6 | 1.1 ± 0.3 × 103 | 8 ± 9 × 108 | 2.0 ± 0.5 |
| SiO2 | 6 ± 2 × 1011 | 3.9 × 10−6 | 0.5 ± 0.4 × 103 | 1.2 ± 0.6 × 1012 | 2.2 ± 0.4 |
1 Internalization threshold concentration Ce*
The value of the concentration Ce* varies by five orders of magnitude when the size of the NPs increases from 20 to 1000 nm, whereas the volume fraction, Φe* = Ce*(4/3)πrp3, is nearly the same for all types of NPs (∼1.5 × 10−6). Hence, cells internalize a maximum volume of NPs, whatever the NP size.13 From Ce* = (rp2/l2)Cis we deduce the characteristic length l, assuming that a cell digests a maximum volume fraction of particles equal to 2 × 10−3 derived from ref. 14. l2/rp2 is nearly constant (∼103) for all types of NPs, which leads to an estimation of τex/τon (of order 106 for rp/R = 10−3).
2 Surface saturation threshold concentration Ce**
For all types of NPs, Ce** ∼ 2Ce*, showing that k2/k1 ≪ Ce*.
3 Sticking efficiency parameter Gs
Comparison between Carbo20, Carbo200 and Carbo1000 suggests that the cell–cell adhesion increases as the NPs' size decreases. Also a comparison between Amine200 and Carbo200 indicates that the charge has no influence on the sticking efficiency of NPs. The adhesive effect of SiO2 used in [4] and [5] is not as strong as that of Carbo20 (Table S1b, ESI†). As discussed above, Gs = 1 + n*U/kBT. Assuming U ∼ kBT, we find that n* is of order of few NPs.
To demonstrate that NPs mimic cellular adhesion molecules (CAMs) present on the surface of cohesive cells, we monitored via the same protocol, but without added NPs, the aggregation of LCAM cells, which are S180 cells transfected to express the highest level of cadherins.6 The time dependence of N, the number of cell groups in a droplet (Fig. S3, ESI†), was determined and the experimental data were fitted with eqn (1) yielding the parameters N0(65 ± 5 cells mm−2) and PLCAMK[(4.2 ± 0.6) × 10−12 m2 s−1]. Using the Pcontrol value obtained for S180 cells in the absence of NPs, we find GLCAM = PLCAM/Pcontrol = 2.1 ± 0.3. The GLCAM values are of the same order of magnitude as G induced by NPs (Table 1). Hence, the methods developed for NPs could be applied also to quantify the adhesion between cells expressing different levels of CAMs on their membrane or differing by other types of modification of the cell membrane. This technique complements very well the classical dual pipette assay6 used to measure cell–cell adhesion via detachment, because in the case of soft objects like cells, the energy of detachment can be a few orders of magnitude larger than the Dupré equilibrium adhesion energy.15
In conclusion, using a simple experimental protocol we have established that nanostickers are able to glue cells together and to increase the cohesion of the cells inside an aggregate. As metastasis is often related to a decrease of cell–cell adhesion, nanostickers will reduce the escape of cells from tumors. They will also slow down the spreading of tumors, which results from a competition between cell–substrate and cell–cell adhesion. Experiments on the spreading of hybrid nanoparticle–cell aggregates and the characterization of their mechanical properties will allow us in the near future to demonstrate the role of nanostickers in the limitation of cancer proliferation. Moreover, beyond implementations in surgery already foreseen by Leibler et al., hybrid cell/NP aggregates are unique constructs that may find applications in tissue engineering and cellular therapy. Most experiments on hybrid aggregates are related to mixtures of two types of cells leading to phase separation.16 Hybrid aggregates of dead and living matter will open a new interesting field.
Acknowledgements
The authors thank Dr J. Niskanen (University of Helsinki, Finland) for carrying out zeta measurements. This work was supported by the WPI-Program of the Ministry of Education, Culture, Sports, Science and Technology (MEXT), Japan, and by the NIMS “Nanotechnology Platform Project” also supported by MEXT. B.B. gratefully acknowledges financial support of his stay in MANA, NIMS, by the Ecole Normale Superieure de Cachan (France).References
- O. Rabin, J. Manuel Perez, J. Grimm, G. Wojtkiewicz and R. Weissleder, Nat. Mater., 2006, 5, 118–122 CrossRef CAS PubMed.
- A. Ito, M. Shinkai, H. Honda and T. Kobayashi, J. Biosci. Bioeng., 2005, 100, 1–11 CrossRef CAS PubMed.
- X. Wang, Y. Wang, H. He, X. Chen, X. Sun, Y. Sun, G. Zhou, H. Xu and F. Huang, J. Mater. Chem. B, 2016, 4, 779–784 RSC.
- S. Rose, A. Prevoteau, P. Elzière, D. Hourdet, A. Marcellan and L. Leibler, Nature, 2014, 505, 382–385 CrossRef CAS PubMed.
- A. Meddahi-Pellé, A. Legrand, A. Marcellan, L. Louedec, D. Letourneur and L. Leibler, Angew. Chem., Int. Ed. Engl., 2014, 53, 6369–6373 CrossRef PubMed.
- Y.-S. Chu, W. A. Thomas, O. Eder, F. Pincet, E. Perez, J. P. Thiery and S. Dufour, J. Cell Biol., 2004, 167, 1183–1194 CrossRef CAS PubMed.
- S. Douezan, K. Guevorkian, R. Naouar, S. Dufour, D. Cuvelier and F. Brochard-Wyart, Proc. Natl. Acad. Sci. U. S. A., 2011, 108, 7315–7320 CrossRef CAS PubMed.
- S. Douezan, J. Dumond and F. Brochard-Wyart, Soft Matter, 2012, 8, 4578 RSC.
- P. Marmottant, A. Mgharbel, J. Käfer, B. Audren, J.-P. Rieu, J.-C. Vial, B. van der Sanden, A. F. M. Marée, F. Graner and H. Delanoë-Ayari, Proc. Natl. Acad. Sci. U. S. A., 2009, 106, 17271–17275 CrossRef CAS PubMed.
- S. Douezan and F. Brochard-Wyart, Soft Matter, 2012, 8, 784 RSC.
- P.-G. de Gennes, P.-H. Puech and F. Brochard-Wyart, Langmuir, 2003, 19, 7112–7119 CrossRef CAS.
- G. I. Bell, M. Dembo and P. Bongrand, Biophys. J., 1984, 45, 1051–1064 CrossRef CAS PubMed.
- N. Oh and J. H. Park, Int. J. Nanomed., 2014, 9(1), 51–63 Search PubMed.
- C. Wilhelm, F. Gazeau, J. Roger, J. N. Pons and J.-C. Bacri, Langmuir, 2002, 18, 8148–8155 CrossRef CAS.
- P. G. de Gennes, Langmuir, 1996, 12, 4497–4500 CrossRef CAS.
- W. Song, C.-K. Tung, Y.-C. Lu, Y. Pardo, M. Wu, M. Das, D.-I. Kao, S. Chen and M. Ma, Soft Matter, 2016, 12, 5739–5746 RSC.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c6sm01450j |
| ‡ These authors contributed equally to the manuscript. |
| This journal is © The Royal Society of Chemistry 2016 |