Open Access Article
Open Access ArticleParallel folding topology-selective label-free detection and monitoring of conformational and topological changes of different G-quadruplex DNAs by emission spectral changes via FRET of mPPE-Ala–Pt(II) complex ensemble†
Kevin
Chan
,
Clive
Yik-Sham Chung
and
Vivian
Wing-Wah Yam
*
Institute of Molecular Functional Materials (Areas of Excellence Scheme, University Grants Committee (Hong Kong)), Department of Chemistry, The University of Hong Kong, Pokfulam Road, Hong Kong, P. R. China. E-mail: wwyam@hku.hk
First published on 26th January 2016
Abstract
The formation of supramolecular assemblies between [Pt(bzimpy-Et){C![[triple bond, length as m-dash]](https://www.rsc.org/images/entities/char_e002.gif) CC6H4(CH2NMe3-4)}]Cl2 (1) and mPPE-Ala and the FRET properties of the ensemble have been revealed from the UV-vis absorption, steady-state emission and time-resolved emission decay studies. The two-component mPPE-Ala–1 ensemble has been employed in a “proof-of-principle” concept for label-free detection of G-quadruplex DNAs with the intramolecular propeller parallel folding topology, such as c-myc, in aqueous buffer solution. By the modulation of the aggregation/deaggregation of the polymer–metal complex aggregates and hence the FRET from the mPPE-Ala donor to the aggregated 1 as acceptor, the ensemble has been demonstrated for sensitive and selective label-free detection of c-myc via the monitoring of emission spectral changes of the ensemble. Ratiometric emission of the ensemble at 461 and 662 nm has been shown to distinguish the intramolecular propeller parallel G-quadruplex folding topology of c-myc from other G-quadruplex-forming sequences of different folding topologies, owing to the strong and specific interactions between c-myc and 1 as suggested by the UV-vis absorption and UV melting studies. In addition, the formation of high-order intermolecular multimeric G-quadruplexes from c-myc under molecular crowding conditions has been successfully probed by the ratiometric emission of the ensemble. The conformational and topological transition of human telomeric DNA from the mixed-hybrid form to the intramolecular propeller parallel form, as observed from the circular dichroism spectroscopy, has also been monitored by the ratiometric emission of the ensemble. The ability of the ensemble to detect these conformational and topological transitions of G-quadruplex DNAs has been rationalized by the excellent selectivity and sensitivity of the ensemble towards the intramolecular propeller parallel G-quadruplex DNAs and their high-order intermolecular multimers, which are due to the extra stabilization gained from Pt⋯Pt and π–π interactions in addition to the electrostatic and hydrophobic interactions found in the polymer–metal complex aggregates.
CC6H4(CH2NMe3-4)}]Cl2 (1) and mPPE-Ala and the FRET properties of the ensemble have been revealed from the UV-vis absorption, steady-state emission and time-resolved emission decay studies. The two-component mPPE-Ala–1 ensemble has been employed in a “proof-of-principle” concept for label-free detection of G-quadruplex DNAs with the intramolecular propeller parallel folding topology, such as c-myc, in aqueous buffer solution. By the modulation of the aggregation/deaggregation of the polymer–metal complex aggregates and hence the FRET from the mPPE-Ala donor to the aggregated 1 as acceptor, the ensemble has been demonstrated for sensitive and selective label-free detection of c-myc via the monitoring of emission spectral changes of the ensemble. Ratiometric emission of the ensemble at 461 and 662 nm has been shown to distinguish the intramolecular propeller parallel G-quadruplex folding topology of c-myc from other G-quadruplex-forming sequences of different folding topologies, owing to the strong and specific interactions between c-myc and 1 as suggested by the UV-vis absorption and UV melting studies. In addition, the formation of high-order intermolecular multimeric G-quadruplexes from c-myc under molecular crowding conditions has been successfully probed by the ratiometric emission of the ensemble. The conformational and topological transition of human telomeric DNA from the mixed-hybrid form to the intramolecular propeller parallel form, as observed from the circular dichroism spectroscopy, has also been monitored by the ratiometric emission of the ensemble. The ability of the ensemble to detect these conformational and topological transitions of G-quadruplex DNAs has been rationalized by the excellent selectivity and sensitivity of the ensemble towards the intramolecular propeller parallel G-quadruplex DNAs and their high-order intermolecular multimers, which are due to the extra stabilization gained from Pt⋯Pt and π–π interactions in addition to the electrostatic and hydrophobic interactions found in the polymer–metal complex aggregates.
Introduction
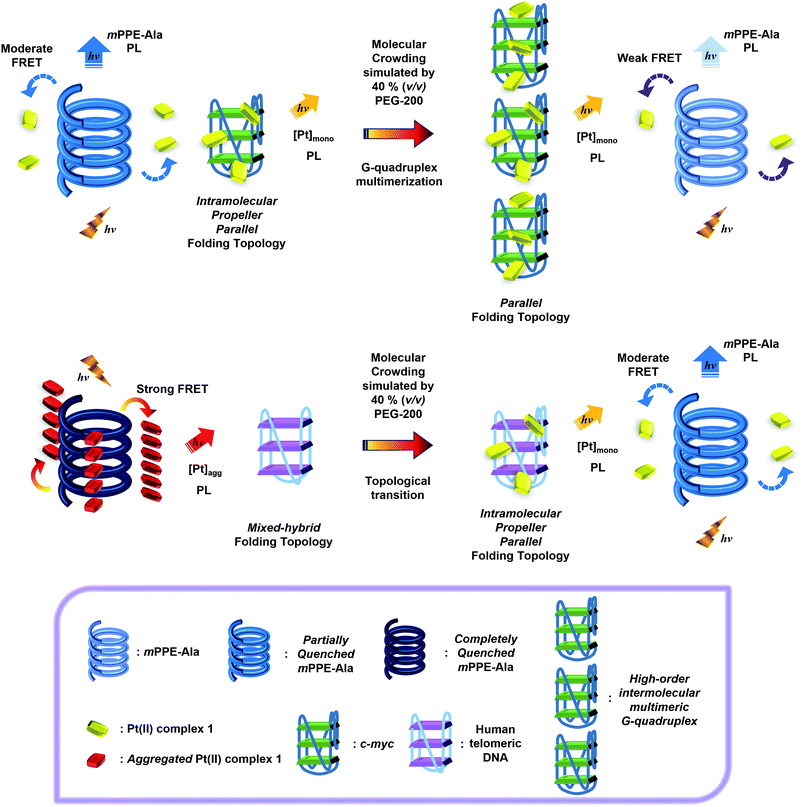
Square-planar d8 platinum(II) polypyridine complexes have been reported to display a strong tendency towards the formation of highly-ordered structures in the solid state via metal–metal and aromatic ligand π–π stacking interactions and to exhibit intriguing spectroscopic properties.1–10 A particularly interesting class of platinum(II) complexes is the alkynylplatinum(II) terpyridine complexes. With enhanced solubility in common organic solvents and/or aqueous media, these complexes can undergo self-assembly/disassembly in solutions.3,4b,c,6a,c–f,8 Since their self-assembly/disassembly processes can be readily modulated by changes in the microenvironment,3–7,8c alteration of the non-covalent interactions between the cationic alkynylplatinum(II) terpyridine complexes and biomolecules, such as proteins, single-stranded nucleic acids, aptamers and heparin, has been found to result in remarkable UV-vis absorption and near-infrared (NIR) emission spectral changes.9c–e In particular, such NIR emission spectral changes are advantageous for non-invasive fluorescence microscopy,11 enabling this class of complexes to serve as spectroscopic reporters for label-free detection of biomolecules and/or probing of enzymatic activities.9c–g,10 However, the underlying detection principles are mainly based on the perturbation of the self-assembly/disassembly processes of the metal complexes via non-covalent interactions, which may lead to a loss in selectivity towards target analytes in complex biological fluids due to the non-specific interactions between the metal complexes and other biomolecules present in the physiological milieu.Significant improvement in the selectivity for biosensing has been accomplished with the use of two-component supramolecular ensembles based on the alkynylplatinum(II) terpyridine complexes and conjugated polyelectrolytes (CPEs).10 CPEs are π-conjugated polyelectrolytes which show strong absorption and fluorescence properties similar to those of conjugated polymers.12–18 With the amplified fluorescence quenching properties,12a CPEs have demonstrated their potential applications in chemo- and biosensing with excellent selectivity based on the changes in their luminescence properties upon their interactions with analytes.12b–f,14 Yet, non-specific interactions with miscellaneous macromolecules present in physiological milieu would be a problem for CPEs to achieve good selectivity towards the target analytes.15 This challenge for CPEs in biosensing, which would also possibly be observed in the studies of platinum(II) complexes, can be overcome by the use of two-component ensembles of alkynylplatinum(II) terpyridine complexes and CPEs.10 An efficient Förster resonance energy transfer (FRET) from CPEs to alkynylplatinum(II) terpyridine complexes has been shown to occur only upon the formation of polymer–metal complex aggregates with Pt⋯Pt, electrostatic and π–π stacking interactions due to the variation in the electronic absorption properties of the monomeric and the aggregated platinum(II) complexes.10 Addition of target analytes has been found to result in drastic emission spectral changes of the ensemble, constituting a highly sensitive detection.10 In addition to electrostatic and hydrophobic interactions,10 extra stabilization from Pt⋯Pt and π–π interactions gives rise to the formation of stable polymer–metal complex aggregates, thereby non-specific interactions from miscellaneous macromolecules would not be strong enough to destroy the aggregates and an improved selectivity towards target analytes could be achieved.
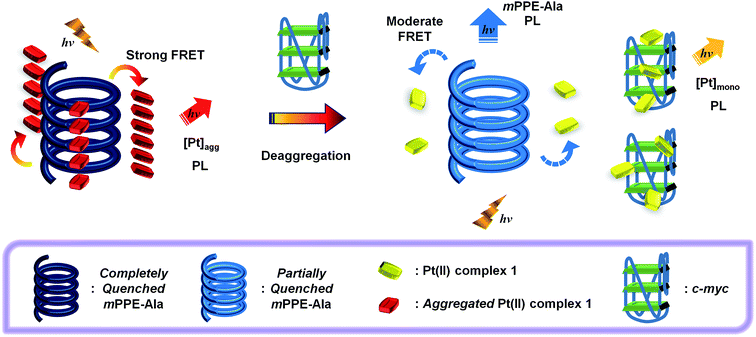
More recently, the photophysical properties of two-component ensembles of alkynylplatinum(II) complexes with 2,6-bis(benzimidazol-2′-yl)pyridine (bzimpy) as the tridentate N-donor ligand and CPEs have been studied.19 Despite the fact that alkynylplatinum(II) bzimpy complexes can undergo self-assembly in solutions similar to that of alkynylplatinum(II) terpyridine complexes,19–21 the energy levels of the intraligand and the charge transfer excited states of this class of complexes are significantly different from those of the terpyridine counterparts.20a In addition to FRET, the photophysical properties of the two-component ensembles were also governed by photo-induced charge transfer and Dexter triplet energy back-transfer, which are related to the HOMO–LUMO energy gaps and the triplet energies of individual components.19 Therefore, the circumspect and sagacious design and choice of their chemical structures would allow the fine-tuning of the extent of fluorescence quenching of the CPEs and photo-sensitization of the triplet metal–metal-to-ligand charge transfer (3MMLCT) emission of the platinum(II) complexes. On the other hand, CPEs of different conformations have been found to show different binding constants with platinum(II) complexes owing to the differences in spatial interactions among the two chemical species.19 Through the optimization of the photophysical properties and excited state energies, as well as the conformations and supramolecular architectures of the CPEs and alkynylplatinum(II) bzimpy complexes, the two-component ensemble of meta-linked mPPE-Ala and [Pt(bzimpy-Et){C![[triple bond, length as m-dash]](https://www.rsc.org/images/entities/char_e002.gif) CC6H4(CH2NMe3-4)}]Cl2 (1) (Scheme 1) has been shown to exhibit very efficient FRET and photo-sensitization of the 3MMLCT emission of the aggregated alkynylplatinum(II) bzimpy complexes.19 Moreover, stronger interactions between the CPE and alkynylplatinum(II) bzimpy complexes have been observed, as compared to the reported two-component ensembles between CPEs and the terpyridine counterparts.10 All these features suggest that the ensemble of mPPE-Ala and 1 would exhibit superior selectivity and sensitivity for label-free detection of target analytes.
CC6H4(CH2NMe3-4)}]Cl2 (1) (Scheme 1) has been shown to exhibit very efficient FRET and photo-sensitization of the 3MMLCT emission of the aggregated alkynylplatinum(II) bzimpy complexes.19 Moreover, stronger interactions between the CPE and alkynylplatinum(II) bzimpy complexes have been observed, as compared to the reported two-component ensembles between CPEs and the terpyridine counterparts.10 All these features suggest that the ensemble of mPPE-Ala and 1 would exhibit superior selectivity and sensitivity for label-free detection of target analytes.
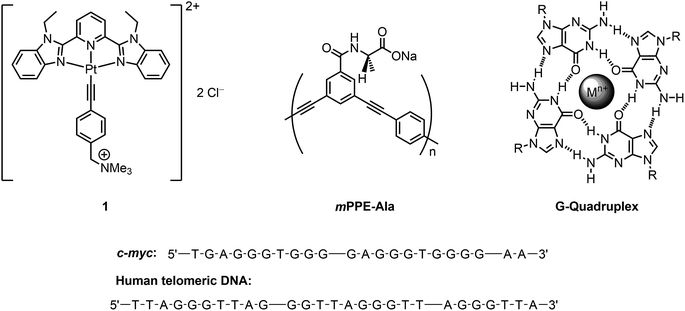 | ||
| Scheme 1 (Top) Chemical structures of the water-soluble platinum(II) bzimpy complex 1, mPPE-Ala and G-quadruplex. (Bottom) Structural formulae of c-myc and human telomeric DNA. | ||
G-Quadruplex DNAs are of widespread interest as they are specifically enriched in the telomere, oncogene promoters, growth control genes across the chromosomes, and transcriptome of human cells.22 It is noteworthy that the folded structures of G-quadruplex DNAs are highly dynamic in nature. The DNAs can fold into different topologies at different temperatures, pHs and concentrations of Na+ and K+ ions, as well as in the presence of a diversity of biomolecules and cell organelles which is generally known as “molecular crowding (MC) conditions”.23,24 For example, human telomeric DNA can readily fold into an intramolecular propeller parallel G-quadruplex structure under MC conditions24c which acts as an important target for anti-cancer therapy and other diseases,25,26 whereas formation of double-stranded structure with its complementary sequence can be observed under physiological conditions.27 As a result, there is an urge for not only the sensitive detection of G-quadruplex DNAs, but also the determination of their conformations and folding topologies, as well as their dynamic conformational and topological transitions. Although a number of luminescence probes for G-quadruplex DNAs have been reported,28–30 further improvements on the sensitivities of the assay methods would be desirable as micro-molar range of G-quadruplex DNAs was required to initiate the changes in the luminescence properties of these probes for signaling. Moreover, none of them have been demonstrated for the detection of conformational and topological changes of G-quadruplex DNAs under MC conditions. This might be attributed to the high threshold of the luminescence probes to achieve a high specificity and selectivity towards a particular folding topology of G-quadruplex DNAs. In view of the high binding affinity of square-planar metal complexes towards DNAs,31 it is anticipated that a particular folding topology of G-quadruplex DNAs can interact with 1 strongly, resulting in the disassembly of mPPE-Ala–1 aggregates. Based on the very efficient FRET and photo-sensitization of 3MMLCT emission of 1, the disassembly should result in remarkable emission spectral changes and allow the detection of G-quadruplex DNAs with that particular folding topology in a highly sensitive manner. This, together with the large association constant of mPPE-Ala and 1, would require specific interactions for the deaggregation of this two-component ensemble. Thus, a highly selective detection of a specific folding topology and the monitoring of conformational and topological changes of G-quadruplex DNAs are anticipated. Herein, we present the FRET study of the two-component mPPE-Ala–1 ensemble in the presence of G-quadruplex DNAs, and demonstrate a “proof-of-principle” concept for the label-free detection of intramolecular propeller parallel folding topology of G-quadruplex DNAs by monitoring the changes in the luminescence properties of the ensemble. Exploitation of the ensemble in probing the conformational and topological changes of G-quadruplex DNAs under MC conditions has also been investigated.
Results and discussion
Induced self-assembly and FRET studies of platinum(II) complex with mPPE-Ala
The photophysical and self-assembly properties of the two-component ensemble formed by mPPE-Ala and 1 in aqueous buffer solution (5 mM Tris–HCl, 50 mM KCl, pH 7.4) were investigated. A buffer of high K+ ion content was employed in the present study because high concentration of monovalent cations, especially K+ ions, has been reported to stabilize G-quadruplex DNAs to a greater extent and similar buffer solutions were employed in the reported studies of G-quadruplex DNAs in the literature.10,30b–e The UV-vis absorption spectra, resonance light scattering (RLS) spectra, steady-state emission spectra and time-resolved emission decay profiles of mPPE-Ala with different concentrations of 1 in aqueous buffer solutions are shown in Fig. S1–S4 in the ESI† respectively. Upon addition of 1 to an aqueous buffer solution of mPPE-Ala, the electronic absorption spectra of mPPE-Ala showed a red shift in the absorption band centered at 361 nm to 364 nm, along with the emergence of an intense high-energy absorption band at ca. 287 nm and less intense absorption tails at ca. 420–570 nm (Fig. S1†). In addition, an enhancement in the intensity of RLS maxima at ca. 452 and 550 nm was found with increasing concentration of 1 (Fig. S2†), which corresponds to the electronic absorption of mPPE-Ala and aggregated forms of 1 respectively. Concomitant with the remarkable spectral changes in the UV-vis absorption spectra, a progressive decline in the emission intensity of mPPE-Ala at ca. 461 nm and a significant growth of emission intensity at ca. 662 nm were observed upon the addition of 1 (Fig. S3a†).32 Similar spectral changes have been reported in the related studies of mPPE-Ala with 1 in aqueous buffer solution of low Na+ ion content.19 These suggest the lower-energy emission to be assigned as originated from an excited state of 3MMLCT 3[dσ*(Pt) → π*(bzimpy)] character, indicative of the formation of polymer–metal complex aggregates through electrostatic, Pt⋯Pt and π–π stacking interactions.Stern–Volmer (SV) equation has been employed for the analysis of the quenching of mPPE-Ala fluorescence by 1 (Fig. S3b†) and the Stern–Volmer quenching constants (KSV) obtained from the steady-state emission spectra at different concentrations of 1 are tabulated in Table S1.† It was found that the fluorescence quenching of mPPE-Ala was more efficient at high concentrations of 1, i.e., >5 μM, as shown from the upward deviation of the SV plot from linearity. The origin of this effective quenching of mPPE-Ala emission by 1 was investigated by time-resolved emission spectroscopic studies (Fig. S4 and Table S1†). The lifetimes of the emission of mPPE-Ala at λ = 461 nm in the absence of 1 were triexponential, consisting of a longer-lived component (τ1 = 9.07 ns) and two short-lived components (τ2 = 2.10 ns and τ3 = 0.71 ns). Upon the addition of 1, increases in emission lifetime and amplitude of the long-lived component τ1 were found, while the short-lived component τ3 showed obvious decreases in its amplitude and emission lifetime (from 0.71 ns to 0.19 ns). The obvious decline in τ3, together with the formation of polymer–metal complex aggregates as indicated by the spectroscopic studies (Fig. S1–S3†), suggest the interchain exciton diffusion of mPPE-Ala and an efficient FRET from mPPE-Ala to the metal complex aggregates. The latter was further supported by the good spectral overlap between the electronic absorption spectrum of the aggregated forms of 1 and the emission spectrum of mPPE-Ala (Fig. S5†). The efficient FRET process, which furnished an additional quenching pathway on top of the ground state static quenching found in the ensemble at low concentrations of 1, would account for the increased KSV value, the drop in 1[π → π*] fluorescence of mPPE-Ala and the growth of 3MMLCT emission at ca. 662 nm of 1 in the steady-state emission spectra (Fig. S3†).
It is worthwhile to note that a relatively large KSV value was found for the ensemble at low concentrations of 1 (<5 μM; Table S1†) when compared to that in the previous studies on CPEs and alkynylplatinum(II) terpyridine complexes.10 As the fluorescence quenching at low concentrations of 1 is mainly attributed to the ground-state static quenching, a large KSV value indicates a large association constant of 1 with mPPE-Ala,14 and hence stronger interactions between mPPE-Ala and 1 are anticipated.
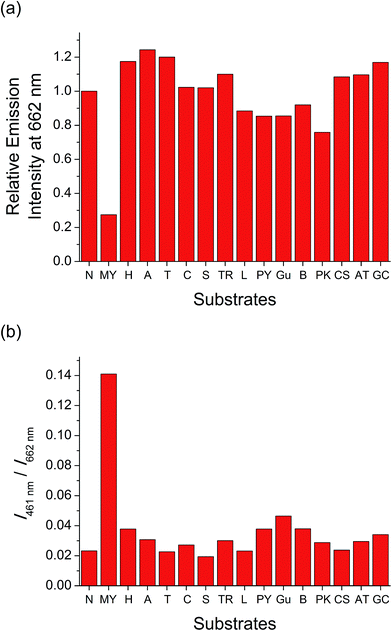
Luminescence properties of two-component mPPE-Ala–1 ensemble in the presence of biomolecules
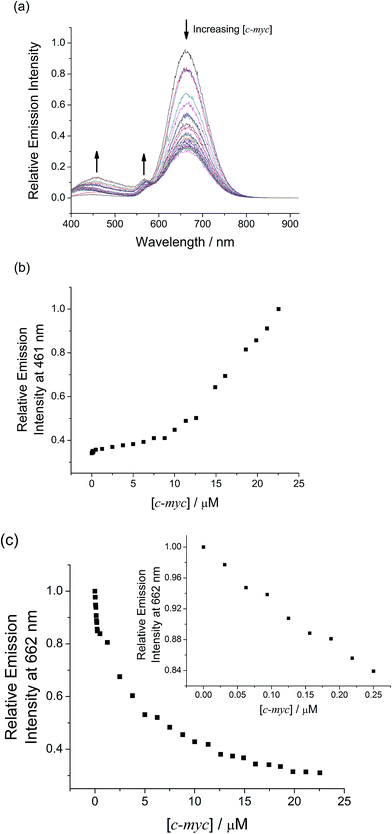
The strong interactions between mPPE-Ala and 1 are believed to be capable of minimizing the interferences from non-specific interactions. Therefore, such attractive feature would be advantageous for achieving highly selective detection of biomolecules based on the self-assembly/disassembly processes of the ensemble. This has prompted us to investigate the changes in the luminescence properties of the two-component mPPE-Ala–1 ensemble towards various biological substrates in aqueous buffer solution (Fig. 1). Among the fifteen substrates tested, only c-myc resulted in significant changes in the emission intensity of the ensemble at 662 nm. Pre-formed double-stranded DNAs (dsDNAs; poly(dA)25–poly(dT)25 and poly(dG)25–poly(dC)25), which should exhibit stronger electrostatic and/or π–π stacking interactions with the ensemble than c-myc, were found to show negligible effect on the emission of the ensemble at 662 nm.A more detailed spectroscopic study of the two-component ensemble with the G-quadruplex structure of c-myc in aqueous buffer solution was performed. The electronic absorption spectra of the ensemble showed a drop in the absorption at ca. 295, 330, 432 and 525 nm with increasing concentration of c-myc (Fig. S6†). On the other hand, the ensemble showed significant changes in the emission spectrum upon the addition of c-myc (Fig. 2). A drop in the emission intensity at 662 nm with a concomitant growth of the emission at both 461 and 568 nm was observed. Based on the drop of the MMLCT absorption at 525 nm and the decrease in 3MMLCT emission intensity of aggregated 1, the deaggregation of the ensemble occurred in the presence of c-myc, resulting in a smaller spectral overlap (Fig. S5†) and decrease in FRET efficiency. Therefore, the recovery of mPPE-Ala fluorescence at 461 nm was observed. The growth of the emission at 568 nm with increasing concentration of c-myc was likely ascribed to the emission of the non-aggregated 1 upon the deaggregation of the ensemble (Scheme 2).19
Specific binding and stabilization of G-quadruplex structure of c-myc by alkynylplatinum(II) bzimpy complex
The unique emission spectral changes of the two-component mPPE-Ala–1 ensemble with c-myc suggest the presence of strong and specific interactions between the ensemble and c-myc. In order to gain more insights into these specific interactions, the conformation of c-myc in aqueous buffer solution was investigated by circular dichroism (CD) measurement (Fig. S7†). The CD spectrum displayed a positive peak at ca. 263 nm followed by a negative peak at ca. 243 nm, which are characteristic of intramolecular propeller parallel folding topology of G-quadruplex DNAs,33 suggesting the formation of G-quadruplex structure of c-myc in aqueous buffer solution.The interactions between 1 and the G-quadruplex structure of c-myc were investigated by UV-vis spectroscopic study using the Scatchard equation34a (Fig. S8†) and the UV melting study28a,34b (Fig. S9 and S10†). The addition of c-myc to an aqueous buffer solution of 1 led to a 14% and 18% of hypochromism at 285 and 327 nm respectively in the UV-vis absorption spectrum without significant bathochromic shift (Fig. S9†). The hypochromic phenomenon is attributed to the strong electronic interactions between the platinum(II) complex and DNA nucleobases. The binding constant of 1 on the G-quadruplex structure of c-myc in the absence of mPPE-Ala is 8.46 × 107 M−1, as determined from the Scatchard equation (Fig. S8†). These, together with the fact that pre-formed duplex DNAs did not show significant effect on the emission intensity of the ensemble at 662 nm (Fig. 1), suggest that the specific interactions between the ensemble and c-myc should not be only ascribed to the guanine-rich (G-rich) sequence that is similar to the reported binding of cisplatin to DNAs.35 Instead, more unique interactions should be associated with the ensemble and the G-quadruplex structure of c-myc in aqueous buffer solutions.
The binding of 1 to the G-quadruplex structure of c-myc was further evaluated by the changes in the G-quadruplex melting temperature (Tm). The UV melting study of c-myc in aqueous buffer solution revealed a significant increase in Tm in the presence of 1 (>96 °C) when compared to the Tm without 1 (91 °C) (Fig. S9 and S10 and Table S2†). This indicates the strong binding as well as the stabilization of the G-quadruplex structure of c-myc by 1, which is consistent with the high binding affinity determined by the Scatchard equation (Fig. S8†). Together with the fact that a number of square-planar platinum(II) complexes have been reported to bind to G-tetrad as well as the loop adenines of the G-quadruplex DNAs through electrostatic and π–π interactions with high specificity and binding affinity over pre-formed duplex DNAs,29,30b the cationic 1 should interact strongly with the intramolecular propeller parallel G-quadruplex folding topology of c-myc. Owing to such strong and specific interactions between 1 and c-myc, the mPPE-Ala–1 ensemble would undergo deaggregation in the presence of c-myc (Scheme 2).
Label-free detection of G-quadruplex structure of c-myc by emission spectral changes in the visible region
The deaggregation of two-component mPPE-Ala–1 ensemble and hence the decrease in FRET efficiency only in the presence of c-myc has led to remarkable emission spectral changes at two distinguishable wavelengths, 461 and 662 nm (Fig. 2). Such two well distinguishable emissions allow the detection of c-myc by ratiometric emissions of the ensemble at 461 and 662 nm, I461/I662 (Fig. 1b). This provides an internal in situ self-calibration for minimizing inherent interferences, such as light scattering of the sample and fluctuations in excitation source power,36 which are usually inevitable in single-intensity-based measurements. The detection limit37 of c-myc was found to be 16.7 nM, suggesting the high sensitivity of the ensemble for label-free detection of c-myc in aqueous buffer solutions.Competition experiments on the detection of the G-quadruplex structure of c-myc by the ensemble in the presence of various biological substrates have also been performed (Fig. 3). Only small discrepancies in the emission intensity at 662 nm were found when compared to the case without interfering biological substrates. Interestingly, further addition of the complementary sequence of c-myc (c-myc-c) to the solution of the ensemble and c-myc did not give any observable emission spectral changes (Fig. S11†). This indicates that the ensemble exhibits a high binding affinity and selectivity towards the G-quadruplex structure of c-myc in aqueous buffer solution, by which c-myc-c could not interrupt the strong and specific interactions between the ensemble and the G-quadruplex structure of c-myc. All these features suggest that the ensemble should be a potential candidate for selective and sensitive detection of G-quadruplex DNAs with intramolecular propeller parallel folding topology such as c-myc in aqueous buffer solutions.
The selectivity in the detection of intramolecular propeller parallel G-quadruplex folding topology of c-myc by emission spectral changes of the ensemble has been further investigated using DNAs with duplex structures. In contrast to the significant emission spectral changes of the ensemble in the presence of intramolecular propeller parallel G-quadruplex folding topology of c-myc, pre-formed duplex structures of c-myc or human telomeric DNA with their respective complementary sequences, were found to show less significant effect on the emission properties of the ensemble (Fig. S12 and S13†). This confirms the high selectivity of the ensemble towards G-quadruplex DNAs over duplex DNAs.
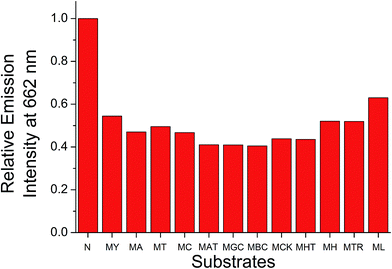
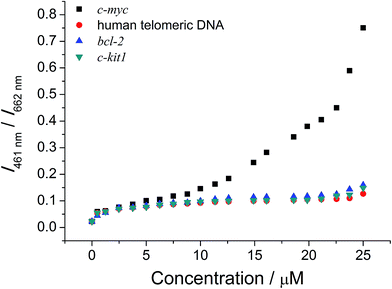
The ability of the ensemble in differentiating intramolecular propeller parallel G-quadruplex folding topology of c-myc from other G-quadruplex folding topologies has also been studied. B-Cell lymphoma 2 (bcl-2), c-kit1 and human telomeric DNA have been known to exist as hybrid,38a,b snap-back parallel,38a,c and mixed-hybrid29,38a,d,e folding topologies of the G-quadruplex structures, respectively, in aqueous buffer solutions. The emission spectra of the ensemble in the presence of these G-quadruplex DNAs are shown in Fig. S14–S16†. With increasing concentrations of these DNAs, the ensemble was found to show a decrease in emission intensity at 662 nm and a growth of emission at 461 nm, but the changes were smaller in extent than those found in the case of c-myc. These, together with the smaller increase in Tm of these G-quadruplex DNAs by 1 (Table S2†), suggest the weaker interactions between 1 and bcl-2, c-kit1 or human telomeric DNA. Such weaker interactions could be rationalized by the difference in steric hindrance caused by the different loop structures and terminal nucleotides of the G-quadruplex-forming sequences.38a For example, the snap-back parallel folding topology adopted by the G-quadruplex structure of c-kit1,38a,c unlike the conventional intramolecular propeller parallel folding topology adopted by the G-quadruplex structure of c-myc,38a,f,39 would prevent the cationic platinum(II) complexes from end-stacking onto the bottom G-quartet of the G-quadruplex,38a thus weakening the binding of the G-quadruplex structure of c-kit1 with 1. As a result, the ensemble would undergo less significant deaggregation in the presence of these G-quadruplex DNAs, and hence smaller emission spectral changes were observed as compared to the study of the ensemble with c-myc. This feature would allow the successful differentiation of c-myc from other G-quadruplex DNAs by the ratiometric monitoring of I461/I662 (Fig. 4), with interference of less than 22% at 25 μM of the DNAs. The self-calibrating peculiarity brought about by such ratiometric measurement further suggests the potential application of the ensemble for sensitive and selective detection of G-quadruplex DNAs with intramolecular propeller parallel folding topology.
It is noteworthy that the ensemble would not result in significant unwinding of the duplex structure of c-myc and its complementary sequence, which has been found in some of the reported G-quadruplex-binding ligands,40 as supported by the CD measurements of the duplex DNA with increasing concentration of 1 (Fig. S17†). This would be a pre-eminent advantage for the ensemble to be a luminescence probe for the G-quadruplex structure of c-myc because the ensemble is side reaction-free from the pre-formed duplex counterpart of the target substrate. On the other hand, although 1 showed strong and specific binding onto the intramolecular propeller parallel G-quadruplex folding topology of c-myc per se, it would not be a suitable luminescence probe for the detection of this DNA structure because of its complicated emission spectral changes upon the addition of c-myc (Fig. S18†).41 The working range of 1 alone as the luminescence probe for c-myc is small, while that of the ensemble is found to be much larger due to the fact that the luminescence properties of the ensemble are governed by both the self-assembly/disassembly processes and FRET from mPPE-Ala to 1. As a result, the ensemble can demonstrate a “proof-of-principle” concept for the luminescence detection of intramolecular propeller parallel G-quadruplex folding topology of c-myc with superior selectivity and sensitivity.
Probing the conformational and topological transitions of G-quadruplex DNAs by the two-component mPPE-Ala–1 ensemble
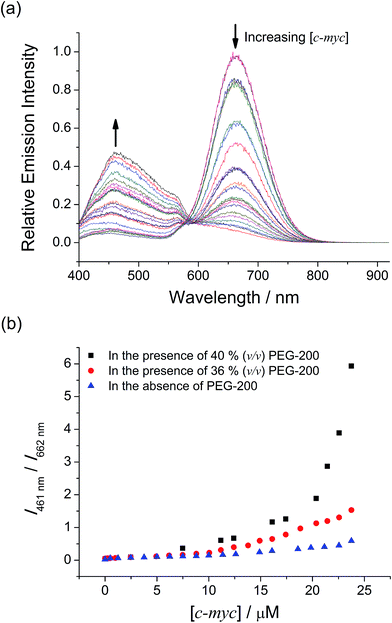
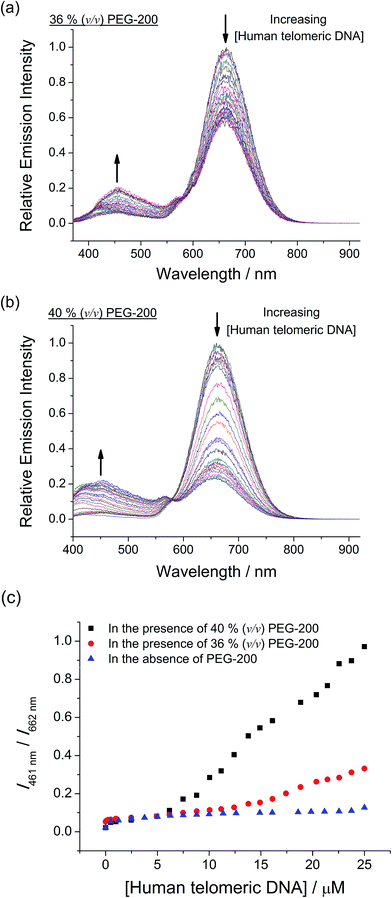
With the excellent selectivity and sensitivity towards the intramolecular propeller parallel G-quadruplex folding topology of c-myc, it is envisaged that the two-component mPPE-Ala–1 ensemble could be utilized for probing the conformational and topological changes of G-quadruplex DNAs, which should give important information for the studies of aging, cancer, HIV and other diseases.26 Small-molecule crowding reagents, such as polyethylene glycol (PEG) with a molecular weight of 200 (PEG-200), are commonly used for mimicking the macromolecular crowded intracellular environment. G-Quadruplex DNAs have been reported to exhibit very different folding topologies in the presence of PEG-200 due to the different thermodynamic and kinetic properties of the nucleic acids. Therefore, the emission properties of the ensemble and c-myc in aqueous buffer solution with different volume percentages of PEG-200 (0, 36 and 40% respectively, v/v) have been examined. The ensemble showed a significant drop in emission intensity at 662 nm with a concomitant growth of the emission at 461 nm (Fig. 5a, S19 and S20†), and the changes were found to be to a larger extent with higher volume percentages of PEG-200 (Fig. 5b). Since PEG-200 of up to 40% (v/v) only exerted small effects on the emission properties of the ensemble (Fig. S21†),42 the drastic emission changes of the ensemble and c-myc in the presence of PEG-200 suggest the possible conformational and topological changes of c-myc.To rationalize the emission spectral changes of the ensemble, the conformation of c-myc in aqueous buffer solution in the presence of 40% (v/v) PEG-200 was studied by CD spectroscopy at room temperature (Fig. S22†). In the absence of PEG-200, the CD spectrum displayed a positive peak at ca. 263 nm and a negative peak at ca. 242 nm, which are characteristic of intramolecular propeller parallel G-quadruplexes.33 On the other hand, once c-myc was incubated at 37 °C in the presence of 40% (v/v) PEG-200, the positive band was found to increase in magnitude and red-shifted. With reference to the reported studies on G-quadruplex DNAs,43a incubation of c-myc at 37 °C for a few hours in the presence of 40% (v/v) of PEG-200 would result in the formation of high-order G-quadruplex topologies via the stacking of the G-tetrads into an axial extension of repeated G-quadruplex superstructures, which are commonly known as multimeric G-quadruplexes or G-wires.23b,c,24c,43b,c Therefore, the CD spectral changes of c-myc in the presence of PEG-200 were likely attributed to the formation of high-order intermolecular multimeric species of intramolecular propeller parallel G-quadruplexes of c-myc.
Based on the findings on the conformational and topological changes of c-myc from the CD spectroscopic study, the emission spectral changes of the ensemble with different concentrations of c-myc in the presence of different volume percentages of PEG-200 (Fig. 5a, S19 and S20†) may be construed as follows. According to the study of the ensemble with the G-quadruplex structure of c-myc aforementioned, the emission spectral changes at 662 nm would likely be attributed to the deaggregation of the polymer–metal complex aggregates with Pt⋯Pt, electrostatic and π–π stacking interactions because of the strong and specific interactions of 1 with the G-quadruplex structure of c-myc. This would lead to the decrease in FRET from the mPPE-Ala donor to the aggregated 1 moieties. Upon an increase in volume percentages of PEG-200, the G-quadruplex of c-myc converted itself from unimolecular form to high-order intermolecular multimeric form via the stacking of the G-tetrads into an axial extension of repeated G-quadruplex superstructures.23b,c,24c,43b,c The higher space charge density of the high-order intermolecular multimeric G-quadruplex would then allow stronger electrostatic interactions with the cationic 1 moieties, which favored a more effective and efficient deaggregation of the ensemble. The deaggregation of the ensemble would lead to a decrease in FRET, resulting in the recovery of mPPE-Ala fluorescence which accounts for the growth of emission at 461 nm (Scheme 3). With higher volume percentage of PEG-200, more high-order intermolecular multimeric G-quadruplexes of c-myc would be formed in the solution as revealed from the increase in CD signal,43a leading to more significant emission spectral changes of the ensemble and c-myc in the presence of 40% (v/v) PEG-200, as compared to the case of 36% (v/v) PEG-200 (Fig. 5b). From the ratiometric emission of the ensemble at 461 and 662 nm, I461/I662, sensitive detection of multimeric G-quadruplexes of c-myc can be achieved with less than 12.6% interference from the unimolecular form of the intramolecular propeller parallel G-quadruplex folding topology of c-myc (Fig. 5b). As a result, the ratiometric emission of the ensemble could be a more desirable alternative to CD spectroscopy for providing structural information of the G-quadruplex DNAs, as the latter was found to show only small differences in the CD spectra of the unimolecular form and high-order intermolecular multimeric form of G-quadruplex DNAs.
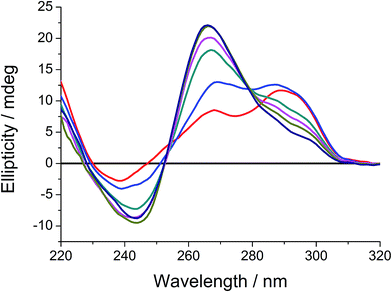
In addition to the formation of high-order structures, G-quadruplex DNAs can adopt different folding topologies under different conditions. For example, as water interactions play an important role in the structural stability and dynamics of nucleic acids,23b,c,24 the G-quadruplex structure of human telomeric DNA has been reported to undergo a conformational and topological transition from mixed-hybrid form to intramolecular propeller parallel form in the presence of PEG-200.24c CD spectroscopic study of the pre-formed mixed-hybrid G-quadruplex structure of human telomeric DNA with different volume percentages of PEG-200 showed similar findings as the previous studies on human telomeric DNA in different buffer solutions (Fig. 6).24c In the absence of PEG-200, the CD spectrum of G-quadruplex structure of human telomeric DNA showed a small negative peak at ca. 239 nm and a positive peak at ca. 290 nm with a shoulder at 268 nm, indicating the mixed-hybrid folding topology of G-quadruplex DNA.33 With increasing volume percentage of PEG-200, an emergence of a positive peak at 266 nm and a diminution of the peak at 290 nm were found. Further increase in volume percentage of PEG-200 to 40% (v/v) resulted in a spectrum similar to those of intramolecular propeller parallel G-quadruplexes DNAs.33 This suggests that a conformational and topological change of the G-quadruplex structure of human telomeric DNA from mixed-hybrid folding topology to intramolecular propeller parallel folding topology with increasing volume percentage of PEG-200 has occurred.
With the excellent selectivity and sensitivity of the ensemble for the luminescence detection of intramolecular propeller parallel G-quadruplex DNAs, attempts have been made to monitor the conformational and topological changes of human telomeric DNA in the presence of different volume percentages of PEG-200 by the ensemble. In the absence of PEG-200, the ensemble showed a decrease in emission intensity at 662 nm, with only infinitesimal changes in emission at 461 nm (Fig. S16†). Interestingly, an increase in volume percentage of PEG-200 resulted in a more significant decrease in emission intensity at 662 nm, with a concomitant growth of the emission at 461 nm (Fig. 7a and b). This led to a larger value of I461/I662 of the ensemble towards human telomeric DNA in the presence of PEG-200, as compared to the case without PEG-200 (Fig. 7c). The small emission spectral changes of the ensemble towards human telomeric DNA in the absence of PEG-200 were attributed to the relatively weak interactions between 1 and the mixed-hybrid G-quadruplex folding topology of human telomeric DNA (Fig. S16†), as supported by the smaller change in Tm (Table S2†). On the other hand, with an increasing volume percentage of PEG-200, human telomeric DNA was found to show a change in conformation and folding topology from mixed-hybrid folding topology to intramolecular propeller parallel folding topology (Fig. 6). With the strong interactions between 1 and intramolecular propeller parallel G-quadruplex DNAs, the ensemble would undergo deaggregation, leading to a decrease in FRET efficiency. As a result, a significant decrease in 3MMLCT emission of 1 at 662 nm and a growth of mPPE-Ala fluorescence at 461 nm were observed in the presence of human telomeric DNA and PEG-200 (Scheme 3 and Fig. 7, S23 and S24†). Notably, the decrease in emission intensity at 662 nm, which is likely due to the deaggregation of the polymer–metal complex aggregates with Pt⋯Pt, electrostatic and π–π stacking interactions through the interactions with the G-quadruplex structure of human telomeric DNA in the presence of 40% (v/v) PEG-200, is comparable to that obtained from the G-quadruplex structure of c-myc in the absence of PEG-200 (Fig. 2). This further supports the high selectivity of the ensemble in the detection of the intramolecular propeller parallel folding topology of G-quadruplex DNAs.
The ratiometric emission of the ensemble was found to show good differentiation of the mixed-hybrid form from the intramolecular propeller parallel form of human telomeric DNA, with a 7.7-fold difference at 25 μM of DNA. In view of the fact that CD spectroscopy alone could not define the structural properties of G-quadruplex DNAs unambiguously, the ratiometric emission of the ensemble could be a valuable tool for providing more information on the conformations and folding topologies of G-quadruplex DNAs, and even the real-time monitoring of conformational and topological transitions of the G-quadruplex by luminescence spectroscopy.
Conclusions
In the present study, a water-soluble cationic alkynylplatinum(II) bzimpy complex 1 has been shown to assemble onto mPPE-Ala through Pt⋯Pt, electrostatic and π–π stacking interactions, leading to the formation of polymer–metal complex aggregates of large KSV value and efficient FRET with a growth of 3MMLCT emission. The two-component mPPE-Ala–1 ensemble has been demonstrated to show a high selectivity towards the detection of G-quadruplex structure of c-myc in aqueous buffer solutions. The high selectivity has been rationalized by the strong and specific interactions between the cationic square-planar platinum(II) complex and the intramolecular propeller parallel G-quadruplex folding topology of c-myc as supported by the high binding affinity determined by the Scatchard equation and UV melting study. This results in the deaggregation of the ensemble only in the presence of c-myc, leading to the decrease in FRET efficiency and obvious emission spectral changes. Together with the high sensitivity as indicated by the low detection limit (16.7 nM) and the self-calibrating feature brought about by the ratiometric emissions, these have allowed the ensemble to serve as an attractive candidate in demonstrating a “proof-of-principle” concept for label-free detection of G-quadruplex structure of c-myc with high sensitivity and selectivity. The ratiometric emission of the ensemble at 461 and 662 nm has been further utilized to differentiate the G-quadruplex structure of c-myc from the structures of other DNAs. The success in the differentiation has been explained by the specificity of 1 towards G-quadruplex DNAs with the intramolecular propeller parallel folding topology.Furthermore, the ensemble can monitor the conformational and topological transitions of c-myc from the intramolecular propeller parallel folding topology to the high-order intermolecular multimeric species, due to the higher space charge density of the multimeric G-quadruplexes which should exert stronger interactions on 1 and hence favor the deaggregation of the mPPE-Ala–1 ensemble. In addition, with the excellent selectivity and sensitivity of the ensemble towards G-quadruplex DNAs with intramolecular propeller parallel folding topology, the conformational and topological transition of mixed-hybrid form of human telomeric DNA to intramolecular propeller parallel form can be probed by the ratiometric emission of the ensemble. In view of the difficulties of CD spectroscopy alone for unambiguous structural determination of G-quadruplex DNAs, the ensemble reported herein can help to identify the intramolecular propeller parallel folding topology and the high-order intermolecular multimeric species by the changes in its luminescence properties. Also, the present work should open up a new avenue for the design and construction of highly selective and sensitive luminescence probes and initiate further studies on the real-time monitoring of the dynamic folding processes of DNAs by luminescence probes.
Acknowledgements
V. W.-W. Y. acknowledges support from the University Grants Committee Areas of Excellence Scheme (AoE/P-03/08), the Research Grants Council of Hong Kong Special Administrative Region, China (HKU 7051/13P), and the National Basic Research Program of China (973 Program; 2013CB834701). K. C. acknowledges the receipt of a Postgraduate Studentship administered by The University of Hong Kong. Prof. W.-K. Chan and Mr T. S.-Y. Li at The University of Hong Kong are thanked for their technical assistance in the use of gel permeation chromatography (GPC) system. Dr M. C.-L. Yeung is acknowledged for her helpful discussions and her help in finalizing the manuscript.Notes and references
- (a) R. S. Osborn and D. Rogers, J. Chem. Soc., Dalton Trans., 1974, 1002–1004 RSC; (b) K. W. Jennette, J. T. Gill, J. A. Sadownick and S. J. Lippard, J. Am. Chem. Soc., 1976, 98, 6159–6168 CrossRef CAS; (c) Extended Linear Chain Compounds, ed. J. S. Miller, Plenum Press, New York, 1982, vol. 1 Search PubMed; (d) V. M. Miskowski and V. H. Houlding, Inorg. Chem., 1991, 30, 4446–4452 CrossRef CAS; (e) V. H. Houlding and V. M. Miskowski, Coord. Chem. Rev., 1991, 111, 145–152 CrossRef CAS; (f) R. Büchner, C. T. Cunningham, J. S. Field, R. J. Haines, D. R. McMillin and G. C. Summerton, J. Chem. Soc., Dalton Trans., 1999, 711–717 RSC; (g) V. W. W. Yam, K. M. C. Wong and N. Zhu, J. Am. Chem. Soc., 2002, 124, 6506–6507 CrossRef CAS PubMed.
- (a) R. H. Herber, M. Croft, M. J. Coyer, B. Bilash and A. Sahiner, Inorg. Chem., 1994, 33, 2422–2426 CrossRef CAS; (b) W. B. Connick, L. M. Henling, R. E. Marsh and H. B. Gray, Inorg. Chem., 1996, 35, 6261–6265 CrossRef CAS; (c) W. B. Connick, R. E. Marsh, W. P. Schaefer and H. B. Gray, Inorg. Chem., 1997, 36, 913–922 CrossRef CAS; (d) R. Büchner, C. T. Cunningham and D. R. McMillin, Inorg. Chem., 1997, 36, 3952–3956 CrossRef; (e) V. W. W. Yam, R. P. L. Tang, K. M. C. Wong and K. K. Cheung, Organometallics, 2001, 20, 4476–4482 CrossRef CAS; (f) A. J. Goshe, I. M. Steele and B. Bosnich, J. Am. Chem. Soc., 2003, 125, 444–451 CrossRef CAS PubMed; (g) T. J. Wadas, Q. M. Wang, Y. J. Kim, C. Flaschenreim, T. N. Blanton and R. Eisenberg, J. Am. Chem. Soc., 2004, 126, 16841–16849 CrossRef CAS PubMed.
- (a) S. Y. L. Leung, A. Y. Y. Tam, C. H. Tao, H. S. Chow and V. W. W. Yam, J. Am. Chem. Soc., 2012, 134, 1047–1056 CrossRef CAS PubMed; (b) S. Y. L. Leung, W. H. Lam and V. W. W. Yam, Proc. Natl. Acad. Sci. U. S. A., 2013, 110, 7986–7991 CrossRef CAS PubMed; (c) S. Y. L. Leung and V. W. W. Yam, Chem. Sci., 2013, 4, 4228–4234 RSC.
- (a) V. W. W. Yam, K. H. Y. Chan, K. M. C. Wong and N. Zhu, Chem.–Eur. J., 2005, 11, 4535–4543 CrossRef PubMed; (b) A. Y. Y. Tam, K. M. C. Wong, N. Zhu, G. Wang and V. W. W. Yam, Langmuir, 2009, 25, 8685–8695 CrossRef CAS PubMed; (c) A. Y. Y. Tam, K. M. C. Wong and V. W. W. Yam, Chem.–Eur. J., 2009, 15, 4775–4778 CrossRef CAS PubMed.
- (a) K. M. C. Wong and V. W. W. Yam, Coord. Chem. Rev., 2007, 251, 2477–2488 CrossRef CAS; (b) M. C. L. Yeung, B. W. K. Chu and V. W. W. Yam, ChemistryOpen, 2014, 3, 172–176 CrossRef CAS PubMed.
- (a) V. W. W. Yam, K. H. Y. Chan, K. M. C. Wong and B. W. K. Chu, Angew. Chem., Int. Ed., 2006, 45, 6169–6173 CrossRef CAS PubMed; (b) K. H. Y. Chan, H. S. Chow, K. M. C. Wong, M. C. L. Yeung and V. W. W. Yam, Chem. Sci., 2010, 1, 477–482 RSC; (c) C. Y. S. Chung and V. W. W. Yam, Chem.–Eur. J., 2013, 19, 13182–13192 CrossRef CAS PubMed; (d) H. L. Au-Yeung, S. Y. L. Leung, A. Y. Y. Tam and V. W. W. Yam, J. Am. Chem. Soc., 2014, 136, 17910–17913 CrossRef CAS PubMed; (e) C. Po, Z. Ke, A. Y. Y. Tam, H. F. Chow and V. W. W. Yam, Chem.–Eur. J., 2013, 19, 15735–15744 CrossRef CAS PubMed; (f) A. Y. Y. Tam, K. M. C. Wong, G. Wang and V. W. W. Yam, Chem. Commun., 2007, 2028–2030 RSC.
- (a) K. M. C. Wong, W. S. Tang, X. X. Lu, N. Zhu and V. W. W. Yam, Inorg. Chem., 2005, 44, 1492–1498 CrossRef CAS PubMed; (b) C. Y. S. Chung, S. P. Y. Li, M. W. Louie, K. K. W. Lo and V. W. W. Yam, Chem. Sci., 2013, 4, 2453–2462 RSC.
- (a) Y. Tanaka, K. M. C. Wong and V. W. W. Yam, Chem. Sci., 2012, 3, 1185–1191 RSC; (b) Y. Tanaka, K. M. C. Wong and V. W. W. Yam, Angew. Chem., Int. Ed., 2013, 52, 14117–14120 CrossRef CAS PubMed; (c) A. K. W. Chan, W. H. Lam, Y. Tanaka, K. M. C. Wong and V. W. W. Yam, Proc. Natl. Acad. Sci. U. S. A., 2015, 112, 690–695 CrossRef CAS PubMed.
- (a) C. Yu, K. M. C. Wong, K. H. Y. Chan and V. W. W. Yam, Angew. Chem., Int. Ed., 2005, 44, 791–794 CrossRef CAS PubMed; (b) C. Yu, K. H. Y. Chan, K. M. C. Wong and V. W. W. Yam, Chem.–Eur. J., 2008, 14, 4577–4584 CrossRef CAS PubMed; (c) C. Yu, K. H. Y. Chan, K. M. C. Wong and V. W. W. Yam, Proc. Natl. Acad. Sci. U. S. A., 2006, 103, 19652–19657 CrossRef CAS PubMed; (d) M. C. L. Yeung, K. M. C. Wong, Y. K. T. Tsang and V. W. W. Yam, Chem. Commun., 2010, 46, 7709–7711 RSC; (e) M. C. L. Yeung and V. W. W. Yam, Chem.–Eur. J., 2011, 17, 11987–11990 CrossRef CAS PubMed; (f) M. C. L. Yeung and V. W. W. Yam, Chem. Sci., 2013, 4, 2928–2935 RSC; (g) C. Y. S. Chung and V. W. W. Yam, Chem.–Eur. J., 2014, 20, 13016–13027 CrossRef CAS PubMed; (h) C. Y. S. Chung, S.-i. Tamaru, S. Shinkai and V. W. W. Yam, Chem.–Eur. J., 2015, 21, 5447–5558 CrossRef CAS PubMed.
- (a) C. Y. S. Chung and V. W. W. Yam, J. Am. Chem. Soc., 2011, 133, 18775–18784 CrossRef CAS PubMed; (b) C. Y. S. Chung and V. W. W. Yam, Chem. Sci., 2013, 4, 377–387 RSC.
- (a) H. R. Kermis, Y. Kostov, P. Harms and G. Rao, Biotechnol. Prog., 2002, 18, 1047–1053 CrossRef CAS PubMed; (b) E. Soini and I. Hemmilä, Clin. Chem., 1979, 25, 353–361 CAS; (c) P. W. Barone, S. Baik, D. A. Heller and M. S. Strano, Nat. Mater., 2005, 4, 86–92 CrossRef CAS PubMed; (d) L. Yuan, W. Lin, K. Zheng, L. He and W. Huang, Chem. Soc. Rev., 2013, 42, 622–661 RSC.
- (a) Q. Zhou and T. M. Swager, J. Am. Chem. Soc., 1995, 117, 12593–12602 CrossRef CAS; (b) C. Tan, M. R. Pinto and K. S. Schanze, Chem. Commun., 2002, 446–447 RSC; (c) S. A. Kushon, K. D. Ley, K. Bradford, R. M. Jones, D. McBranch and D. Whitten, Langmuir, 2002, 18, 7245–7249 CrossRef CAS; (d) M. R. Pinto and K. S. Schanze, Proc. Natl. Acad. Sci. U. S. A., 2004, 101, 7505–7510 CrossRef CAS PubMed; (e) S. Kumaraswamy, T. Bergstedt, X. Shi, F. Rininsland, S. Kushon, W. Xia, K. Ley, K. Achyuthan, D. McBranch and D. Whitten, Proc. Natl. Acad. Sci. U. S. A., 2004, 101, 7511–7515 CrossRef CAS PubMed; (f) F. Rininsland, W. Xia, S. Wittenburg, X. Shi, C. Stankewicz, K. Achyuthan, D. McBranch and D. Whitten, Proc. Natl. Acad. Sci. U. S. A., 2004, 101, 15295–15300 CrossRef CAS PubMed; (g) Y. Liu, K. Ogawa and K. S. Schanze, Anal. Chem., 2008, 80, 150–158 CrossRef CAS PubMed.
- (a) B. S. Gaylord, A. J. Heeger and G. C. Bazan, Proc. Natl. Acad. Sci. U. S. A., 2002, 99, 10954–10957 CrossRef CAS PubMed; (b) B. S. Gaylord, A. J. Heeger and G. C. Bazan, J. Am. Chem. Soc., 2003, 125, 896–900 CrossRef CAS PubMed; (c) S. Wang, B. S. Gaylord and G. C. Bazan, J. Am. Chem. Soc., 2004, 126, 5446–5451 CrossRef CAS PubMed.
- (a) L. Chen, D. W. McBranch, H. L. Wang, R. Helgeson, F. Wudl and D. Whitten, Proc. Natl. Acad. Sci. U. S. A., 1999, 96, 12287–12292 CrossRef CAS PubMed; (b) J. Wang, D. Wang, E. K. Miller, D. Moses, G. C. Bazan and A. J. Heeger, Macromolecules, 2000, 33, 5153–5158 CrossRef CAS; (c) C. Fan, K. W. Plaxco and A. J. Heeger, J. Am. Chem. Soc., 2002, 124, 5642–5643 CrossRef CAS PubMed.
- S. W. Thomas III, G. D. Joly and T. M. Swager, Chem. Rev., 2007, 107, 1339–1386 CrossRef PubMed.
- (a) F. He, Y. Tang, S. Wang, Y. Li and D. Zhu, J. Am. Chem. Soc., 2005, 127, 12343–12346 CrossRef CAS PubMed; (b) F. He, Y. Tang, M. Yu, F. Feng, L. An, H. Sun, S. Wang, Y. Li, D. Zhu and G. C. Bazan, J. Am. Chem. Soc., 2006, 128, 6764–6765 CrossRef CAS PubMed.
- (a) X. Zhao and K. S. Schanze, Langmuir, 2006, 22, 4856–4862 CrossRef CAS PubMed; (b) Y. Q. Huang, Q. L. Fan, S. B. Li, X. M. Lu, F. Cheng, G. W. Zhang, Y. Chen, L. H. Wang and W. Huang, J. Polym. Sci., Part A: Polym. Chem., 2006, 44, 5424–5437 CrossRef CAS; (c) Y. Q. Huang, Q. L. Fan, X. F. Liu, N. N. Fu and W. Huang, Langmuir, 2010, 26, 19120–19128 CrossRef CAS PubMed; (d) B. B. Ni, Q. Yan, Y. Ma and D. Zhao, Coord. Chem. Rev., 2010, 254, 954–971 CrossRef CAS.
- (a) J. C. Nelson, J. G. Saven, J. S. Moore and P. G. Wolynes, Science, 1997, 277, 1793–1796 CrossRef CAS PubMed; (b) S. H. Gellman, Acc. Chem. Res., 1998, 31, 173–180 CrossRef CAS; (c) D. J. Hill, M. J. Mio, R. B. Prince, T. S. Hughes and J. S. Moore, Chem. Rev., 2001, 101, 3893–4011 CrossRef CAS PubMed.
- K. Chan, C. Y. S. Chung and V. W. W. Yam, Chem.–Eur. J., 2015, 21, 16434–16447 CrossRef CAS PubMed.
- (a) A. Y. Y. Tam, W. H. Lam, K. M. C. Wong, N. Zhu and V. W. W. Yam, Chem.–Eur. J., 2008, 14, 4562–4576 CrossRef CAS PubMed; (b) A. Y. Y. Tam, K. M. C. Wong and V. W. W. Yam, J. Am. Chem. Soc., 2009, 131, 6253–6260 CrossRef CAS PubMed.
- (a) C. Po, A. Y. Y. Tam, K. M. C. Wong and V. W. W. Yam, J. Am. Chem. Soc., 2011, 133, 12136–12143 CrossRef CAS PubMed; (b) C. Po, A. Y. Y. Tam and V. W. W. Yam, Chem. Sci., 2014, 5, 2688–2695 RSC; (c) C. Po and V. W. W. Yam, Chem. Sci., 2014, 5, 4868–4872 RSC.
- (a) G. Biffi, D. Tannahill, J. McCafferty and S. Balasubramanian, Nat. Chem., 2013, 5, 182–186 CrossRef CAS PubMed; (b) G. Biffi, M. Di Antonio, D. Tannahill and S. Balasubramanian, Nat. Chem., 2014, 6, 75–80 CrossRef CAS PubMed; (c) A. Henderson, Y. Wu, Y. C. Huang, E. A. Chavez, J. Platt, F. B. Johnson, R. M. Brosh, D. Sen and P. M. Lansdorp, Nucleic Acids Res., 2013, 42, 860–869 CrossRef PubMed.
- (a) H. X. Zhou, G. Rivas and A. P. Minton, Annu. Rev. Biophys., 2008, 37, 375–397 CrossRef CAS PubMed; (b) R. Buscaglia, M. C. Miller, W. L. Dean, R. D. Gray, A. N. Lane, J. O. Trent and J. B. Chaires, Nucleic Acids Res., 2013, 41, 7934–7946 CrossRef CAS PubMed; (c) S.-i. Nakano, D. Miyoshi and N. Sugimoto, Chem. Rev., 2014, 114, 2733–2758 CrossRef CAS PubMed.
- (a) D. Miyoshi, H. Karimata and N. Sugimoto, Angew. Chem., 2005, 117, 3806–3810 ( Angew. Chem., Int. Ed. , 2005 , 44 , 3740–3744 ) CrossRef; (b) Z. y. Kan, Y. Yao, P. Wang, X. h. Li, Y. h. Hao and Z. Tan, Angew. Chem., Int. Ed., 2006, 45, 1629–1632 CrossRef CAS PubMed; (c) Y. Xue, Z. y. Kan, Q. Wang, Y. Yao, J. Liu, Y. h. Hao and Z. Tan, J. Am. Chem. Soc., 2007, 129, 11185–11191 CrossRef CAS PubMed.
- D. E. Gilbert and J. Feigon, Curr. Opin. Struct. Biol., 1999, 9, 305–314 CrossRef CAS PubMed.
- (a) A. M. Zahler, J. R. Williamson, T. R. Cech and D. M. Prescott, Nature, 1991, 350, 718–720 CrossRef CAS PubMed; (b) S. Neidle and G. Parkinson, Nat. Rev. Drug Discovery, 2002, 1, 383–393 CrossRef CAS PubMed; (c) J. L. Mergny, J. F. Riou, P. Mailliet, M. P. Teulade-Fichou and E. Gilson, Nucleic Acids Res., 2002, 30, 839–865 CrossRef CAS PubMed.
- A. T. Phan and J. L. Mergny, Nucleic Acids Res., 2002, 30, 4618–4625 CrossRef CAS PubMed.
- (a) F. X. Han, R. T. Wheelhouse and L. H. Hurley, J. Am. Chem. Soc., 1999, 121, 3561–3570 CrossRef CAS; (b) C. L. Grand, H. Han, R. M. Muñoz, S. Weitman, D. D. Von Hoff, L. H. Hurley and D. J. Bearss, Mol. Cancer Ther., 2002, 1, 565–573 CAS; (c) J. Seenisamy, S. Bashyam, V. Gokhale, H. Vankayalapati, D. Sun, A. Siddiqui-Jain, N. Streiner, K. Shin-ya, E. White, W. D. Wilson and L. H. Hurley, J. Am. Chem. Soc., 2005, 127, 2944–2959 CrossRef CAS PubMed; (d) I. M. Dixon, F. Lopez, A. M. Tejera, J. P. Estève, M. A. Blasco, G. Pratviel and B. Meunier, J. Am. Chem. Soc., 2007, 129, 1502–1503 CrossRef CAS PubMed.
- (a) H. Bertrand, S. Bombard, D. Monchaud, E. Talbot, A. Guedin, J. L. Mergny, R. Grunert, P. J. Bednarski and M. P. Teulade-Fichou, Org. Biomol. Chem., 2009, 7, 2864–2871 RSC; (b) K. Suntharalingam, A. J. P. White and R. Vilar, Inorg. Chem., 2009, 48, 9427–9435 CrossRef CAS PubMed; (c) J. T. Wang, Y. Li, J. H. Tan, L. N. Ji and Z. W. Mao, Dalton Trans., 2011, 40, 564–566 RSC.
- (a) J. Alzeer, B. R. Vummidi, P. J. C. Roth and N. W. Luedtke, Angew. Chem., 2009, 121, 9526–9529 ( Angew. Chem., Int. Ed. , 2009 , 48 , 9362–9365 ) CrossRef; (b) D. L. Ma, C. M. Che and S. C. Yan, J. Am. Chem. Soc., 2009, 131, 1835–1846 CrossRef CAS PubMed; (c) K. Suntharalingam, A. Łęczkowska, M. A. Furrer, Y. Wu, M. K. Kuimova, B. Therrien, A. J. P. White and R. Vilar, Chem.–Eur. J., 2012, 18, 16277–16282 CrossRef CAS PubMed; (d) P. Wu, D. L. Ma, C. H. Leung, S. C. Yan, N. Zhu, R. Abagyan and C. M. Che, Chem.–Eur. J., 2009, 15, 13008–13021 CrossRef CAS PubMed; (e) P. Wang, C. H. Leung, D. L. Ma, S. C. Yan and C. M. Che, Chem.–Eur. J., 2010, 16, 6900–6911 CrossRef CAS PubMed.
- (a) J. K. Barton and S. J. Lippard, Biochemistry, 1979, 18, 2661–2668 CrossRef CAS PubMed; (b) K. E. Erkkila, D. T. Odom and J. K. Barton, Chem. Rev., 1999, 99, 2777–2796 CrossRef CAS PubMed; (c) E. Rüba, J. R. Hart and J. K. Barton, Inorg. Chem., 2004, 43, 4570–4578 CrossRef PubMed; (d) W. Lu, D. A. Vicic and J. K. Barton, Inorg. Chem., 2005, 44, 7970–7980 CrossRef CAS PubMed; (e) B. M. Zeglis, V. C. Pierre and J. K. Barton, Chem. Commun., 2007, 4565–4579 RSC; (f) B. M. Zeglis, J. A. Boland and J. K. Barton, Biochemistry, 2009, 48, 839–849 CrossRef CAS PubMed; (g) A. C. Komor and J. K. Barton, Chem. Commun., 2013, 49, 3617–3630 RSC.
- At low concentration of 1 (i.e., 4 μM), a significant decline in the emission intensity of mPPE-Ala was observed, at which the emission intensity of mPPE-Ala has dropped to ca. 8.5% of the emission intensity without 1. As soon as the complex concentration approaches 15 μM, the quenching of the mPPE-Ala fluorescence by 1 has almost completed, with ca. 1.0% of the original emission intensity detected. Therefore, any enhanced light absorption by 1 at high concentrations should not contribute much to the quenching of the mPPE-Ala fluorescence and thus, the errors imposed by the possible self-absorption of 1 to the KSV values calculated at those concentrations should be negligible.
- R. Giraldo, M. Suzuki, L. Chapman and D. Rhodes, Proc. Natl. Acad. Sci. U. S. A., 1994, 91, 7658–7662 CrossRef CAS.
- (a) C. V. Kumar and E. H. Asuncion, J. Am. Chem. Soc., 1993, 115, 8547–8553 CrossRef CAS; (b) F. Koeppel, J. F. Riou, A. Laoui, P. Mailliet, P. B. Arimondo, D. Labit, O. Petitgenet, C. Hélène and J. L. Mergny, Nucleic Acids Res., 2001, 29, 1087–1096 CrossRef CAS PubMed.
- (a) G. Cohen, W. Bauer, J. Barton and S. Lippard, Science, 1979, 203, 1014–1016 CAS; (b) W. I. Sundquist and S. J. Lippard, Coord. Chem. Rev., 1990, 100, 293–322 CrossRef CAS; (c) E. R. Jamieson and S. J. Lippard, Chem. Rev., 1999, 99, 2467–2498 CrossRef CAS PubMed.
- (a) H. Xu, J. W. Aylott and R. Kopelman, Analyst, 2002, 127, 1471–1477 RSC; (b) P. D. JÖBsis, C. A. Combs and R. S. Balaban, J. Microsc., 2005, 217, 260–264 CrossRef PubMed; (c) A. Ojida, Y. Miyahara, J. Wongkongkatep, S.-i. Tamaru, K. Sada and I. Hamachi, Chem.–Asian J., 2006, 1, 555–563 CrossRef CAS PubMed.
- H. D. Batey, A. C. Whitwood and A. K. Duhme-Klair, Inorg. Chem., 2007, 46, 6516–6528 CrossRef CAS PubMed.
- (a) B. Jin, X. Zhang, W. Zheng, X. Liu, J. Zhou, N. Zhang, F. Wang and D. Shangguan, Anal. Chem., 2014, 86, 7063–7070 CrossRef CAS PubMed; (b) J. Dai, D. Chen, R. A. Jones, L. H. Hurley and D. Yang, Nucleic Acids Res., 2006, 34, 5133–5144 CrossRef CAS PubMed; (c) S. Rankin, A. P. Reszka, J. Huppert, M. Zloh, G. N. Parkinson, A. K. Todd, S. Ladame, S. Balasubramanian and S. Neidle, J. Am. Chem. Soc., 2005, 127, 10584–10589 CrossRef CAS PubMed; (d) S. Neidle and G. N. Parkinson, Curr. Opin. Struct. Biol., 2003, 13, 275–283 CrossRef CAS PubMed; (e) C. C. Chang, C. W. Chien, Y. H. Lin, C. C. Kang and T. C. Chang, Nucleic Acids Res., 2007, 35, 2846–2860 CrossRef CAS PubMed; (f) A. Siddiqui-Jain, C. L. Grand, D. J. Bearss and L. H. Hurley, Proc. Natl. Acad. Sci. U. S. A., 2002, 99, 11593–11598 CrossRef CAS PubMed.
- (a) S. Burge, G. N. Parkinson, P. Hazel, A. K. Todd and S. Neidle, Nucleic Acids Res., 2006, 34, 5402–5415 CrossRef CAS PubMed; (b) D. J. Patel, A. T. Phan and V. Kuryavyi, Nucleic Acids Res., 2007, 35, 7429–7455 CrossRef CAS PubMed.
- (a) J. B. Lepecq and C. Paoletti, J. Mol. Biol., 1967, 27, 87–106 CrossRef CAS PubMed; (b) Q. Guo, M. Lu, L. A. Marky and N. R. Kallenbach, Biochemistry, 1992, 31, 2451–2455 CrossRef CAS PubMed; (c) T. Shida, N. Ikeda and J. Sekiguchi, Nucleosides Nucleotides, 1996, 15, 599–605 CrossRef CAS; (d) F. Rosu, E. de Pauw, L. Guittat, P. Alberti, L. Lacroix, P. Mailliet, J. F. Riou and J. L. Mergny, Biochemistry, 2003, 42, 10361–10371 CrossRef CAS PubMed; (e) J. Reynisson, G. B. Schuster, S. B. Howerton, L. D. Williams, R. N. Barnett, C. L. Cleveland, U. Landman, N. Harrit and J. B. Chaires, J. Am. Chem. Soc., 2003, 125, 2072–2083 CrossRef CAS PubMed; (f) A. Pothukuchy, C. L. Mazzitelli, M. L. Rodriguez, B. Tuesuwan, M. Salazar, J. S. Brodbelt and S. M. Kerwin, Biochemistry, 2005, 44, 2163–2172 CrossRef CAS PubMed; (g) M. J. Hannon, Chem. Soc. Rev., 2007, 36, 280–295 RSC.
- At low concentrations of c-myc (≤1.57 μM), the electrostatic and/or π–π stacking interactions between 1 and c-myc would bring the complex molecules into close proximity, resulting in an increase in 3MMLCT emission intensity (Fig. S18a†). Further increase in concentration of c-myc (>1.57 μM) would lead to an increase in the amount of negative charges in the solution, and hence a “dilution” in the local concentration of platinum(II) complex would be found, resulting in the deaggregation and the diminution of the 3MMLCT emission (Fig. S18b and S18c†).
- An increase in emission intensities of mPPE-Ala and 1, respectively, was also found upon the addition of PEG-200 (Fig. S21a and S21b†). This may be due to the reduction of their polar interactions with water through the release of interfacial water molecules and the rigidifcation of the molecules under MC conditions. Therefore, the excited state molecules would be less readily quenched via the non-radiative deactivation pathways (from ref. 15 and 44).
- (a) R. Hänsel, F. Löhr, S. Foldynová-Trantírková, E. Bamberg, L. Trantírek and V. Dötsch, Nucleic Acids Res., 2011, 39, 5768–5775 CrossRef PubMed; (b) J. Lin, Y. Y. Yan, T. M. Ou, J. H. Tan, S. L. Huang, D. Li, Z. S. Huang and L. Q. Gu, Biomacromolecules, 2010, 11, 3384–3389 CrossRef CAS PubMed; (c) L. Petraccone, in Quadruplex Nucleic Acids, ed. J. B. Chaires and D. Graves, Springer Berlin Heidelberg, 2013, vol. 330, pp. 23–46 Search PubMed.
- (a) L. Chen, S. Xu, D. McBranch and D. Whitten, J. Am. Chem. Soc., 2000, 122, 9302–9303 CrossRef CAS; (b) J. J. Lavigne, D. L. Broughton, J. N. Wilson, B. Erdogan and U. H. F. Bunz, Macromolecules, 2003, 36, 7409–7412 CrossRef CAS; (c) J. Dalvi-Malhotra and L. Chen, J. Phys. Chem. B, 2005, 109, 3873–3878 CrossRef CAS PubMed; (d) T. Zhang, H. Fan, J. Zhou, G. Liu, G. Feng and Q. Jin, Macromolecules, 2006, 39, 7839–7843 CrossRef CAS; (e) H. A. Al Attar and A. P. Monkman, J. Phys. Chem. B, 2007, 111, 12418–12426 CrossRef CAS PubMed; (f) H. Guan, M. Cai, L. Chen, Y. Wang and Z. He, Luminescence, 2010, 25, 311–316 CrossRef CAS PubMed; (g) A. T. Ngo and G. Cosa, Langmuir, 2010, 26, 6746–6754 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available: Experimental details, electronic absorption spectra and resonance light scattering spectra of mPPE-Ala at different concentrations of 1; emission spectra and time-resolved emission decay profiles of mPPE-Ala at different concentrations of 1; parameters obtained from the emission spectra and time-resolved emission decay profiles of mPPE-Ala at different concentrations of 1; electronic absorption spectra of 1 at different concentrations of c-myc; circular dichroism (CD) spectrum of c-myc; electronic absorption spectra and UV melting profile of c-myc at different temperatures in the absence and in the presence of 1; table listing the UV melting temperatures of c-myc, bcl-2, c-kit1 and human telomeric DNA in the absence and in the presence of 1; emission spectra of mPPE-Ala–1 ensemble at different concentrations of c-myc-c, pre-formed duplex of c-myc and c-myc-c, pre-formed duplex of human telomeric DNA and complementary sequence of human telomeric DNA, bcl-2, c-kit1 and human telomeric DNA; CD spectra of pre-formed duplex of c-myc and c-myc-c at different concentrations of 1; emission spectra of 1 at different concentrations of c-myc; emission spectra of mPPE-Ala–1 ensemble at different concentrations of c-myc in the presence of 36% (v/v) PEG-200; changes in relative emission intensities of mPPE-Ala–1 ensemble at different concentrations of c-myc at different volume percentages of PEG-200; emission spectra of mPPE-Ala, 1 and mPPE-Ala–1 ensemble at different volume percentages of PEG-200; CD spectra of c-myc at different volume percentages of PEG-200; changes in relative emission intensities of mPPE-Ala–1 ensemble at different concentrations of human telomeric DNA at different volume percentages of PEG-200. See DOI: 10.1039/c5sc04563k |
| This journal is © The Royal Society of Chemistry 2016 |