A cellular model for screening of lactobacilli that can enhance tight junctions
Qi Xuab,
Xiangfei Liab,
Eryin Wangab,
Yufeng Heab,
Boxing Yinc,
Dongsheng Fangc,
Gang Wang*ab,
Jianxin Zhaoab,
Hao Zhangab and
Wei Chenabd
aState Key Laboratory of Food Science and Technology, School of Food Science and Technology, Jiangnan University, Wuxi 214122, P. R. China. E-mail: wanggang@jiangnan.edu.cn; Fax: +86-510-85912155; Tel: +86-510-85912155
bInternational Joint Research Laboratory for Probiotics, Jiangnan University, Wuxi 214122, P. R. China
cKangyuan Dairy Co., Ltd., Yangzhou University, Yangzhou 225004, P. R. China
dBeijing Innovation Centre of Food Nutrition and Human Health, Beijing Technology and Business University (BTBU), Beijing 100048, P. R. China
First published on 18th November 2016
Abstract
It is widely believed that the dysfunction of tight junctions (TJs) in the gut barrier is one of the most important pathogenic factors in inflammatory bowel disease (IBD). More and more studies show that probiotics can be engaged as a substitute for traditional medicine such as antibiotics to treat IBD. However, there are very few efficient cellular models that can rapidly screen effective probiotics for TJ regulation. Co-incubation of probiotics and PT–gliadin has been used by some researchers to study the TJ regulatory function of bacteria. In this study, a modification was performed to overcome the deficiency of this method, i.e., that the indirect effect of probiotics on TJ restoration cannot be excluded. In a sectional incubation model, some of the Lactobacillus strains proved to be effective in TJ regulation, which is similar to the result obtained with an animal model. Principal component analysis showed that the modified sectional incubation model reflected the regulatory mode of probiotics in the gut more accurately than that by the co-incubation model. This study provides a new strategy that may improve the efficiency of rapid screening of available probiotics for intervention in IBD.
Introduction
As the pace of life quickens, more and more people tend to suffer from inflammatory bowel disease (IBD) including Crohn's disease and ulcerative colitis, which are common enteropathies in Europe and America.1,2 It has been reported that the dysfunction of tight junctions (TJs) controlling the paracellular pathway in the gut barrier is one of the most important pathogenic factors in intestinal disorders and the maintenance of gut barrier integrity to preserve human health.3–5 Because the precise etiology of IBD is currently unknown, several kinds of antibiotic drugs, corticosteroids and immuno-modulators have been used to treat IBD in spite of their strong side effects. Thus, researchers have tried to discover long-term therapies that are harmless to patients.6–9 In recent studies, Lactobacillus plantarum was proposed to enhance the gut barrier by increasing the scaffold protein zonula occludens (ZO) and the transmembrane protein occludin, and at the same time, probiotics have been promoted to modify the localization of TJ proteins and prevent the loss of claudin-1 and -2.10,11 Although some researchers have suggested that L. rhamnosus GG (LGG) may increase claudin 3 expression to strengthen gut barrier function, others have suggested that barrier dysfunction is caused by the increased claudin-2 and decreased occludin expression in epithelial TJs, which causes weaker anastomoses between neighboring cells. Thus, there is no precise evidence for how the different kinds of lactobacilli influence the postnatal expression and localization of claudins.12,13 However, most of these studies focused on the effects of a single lactic acid bacterial strain or a mixed system such as VSL#3 on the gut barrier, which could lead readers to believe that all strains of the same bacterial genus have the same functions.Several animal models exist for exploring the pathogenesis of IBD, such as mouse, broiler chicken and piglet models.14–16 To simplify the research procedures and improve the screening efficiency of probiotics effective for coeliac disease, T-84 cells, HT-29 cells, Caco-2 cells and other typical intestinal cell lines are widely used for in vitro research.17–19 Especially, HT-29 cells are always used as a cellular model because of certain characteristics of intestinal epithelial cells, including a mucous layer that does not form in a Caco-2 monolayer model.20 Furthermore, co-incubation of lactobacilli and HT-29 cells is studied with the advantage of requiring less time to cultivate wells compared to other intestinal cell lines.21,22 It is well known in Western countries that one of the most common food-related disorders is celiac disease, an autoimmune-mediated intestinal disorder induced by gluten ingestion from wheat gliadin, whose high proline- and glutamine-content peptide fragments are responsible for damaging intestinal tissue.23–25 Because of the increased permeability of the epithelial layer, intestinal epithelial cell lines are considered leaky when exposed to a peptic-tryptic digest of gliadin (PT–gliadin), and this cellular model has been used to investigate the restorative capacity of certain probiotics following TJ damage.26,27
The most used method for evaluating bacteria whose protective effect on intestinal epithelial cells is stimulated by PT–gliadin is to co-incubate lactobacilli and PT–gliadin with the cells.28 However, existing research has found that Saccharomyces, Bifidobacterium lactis, L. sanfranciscensis and some other lactobacilli from sourdough starter are fermentative strains for degrading wheat gliadin.29,30 So, with this method, it is doubtful that the protective effects of probiotics on TJs is related to the regulatory role of the bacteria themselves. Therefore, in this study, we propose a novel sectional incubation method that avoids the factor of gliadin degradation by bacteria. With this model, different functions of different bacterial strains on TJ regulation can be displayed clearly, and this cellular model allows more accurate and efficient screening of effective probiotics to characterize their capacity to regulate the intestinal barrier.
Materials and methods
Bacterial strains and culture conditions
All 6 Lactobacillus strains used in this study (Table 1) were stored in the Culture Collection of Food Microorganisms of Jiangnan University (Wuxi, China). LGG (ATCC 53103) was purchased from the American Type Culture Collection, and the other 5 strains were isolated from pickles, yoghurt, or distiller's yeast. These strains were maintained as frozen stocks (−80 °C) in de Man, Rogosa and Sharpe (MRS) broth (Hopebio Company, Qingdao, China) supplemented with 30% (v/v) glycerol. All strains were subcultured at least three times using 2% (v/v) inoculum in MRS broth at 37 °C for 18 h prior to use.| Lactobacillus strain | Characteristic | Source or reference |
|---|---|---|
| a CFM-JU, Culture Collection of Food Microorganisms of Jiangnan University (Wuxi, China). | ||
| L. plantarum (P1) | Yoghurt | CCFM-JU |
| L. fermentum (F1) | Distiller's yeast | CCFM-JU |
| L. plantarum (P2) | Pickles | CCFM-JU |
| L. plantarum (P3) | Pickles | CCFM-JU |
| L. rhamnosus GG ATCC 53103 (LGG) | Healthy human intestinal flora | American type culture collection |
| L. casei (C1) | Pickles | CCFM-JU |
Cell culture
The human colonic cell line HT-29 was obtained from the cell bank of the type culture collection of the Chinese Academy of Sciences, Shanghai, China. HT-29 cells were cultured in Roswell Park Memorial Institute 1640 Glutamax medium (RPMI; Invitrogen; Life Technologies) containing 10% FBS (v/v) and 1% (v/v) antibiotics. Cells were cultured in a 5% CO2 atmosphere at 37 °C and passaged 105 cm−2 when they reached 85% confluence, with 23–60 passages being used. The culture medium was changed every 2 days, and medium without antibiotics was used for the co-culture with lactobacilli.Preparation of PT–gliadin and PT–bovine serum albumin (BSA)
Sixty milligrams of gliadin from wheat (G3375; Sigma-Aldrich) or 60 mg of albumin from bovine serum (A7030; Sigma-Aldrich) was dissolved in 10 mL of 50 mM Na-acetate buffer (pH 4.0). Then, 3 mg pepsin (64007137; Sinopharm Chemical Reagent Co., Ltd.) was added, and the mixture incubated for 2 h at 37 °C under agitation; thereafter, 71 mg of Na2HPO4 was added to the solution, and the pH was adjusted to 7.0 by NaOH. Three milligrams of trypsin (64008860; Sinopharm Chemical Reagent Co., Ltd.) was added, and the reaction mixture was incubated for another 2 h at 37 °C under agitation. The reaction was stopped by heating (>95 °C, 10 min), and both the resulting PT–gliadin mixture and PT–BSA mixture were frozen and lyophilized. Lyophilized fractions were stored at −20 °C. Fractions were added to HT-29 cell monolayers at a concentration of 4 mg mL−1.Treatment of HT-29 monolayers with bacteria
Measurement of transepithelial electrical resistance (TER)
HT-29 cells were plated on Transwell Permeable Supports (3640-Clear, Corning Corporate) and grown until the Transwell inserts were overspread with cells, which were identified as confluent. After the addition of PT–gliadin, PT–BSA or lactobacilli to the medium, TER was measured by a Millicell ERS-2 volt-ohm meter (Millipore Corporate) immediately after changing the media every half hour. Furthermore, there were some groups that were first treated with PT–gliadin for 3 h, and then the PT–gliadin was washed away. After washing, lactobacilli in medium was added for another 3 h incubation. During incubation, TER was measured each hour.31Extraction of total RNA and quantitative polymerase chain reaction (qPCR)
Total isolation of RNA from cultured HT-29 cells and from each colonic tissue sample was performed with TRIzol (Invitrogen, Carlsbad, CA) according to the manufacturer's instructions. cDNA was synthesized from total RNA using Prime Script RT reagent Kit with gDNA Eraser (Takara, Tokyo, Japan). Subsequently, qPCR was performed using SYBR Green super mix (Qiagen, Germany) in a CFX96 Real-Time System (Bio-Rad, Hercules, CA). Each PCR was performed in triplicate and normalized with the housekeeping gene human β-actin or mouse GAPDH. The results were analyzed by use of the 2−ΔΔCq method.32 qPCR was performed with the primer pairs depicted in Table 2.| Gene | Sequence |
|---|---|
| a Most of the sequences were reported by Junki Miyamoto33 and Antonella Orlando.28 | |
| Human β-actin | Forward 5′-TTTTAGGATGGCAAGGGACTT-3′ |
| Reverse 5′-GATGAGTTGGCATGGCTTTA-3′ | |
| Human ZO-1 | Forward 5′-ATCCCTCAAGGAGCCATTC-3′ |
| Reverse 5′-CACTTGTTTTGCCAGGTTTTA-3′ | |
| Human ZO-2 | Forward 5′-AAAGCAGAGCGAACGAAGAG-3′ |
| Reverse 5′-TTTAGTTGCCAGACCCGTTC-3′ | |
| Human occludin | Forward 5′-CCAATGTCGAGGAGTGGG-3′ |
| Reverse 5′-CGCTGCTGTAACGAGGCT-3′ | |
| Human claudin-1 | Forward 5′-AAGTGCTTGGAAGACGATGA-3′ |
| Reverse 5′-CTTGGTGTTGGGTAAGAGGTT-3′ | |
| Human claudin-3 | Forward 5′-AAGGTGTACGACTCGCTGCT-3′ |
| Reverse 5′-GAAGTCCCGGATAATGGTGTT-3′ | |
| Mouse GAPDH | Forward 5′-TCAAGAAGGTGGTGAAGCAG-3′ |
| Reverse 5′-AAGGTGGAAGAGTGGGAGTTG-3′ | |
| Mouse ZO-1 | Forward 5′-CTTCTCTTGCTGGCCCTAAAC-3′ |
| Reverse 5′-TGGCTTCACTTGAGGTTTCTG-3′ | |
| Mouse ZO-2 | Forward 5′-AACGGATGCTGGAAGTTAAT-3′ |
| Reverse 5′-TCTGCTTGCTGTCTCTCAACA-3′ | |
| Mouse occludin | Forward 5′-CACACTTGCTTGGGACAGAG-3′ |
| Reverse 5′-TAGCCATAGCCTCCATAGCC-3′ | |
| Mouse claudin-1 | Forward 5′-GATGTGGATGGCTGTCATTG-3′ |
| Reverse 5′-CCTGGCCAAATTCATACCTG-3′ | |
| Mouse claudin-3 | Forward 5′-AACTGCGTACAAGACGAGACG-3′ |
| Reverse 5′-ATCCCTGATGATGGTGTTGG-3′ | |
Animal model
Six-week-old female C57BL6/J mice34 purchased from Shanghai Laboratory Animal Center (Shanghai, China) were housed in the Animals Housing Unit of Jiangnan University in a controlled environment (temperature, 22 °C ± 2 °C; humidity, 55% ± 5%; lights, 12 h light/dark cycle). All experimental procedures were approved by the Animal Ethics Committee of Jiangnan University, China, and strictly followed the ethical guidelines of the European Community Directive 2010/63/EU. The experimental procedures were as follows: 54 C57BL6/J female mice were randomly divided into 9 groups (n = 6): control, dextran sodium sulfate (DSS) + vehicle, DSS + P1, DSS + F1, DSS + P2, DSS + P3, DSS + LGG, DSS + C1, DSS + sulfasalazine (SASP). From day 1 to 7, all the mice were freely kept with ordinary fodder and water in the animals houses for adaptation. Then, all the mice in the lactobacilli groups were administered with 1 × 109 CFU mL−1 lactobacilli in vehicle (3% sucrose solution) or vehicle only once a day by gavage from day 8 to 21. Acute colitis was induced in the mice in all groups except for the control group by the addition of 2.5% (w/v) DSS (molecular weight 36![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000–50
000–50![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000, MP Biomedicals, Aurora, OH) to their drinking water from day 15 to 21. The SASP group was given standard laboratory chow with 50.0 mg mL−1 mesalazine (Ethypharm Pharmaceutical Co., Ltd., Shanghai, China) in vehicle once a day by gavage from day 15 to 21. At the end of the 3 week experimental period, all the mice were anesthetized (100 mg per kg bw ketamine) and sacrificed. The colonic tissues were removed from the mice, and total RNA from the diseased colon was extracted by TRIzol.
000, MP Biomedicals, Aurora, OH) to their drinking water from day 15 to 21. The SASP group was given standard laboratory chow with 50.0 mg mL−1 mesalazine (Ethypharm Pharmaceutical Co., Ltd., Shanghai, China) in vehicle once a day by gavage from day 15 to 21. At the end of the 3 week experimental period, all the mice were anesthetized (100 mg per kg bw ketamine) and sacrificed. The colonic tissues were removed from the mice, and total RNA from the diseased colon was extracted by TRIzol.
Zonulin determination by ELISA
In the co-incubation cell model, the cell culture medium was quantified every 1 h (ranging from 0 to 3 h) after PT–gliadin and different lactobacilli co-culture, following the protocol of the zonulin enzyme-linked immunosorbent assay (ELISA) kit (SY-ELA7760, Shanghai Win-Win Biological Technology Co., Ltd.). In the sectional incubation cell model, after PT–gliadin (already co-cultured with HT-29 for 3 h) was washed away, lactobacilli suspensions were added to the washed cells. Then, the cell culture medium was quantified every 1 h (ranging from 3.5 to 6.5 h) with the use of the method mentioned above.Immunofluorescence microscopy
HT-29 cells grown on cover glasses were treated with PT–gliadin or PT–BSA for 3 h. After washing, the cells were incubated with different Lactobacillus strains for another 3 h. 1640 medium was used as a control. The cells were then washed with ice-cold PBS and fixed with 4% (w/v) paraformaldehyde for 20 min at room temperature. After washing, the fixed cells were permeabilized with 0.1% Triton X-100 in PBS for 10 min. The cells were blocked in 2% PSA (2% BSA dissolved in PBS) for 30 min and incubated with rabbit polyclonal anti-ZO-1/TJP1 (40-2200, Life Technologies) or rabbit polyclonal anti-occludin (71-1500, Life Technology) or rabbit polyclonal anti-claudin-1 (71-7800, Life Technologies) at 4 °C overnight. After this, the antibodies were washed away by PBST (0.05% Tween-20 dissolved in PBS) followed by 50 min of incubation in the dark with 0.2% FITC-conjugated goat anti-rabbit IgG (H + L) secondary antibody (Alexa Fluor 488 conjugate, A-11008, Life Technologies) and 10 min of incubation with 200 μL of DAPI (Beyotime). A confocal microscope (LEICA TCS SP8, Germany) was used to observe immunofluorescence. Pixel intensity plot profiles were obtained in the ZO-1, occludin and claudin-1 channels and plotted with Microsoft Paint software.Results
Lactobacilli recovered the TER level of monolayer HT-29 cells to different extents to avoid the degradation of PT–gliadin by the strains
Two different treatments of PT–gliadin and lactobacilli were engaged to investigate the role of different strains on the recovery of TER decreased by PT–gliadin. As shown in Fig. 2A, TER was monitored both at baseline and following the addition of PT–gliadin and lactobacilli simultaneously each half hour from 0 h to 3 h, whereas in Fig. 1B, PT–gliadin was added into the HT-29 cell culture medium and then washed off with PBS after 3 h incubation. After washing, lactobacilli were added into RPMI medium and incubated with HT-29 cells for another 3.5 h. TER was monitored both at baseline and following PT–gliadin treatment and lactobacilli treatment each hour from 0 to 6.5 h except from 3.0 h to 3.5 h. TER was reduced in all of the PT–gliadin-treated HT-29 monolayers compared with that in the control or PT–BSA-treated group (over 20% reduction in Fig. 2A and about 40% reduction in Fig. 2B). All of the Lactobacillus strains incubated with HT-29 monolayers together with PT–gliadin showed a similar trend of recovery of the TER level but to different extents (83–92% recovery) from 0.5 h to 3 h (Fig. 2A), which indicated that all of the strains had the potential to alleviate the destruction of integrity caused by the PT–gliadin. However, although most of the Lactobacillus strains also showed a similar recovery function of TER in the HT-29 monolayers decreased by PT–gliadin from 3 h to 6.5 h (about 30% recovery), no detectable TER recovery was observed with L. fermentum F1 when PT–gliadin was washed off beforehand (Fig. 2B). This result suggested that the recovery activity of the TER of L. fermentum F1 in Fig. 2A was probably not because of its direct regulatory effect on cell TJs.Lactobacilli alleviated the zonulin releasing level of monolayer HT-29 cells to different extents to avoid degradation of the PT–gliadin by the strains
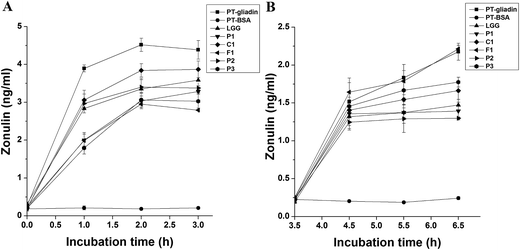
The role of the different strains on the relief of zonulin release caused by PT–gliadin were also investigated with the two different treatments of PT–gliadin and lactobacilli. As shown in Fig. 3A, the different Lactobacillus strains and PT–gliadin (PT–BSA) were added to culture medium of HT-29 cells simultaneously. There were no significant effects of PT–BSA on the secretion of zonulin, whereas the incubation of HT-29 cells with PT–gliadin led to the obvious release of zonulin. All 6 strains showed detectable relief effects on zonulin release caused by PT–gliadin treatment. In another treatment, after washing away the cell culture medium containing PT–gliadin only, zonulin release reached baseline values and then showed a transitory increase (Fig. 3B). All of the strains except for L. fermentum F1 showed obvious relief effects on zonulin release when co-cultivated with HT-29 cells from 3.5 h to 6.5 h. Among these strains, L. plantarum P1 and L. plantarum P2 showed a relatively more prominent effect on the alleviation of zonulin release. Interestingly, LGG, which was reported to be an efficient candidate for cell TJ regulation, was not the best choice. Corresponding with the results seen with TER recovery (Fig. 2), L. fermentum F1 also showed a distinct role in relieving zonulin release, which indicated the indirect effect of this strain on cell TJ regulation.Some lactobacilli restored the protein level of cell TJ-related molecules decreased by PT–gliadin treatment
TJ proteins such as ZO-1, occludin and claudin-1 were located on the apical surface of cells connected to one another in the tissue, such as intestinal epithelium, the integrity of which determines the gut barrier function. Thus, these three proteins in HT-29 monolayers were analyzed with immunofluorescence to determine whether probiotics could affect the localization and the level of these molecules on the apical surface of adjacent HT-29 cell membranes. All of the immunofluorescent patterns were the same, with green labeling the TJ proteins and blue (DAPI staining of DNA) labeling the nuclei. As shown in Fig. 4, 3 h treatment of PT–gliadin significantly decreased the fluorescent staining of ZO-1, occludin and claudin-1 in the TJ compared to the control group, in which regular cultivation without serum or antibiotics was performed. Similarly, adding PT–BSA in the medium for co-culture did not damage the well-balanced TJs of the HT-29 cells, which showed that pepsin and trypsin were totally inactivated and harmless to the cells. After washing away the cell culture medium containing PT–gliadin, the 6 different Lactobacillus strains were incubated with PT–gliadin-treated HT-29 cells for another 3 h. As shown in Fig. 4, all strains except for L. fermentum F1 showed obvious recovery effects on the protein level of claudin-1, which located at the apical surface of HT-29 cells after the 3 h incubation. Also, L. fermentum F1 showed weak recovery effects on the other two TJ proteins, which agrees with the results in Fig. 2 and 3. In addition, LGG showed effects on the restoration of all of the TJ proteins on the cell surface. L. casei C1 maintained ZO-1 and claudin-1 effectively. L. plantarum P3 recovered claudin-1 to a certain extent. Consistent with the results in Fig. 3B, L. plantarum P1 and L. plantarum P2 showed a relatively more prominent effect on the level of ZO-1 protein restoration.Sectional incubation of PT–gliadin and bacteria estimated the effect of lactobacilli on intestinal barrier recovery more efficiently than co-incubation
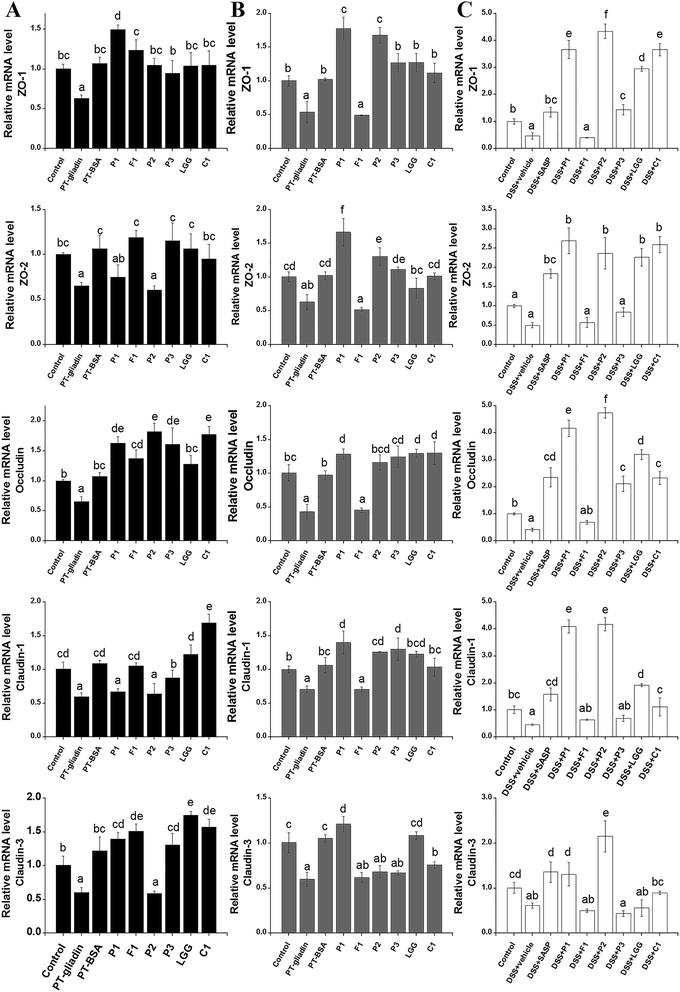
The mRNA level of five different TJ proteins was further examined to investigate the barrier-recovering effects of different lactobacilli in two different treatments of PT–gliadin and lactobacilli (sectional incubation and co-incubation) on HT-29 cells in vitro as shown in Fig. 5A and B. The protective effects of Lactobacillus strains on the intestinal barrier of the animals were also checked in vivo with quantitative PCR on the mRNA levels of TJ proteins of the C57BL6/J mice (Fig. 5C). When PT–gliadin was added to the lumen side of HT-29 cell monolayers grown on permeable supports, it significantly down-regulated the mRNA level of ZO-1, ZO-2, occludin, claudin-1 and claudin-3 compared to the control or PT–BSA in vitro. Similarly, DSS treatment caused about a 50% loss of the relative mRNA levels of the TJ proteins in colon tissues. As shown in the animal experiments (Fig. 5C), L. plantarum P1 and L. plantarum P2 restored the mRNA levels of almost all five of the TJ proteins or even elevated them in a compensatory manner. These two strains identically increased by a factor of nearly 4 (2−ΔΔCq) the relative mRNA levels of ZO-1, occludin and claudin-1 in mice colon tissue. However, L. fermentum F1 showed no obvious recovery effect on any of the five TJ protein mRNA levels. These results were almost identical with those obtained through in vitro experiments by treatment with sectional incubation. In this model, L. plantarum P1 and L. plantarum P2 elevated the mRNA levels of TJ proteins except for that of claudin-3 by L. plantarum P2. L. fermentum F1 failed to recover any of the TJ protein mRNA levels (Fig. 5B). However, the effects of these three strains were quite different in the co-incubation experiments in vitro. As shown in Fig. 5A, L. fermentum F1 showed excellent restorative effects on the mRNA levels of all five of the TJ proteins whereas L. plantarum P1 and L. plantarum P2 exhibited weak restorative effects on ZO-2 and claudin-1. L. plantarum P2 also failed to restore the mRNA level of claudin-3 in the co-incubation experiments in vitro (Fig. 5A).To evaluate which of these two cellular models predicted the efficiency of probiotics on intestinal barrier repair, principal component analysis (PCA) of the relative mRNA levels of TJ proteins in three experimental models was performed. All the values of the relative mRNA levels of TJs proteins were inputted into statistical software SPSS 16. All the experiment groups were set in variables and the extraction parameters were used as the normal order such as λe = 1.35 PC1 and PC2 were presented to show principal component scores of the relative mRNA levels effected by different treatments. PC1 scores were the most important judgment data of similarity, while PC2 scores indicated minor important judgment data of similarity. As shown in Fig. 6, the sectional incubation cell model was more aligned with the animal model than the co-incubation cell model. Although the PC1 score of the co-incubation cell model was closer to the animal model based on the relative mRNA level of occludin than that of the sectional incubation cell model, the PC2 scores of the co-incubation cell model and the sectional model were similar. In addition, the gap between the two scores of the sectional incubation cell model and the animal model was shorter than that between the two scores of the co-incubation cell traditional model and the animal model in all four of the other indexes including the relative mRNA levels of ZO-1, ZO-2, claudin-1 and claudin-3. This showed that the results of the sectional incubation cell model could estimate the effect of lactobacilli on intestinal barrier recovery more efficiently than could the co-incubation cell model.
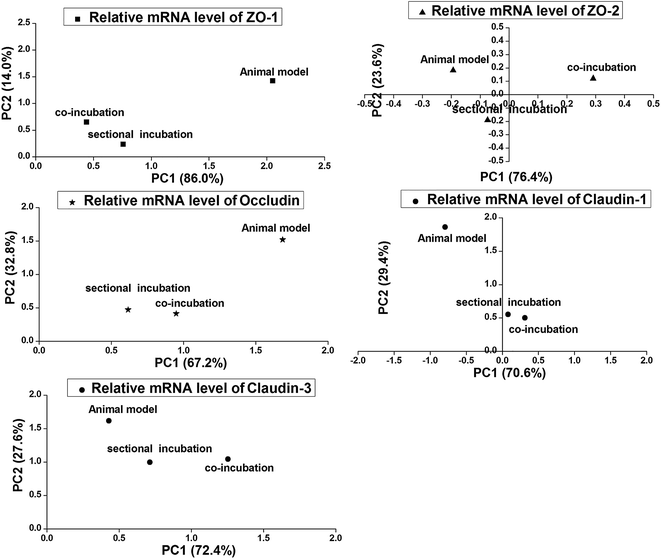 | ||
| Fig. 6 Principal component analysis score in three different models based on the relative mRNA level of TJ proteins. | ||
Discussion
The difficulty in understanding the mechanisms of IBD have led to uncertain therapies that nevertheless provide clear therapeutic effects. Traditional therapies such as antibiotics and immune regulatory agents have different levels of side effects. Recently, probiotics have been proposed to treat IBD instead of traditional medicine due to their biological activity. For example, VSL#3 was reported to increase the integrity of the intestinal mucosa. However, the strain of VSL#3 that can make a critical difference has yet to be identified.36 Some other probiotics have also been reported to be effective in the recovery of animal intestinal barriers and have been applied to animal feeds.37–39 There has been effective animal model for IBD investigation such as the classical model proposed by Cooper, et al.34 in which 2.5% DSS dissolved in water was engaged for 7 days on mice in inducing inflammatory bowel disease. In addition, this model was also engaged to evaluate the recovery effects of probiotics on IBD.6,40 However, animal experiments are not suitable for the large-scale screening of probiotics that can effectively restore the intestinal barrier because of their long duration and high cost. In addition, concerns about animal ethics make it more and more difficult to carry out such experimentation. Thus, the development of an in vitro method that can effectively reflect the effects of probiotics on the regulation of intestinal epithelial cell TJs in a short period of time is urgently needed.Cultivating human intestinal epithelial cells with PT–gliadin has already been proved to be effective in damaging the TJs of cells.41 This model has also been used to check the recovery effects of probiotics on cell TJs.42,43 Probiotics co-incubated with PT–gliadin significantly reduced the damage to cell TJs by PT–gliadin. However, some studies reported that some bacteria isolated from fermented foods can degrade gliadin.44 These results make this model unreliable because the probiotics might alleviate the damage through gliadin degradation rather than by the enhancement effect of probiotics on intestinal TJs. Thus, such factors related to the degradation of gliadin by probiotics must be eliminated. In this study, the co-incubation of cells with PT–gliadin and bacteria was substituted by sectional incubation. In cells treated with PT–gliadin beforehand, the PT–gliadin was completely washed away with PBS before incubating with lactobacilli so that the damaging effects of PT–gliadin on cell TJs would not be decreased because of gliadin degradation induced by the bacteria. The results of TER and TJ protein levels obtained with these two different treatments also confirmed the aforementioned hypotheses. TER has been widely measured in many study to investigate the permeability of the cell monolayer which indicated the extent of intercellular TJs damages.43 It was reported that some kinds of probiotics had the ability to regulate the TER of the cell monolayer.13,45 In this study, when lactobacilli and PT–gliadin were added simultaneously to HT-29 cells, the gliadin-induced decline in the TER level was significantly alleviated to different degrees by all of the Lactobacillus strains. However, in the groups in which lactobacilli were added after the cleaning pretreatment of PT–gliadin, although L. fermentum F1 had no direct effect on restoring the TER level, the other Lactobacillus strains still clearly restored the TER levels to some extent. Similar results were also found in the zonulin release experiments. The impairment of intercellular TJs might result as zonulin release so that the ascending zonulin concentration could be detected and recognized as the symptom of TJ disassembly.46 When lactobacilli were co-incubated with PT–gliadin, the zonulin release of HT-29 cells was alleviated to different degrees, whereas in the sectional incubation experiments, L. fermentum F1, which showed excellent alleviation of zonulin release caused by PT–gliadin pretreatment in the co-incubation experiments, exhibited no restorative ability on the alleviation of protein release. All the results above indicate that L. fermentum F1 may be the bacteria which can reduce the effective concentration of PT–gliadin while has no effect on recovery of cell TJs. Interestingly, three different species of L. plantarum P1, P2 and P3, showed different behaviors in two different models. The most effective bacteria in co-incubation model, L. plantarum P3, showed weakest effect on zonulin release alleviation in sectional incubation model. This also suggests that different effects on cell TJs are indeed exist even in the same species of lactobacilli. Consistent with the results of TER and zonulin release levels, the immunofluorescence experiments showed similar results. In the sectional incubation treatment, L. fermentum F1 showed no obvious recovery effects on the protein levels of ZO-1, occludin and claudin-1 at the apical surface, which were decreased by the PT–gliadin treatment. LGG showed obvious effects on the restoration of the levels of all of the TJ proteins on the cell surface. Consistent with the results in the zonulin release experiments, L. plantarum P1 and L. plantarum P2 showed a relatively more prominent effect on the restoration of the ZO-1 protein level. Also, L. casei C1 and L. plantarum P3 showed different maintenance abilities for TJ proteins such as ZO-1 or claudin-1. All of these results indicate that the significant mitigation ability of L. fermentum F1 on the PT–gliadin treatment was probably due to its capacity to degrade gliadin. Because the sectional incubation model in this study excluded the opportunity of the bacteria and PT–gliadin to interact, it was helpful in choosing the lactobacilli that had a direct effect on repairing cell TJs. These changes also showed that there were differences in the modulation effects of the different kinds of lactobacilli and different strains of L. plantarum that stimulated us to search for the dominant factor in regulating gut barrier function.
Although the results above indicate that false positive results could be avoided with sectional incubation model, it is still questionable that if the results obtained with this model correspond to the real situation of the body. To confirm the advantage of the sectional incubation model, the restorative capacity of the different Lactobacillus strains on TJ protein expression decreased by PT–gliadin was examined with two different models and compared with the results obtained with the animal model. The animal model is the most reliable method with which to investigate the restorative ability of the intestinal barrier by probiotics. It was commonly reported that DSS could induce inflammatory bowel disease in mice and this model was widely adopted to investigate how Lactobacillus strains alleviate intestinal injury.47,48 In these studies, 109 CFU alive bacteria were widely used to keep more than 106 CFU viable bacteria in the intestine to play a beneficial role considering the lethal effect of stomach acid and bile salts.49,50 From the results obtained with three different models, such as the effects of L. fermentum F1 on all the TJs proteins, L. plantarum P1 and L. plantarum P2 on ZO-2 and claudin-1, the effects of L. plantarum P1 and L. plantarum P3 on claudin-3, it could be found that results from sectional incubation model showed closer relevance with that from animal model. However, there were still counterexample such as the effect of L. plantarum P3 on claudin-1, which showed that the result from co-incubation model was closer to that from animal model. In addition, the contradictive effect of L. plantarum P2 on claudin-3 indicated that cellular model could not replace the animal models totally. To evaluate the correlation of data from three models, the principal component analysis was engaged. As we expected, PCA results showed that, comparing with scores of the co-incubation model, all the PC1 scores of the sectional model based on the relative mRNA level of ZO-1, ZO-2, claudin-1 and claudin-3 were closer to that of the animal model. However, the PC1 score of the co-incubation model based on the relative mRNA level of occludin showed closer location to the animal model than that of the sectional model did, without significant differences of PC2 scores. So, from the results of occludin, it seemed that co-incubation model was better. Even so, from the results in Fig. 5, it could be found that all the bacteria except for L. fermentum F1 showed positive effects on recovery of occludin RNA level, which totally agreed with the results from sectional incubation model, only with different extent on restoration. Therefore, the conclusion can be drawn that the sectional incubation model in this study was more authentic than the traditional co-incubation model in evaluating gut barrier function regulated by probiotics. However, even in the sectional incubation model, the modes of TJ restoration by probiotics were not very consistent with those in the animal model, which indicated the mechanisms in vivo were more complicated than those in the cultured cells in vitro. Although the modified extracorporeal model could not thoroughly replace the in vivo experiment, it can be used as a rapid method to some extent to screen probiotics with the potential function of intestinal barrier restoration.
Conclusions
The results of this study proved that the sectional incubation model using PT–gliadin and Lactobacillus strains to treat the cell monolayer in sequence could avoid the false positive data exist in co-incubation model. To the best of our knowledge, this is the first report of an in vitro sectional incubation model that can be used to investigate the ability of lactobacilli to restore the gliadin-induced reduction of TJ proteins. This model, which simulated animal experiments more precisely than the co-incubation model, can further be used to extensively examine the mechanism of probiotics on intestinal injury repair. Further, the modified extracorporeal model developed in this research could improve the efficiency of the rapid screening of available probiotics for intervention in IBD.Acknowledgements
This work was supported by the National Natural Science Foundation of China (No. 31671839, 31301407), the Key Projects in the National Science and Technology Pillar Program during the 12th Five-Year Plan (No. 2012BAD12B08), the National Basic Research Program of China (973 Program No. 2012CB720802), the Program of Introducing Talents of Discipline to Universities (B07029), the Fundamental Research Funds for the Central Universities (JUSRP51501), a project funded by the Priority Academic Program Development of Jiangsu Higher Education Institutions and the Program of Collaborative Innovation Center of Food Safety and Quality Control in Jiangsu Province.References
- H. Ashida, M. Ogawa, M. Kim, H. Mimuro and C. Sasakawa, Nat. Chem. Biol., 2012, 8, 36–45 CrossRef PubMed.
- L. Elli, E. Dolfini and M. T. Bardella, Toxicol. Lett., 2003, 146, 1–8 CrossRef PubMed.
- G. Thuijls, J.-J. de Haan, J. P. M. Derikx, I. Daissormont, M. h. Hadfoune, E. Heineman and W. A. Buurman, Shock, 2009, 31, 164–169 CrossRef PubMed.
- M. J. Puppa, J. P. White, S. Sato, M. Cairns, J. W. Baynes and J. A. Carson, Biochim. Biophys. Acta, Mol. Basis Dis., 2011, 1812, 1601–1606 CrossRef PubMed.
- M. Nébot-Vivinus, C. Harkat, H. Bzioueche, C. Cartier, R. Plichon-Dainese, L. Moussa, H. Eutamene, D. Pishvaie, S. Holowacz, C. Seyrig, T. Piche and V. Theodorou, World J. Gastroenterol., 2014, 20, 6832 CrossRef PubMed.
- J. T. Wang, H. Q. Chen, B. Yang, Z. N. Gu, H. Zhang, W. Chen and Y. Q. Chen, RSC Adv., 2016, 6, 14457–14464 RSC.
- B. G. Levesque, W. J. Sandborn, J. Ruel, B. G. Feagan, B. E. Sands and J.-F. Colombel, Gastroenterology, 2015, 148, 37–51 CrossRef PubMed.
- R. Bibiloni, R. N. Fedorak, G. W. Tannock, K. L. Madsen, P. Gionchetti, M. Campieri, C. De Simone and R. B. Sartor, Am. J. Gastroenterol., 2005, 100, 1539–1546 CrossRef PubMed.
- Z. Weizman, Lancet, 2010, 376, 233 CrossRef.
- J. Karczewski, F. J. Troost, I. Konings, J. Dekker, M. Kleerebezem and R. J. Brummer, Am. J. Physiol.: Gastrointest. Liver Physiol., 2010, 298, 851–859 CrossRef PubMed.
- F. Lutgendorff, R. M. Nijmeijer, P. A. Sandström, L. M. Trulsson, K. E. Magnusson and H. M. Timmerman, PLoS One, 2009, 4, e4512 Search PubMed.
- R. M. Patel, L. S. Myers, A. R. Kurundkar, A. Maheshwari, A. Nusrat and P. W. Lin, Am. J. Pathol., 2012, 180, 626–635 CrossRef PubMed.
- D. L. Boone, D. Corridoni, L. Pastorelli, B. Mattioli, S. Locovei, D. Ishikawa, K. O. Arseneau, M. Chieppa, F. Cominelli and T. T. Pizarro, PLoS One, 2012, 7, e42067 Search PubMed.
- M. Vidal, C. Forestier, N. Charbonnel, S. Henard, C. Rabaud and O. Lesens, J. Clin. Microbiol., 2010, 48, 2595–2598 CrossRef PubMed.
- W. L. Willis and L. Reid, Poult. Sci., 2008, 87, 606–611 CrossRef PubMed.
- H. H. Giang, T. Q. Viet, B. Ogle and J. E. Lindberg, Livest. Sci., 2010, 133, 182–184 CrossRef.
- S. Michail and F. Abernathy, J. Pediatr. Gastroenterol. Nutr., 2003, 36, 385–391 CrossRef PubMed.
- J. Sun, G. W. Le, Y. H. Shi and G. W. Su, Lett. Appl. Microbiol., 2007, 44, 79–85 CrossRef PubMed.
- Y. Sambuy, I. D. Angelis, G. Ranaldi, M. L. Scarino, A. Stammati and F. Zucco, Cell Biol. Toxicol., 2005, 21, 1–26 CrossRef PubMed.
- T.-S. Song, J.-Y. Kim, K.-H. Kim, B.-M. Jung, S.-S. Yun and S.-S. Yoon, Food Sci. Biotechnol., 2010, 19, 19–25 CrossRef.
- A. M. O'Hara, P. O'Regan, A. Fanning, C. O'Mahony, J. MacSharry, A. Lyons, J. Bienenstock, L. O'Mahony and F. Shanahan, Immunology, 2006, 118, 202–215 CrossRef PubMed.
- V. M. Wolters and C. Wijmenga, Am. J. Gastroenterol., 2007, 103, 190–195 CrossRef PubMed.
- M. Calasso, O. Vincentini, F. Valitutti, C. Felli, M. Gobbetti and C. R. Di, Eur. J. Nutr., 2012, 51, 507–512 CrossRef PubMed.
- E. Smecuol, H. J. Hwang, E. Sugai, L. Corso, A. C. Cherñavsky and F. P. Bellavite, J. Clin. Gastroenterol., 2013, 47, 139–147 CrossRef PubMed.
- S. Sibartie, A. M. O. Hara, J. Ryan, Á. Fanning, J. O. Mahony, S. O. Neill, B. Sheil, L. O. Mahony and F. Shanahan, BMC Immunol., 2009, 10, 54 CrossRef PubMed.
- K. A. Donato, M. G. Gareau, Y. J. Wang, P. M. Sherman, H. J. Flint and P. W. O. Toole, Microbiology, 2010, 156, 3288–3297 CrossRef PubMed.
- H. Putaala, T. Salusjärvi and M. Nordström, Res. Microbiol., 2008, 159, 692–698 CrossRef PubMed.
- A. Orlando, M. Linsalata, M. Notarnicola, V. Tutino and F. Russo, BMC Microbiol., 2014, 14, 19 CrossRef PubMed.
- J. M. Laparra and Y. Sanz, J. Cell. Biochem., 2009, 159, 801–807 Search PubMed.
- C. L. Gerez, G. C. Rollan and G. F. Valdez, Lett. Appl. Microbiol., 2006, 42, 459–464 CrossRef PubMed.
- K. Lindfors, T. Blomqvist, K. Juuti-Uusitalo, S. Stenman, J. Venäläinen, M. Mäki and K. Kaukinen, Clin. Exp. Immunol., 2008, 152, 552–558 CrossRef PubMed.
- K. J. Livak and T. D. Schmittgen, Methods, 2001, 25, 402–408 CrossRef PubMed.
- J. Miyamoto, T. Mizukure, S.-B. Park, S. Kishino, I. Kimura, K. Hirano, P. Bergamo, M. Rossi, T. Suzuki, M. Arita, J. Ogawa and S. Tanabe, J. Biol. Chem., 2015, 290, 2902–2918 CrossRef PubMed.
- H. S. Cooper, S. N. Murthy, R. S. Shah and D. J. Sedergran, Lab. Invest., 1993, 69, 238–249 Search PubMed.
- F. Zhao, J. Liu, X. Qu, X. Xu, X. Chen, X. Yang, F. Cao, J. Liang and J. Tian, Phys. Med. Biol., 2014, 59, 7777–7791 CrossRef PubMed.
- T. Kuhbacher, Gut, 2006, 55, 833–841 CrossRef PubMed.
- L. Zhang, Y. Q. Xu, H. Y. Liu, T. Lai, J. L. Ma, J. F. Wang and Y. H. Zhu, Vet. Microbiol., 2010, 141, 142–148 CrossRef CAS PubMed.
- S. E. Higgins, J. P. Higgins, A. D. Wolfenden, S. N. Henderson, A. T. Rodriguez and G. Tellez, Poult. Sci., 2008, 87, 27–31 CrossRef CAS PubMed.
- H. H. Giang, T. Q. Viet, B. Ogle and J. E. Lindberg, Livest. Sci., 2010, 133, 182–184 CrossRef.
- F. E. Macho, B. Pot and C. Grangette, Gut Microbes, 2011, 2, 280–286 CrossRef PubMed.
- E. K. Arendt, A. Moroni and E. Zannini, Microb. Cell Fact., 2011, 10, S15 CrossRef PubMed.
- S. Drago, A. R. El, P. M. Di, C. M. Grazia, A. Tripathi and A. Sapone, Scand. J. Gastroenterol., 2006, 41, 408–419 CrossRef CAS PubMed.
- G. R. Sander, A. G. Cummins and B. C. Powell, Febs Lett., 2005, 579, 4851–4855 CrossRef CAS PubMed.
- M. G. Clemente, Gut, 2003, 52, 218–223 CrossRef CAS PubMed.
- R. C. Anderson, A. L. Cookson, W. C. Mcnabb, P. Zaneta, J. M. Mark, J. K. William and C. R. Nicole, BMC Microbiol., 2010, 10, 1–11 CrossRef PubMed.
- A. Orlando, M. Linsalata, M. Notarnicola, V. Tutino and F. Russo, BMC Microbiol., 2014, 14, 19 CrossRef PubMed.
- R. Mennigen, K. Nolte, E. Rijcken, M. Utech, B. Loeffler, N. Senninger and M. Bruewer, Am. J. Physiol.: Gastrointest. Liver Physiol., 2009, 296, G1140–G1149 CrossRef CAS PubMed.
- L. X. Sang, B. Chang, W. L. Zhang, X. M. Wu, X. H. Li and M. Jiang, World J. Gastroenterol., 2010, 16, 1908–1915 CrossRef PubMed.
- P. Chen, Q. Zhang, H. Dang, X. Liu, F. Tian, J. Zhao, Y. Chen, H. Zhang and W. Chen, Nutrition, 2014, 30, 1061–1068 CrossRef CAS PubMed.
- C. Li, Q. Ding, S. P. Nie, Y. S. Zhang, T. Xiong and M. Y. Xie, J. Agric. Food Chem., 2014, 62, 11884–11891 CrossRef CAS PubMed.
| This journal is © The Royal Society of Chemistry 2016 |