Highly sensitive determination of ethyl carbamate in alcoholic beverages by surface-enhanced Raman spectroscopy combined with a molecular imprinting polymer†
Zhengzong Wuab,
Enbo Xuab,
Jingpeng Liab,
Jie Longab,
Aiquan Jiaoab and
Zhengyu Jin*ab
aThe State Key Laboratory of Food Science and Technology, School of Food Science and Technology, Jiangnan University, Wuxi 214122, China. E-mail: jinlab2008@yahoo.com; Fax: +86-510-85913299; Tel: +86-510-85913299
bCollaborative Innovation Center of Food Safety and Quality Control in Jiangsu Province, Jiangnan University, Wuxi 214122, China
First published on 8th November 2016
Abstract
A simple and reliable method for fast extraction and sensitive detection of ethyl carbamate (EC) in rice wine and fruit brandy based on the integration of molecularly imprinted polymers and surface-enhanced Raman spectroscopy (MIPs–SERS) was developed. Molecularly imprinted polymer microspheres were synthesized as artificial antibodies towards EC. Adsorption tests and thermogravimetric analysis validated the specific selectivity and stability of MIPs respectively. The synthesized MIPs was used as sorbents in solid-phase extraction (SPE) and could selectively separate and enrich EC from rice wine and fruit brandy samples with little interference. A silver dendrite nanostructure was employed as a SERS-active substrate for the enhancement of Raman signals. Spectra processed by principal component analysis (PCA) can clearly differentiate Raman signatures of wine samples with various EC contents. A partial least square (PLS) regression model demonstrated a good fitting effect (the correlation coefficient of calibration and prediction were 0.9614 and 0.9456, and 0.9559 and 0.9393 respectively for rice wine and fruit brandy) between the predicted and the reference data of EC in rice wine and fruit brandy samples. Our results show that the high selectivity of MIPs and the fingerprint Raman identification can be integrated into a novel nano-biosensor for fast and efficient detection of hazardous substances (EC) in complex rice wine and fruit brandy samples.
1. Introduction
EC, also known as carbamic acid ethyl ester, is a by-product of fermentation and storage with widespread occurrence in alcoholic beverages (e.g., wine, beer, Chinese rice wine).1 It has been confirmed to be genotoxic and carcinogenic in many animal species.2 In 2007, IARC reassessed and upgraded the status of EC from a possible human carcinogen (Group 2B) in 1987 to a probable human carcinogen (Group 2A), which suggested a potential carcinogenic risk to humans.3 In recent years, with the improvement of living conditions, more and more people pay much attention to the safety of alcoholic beverages. In this context, ethyl carbamate (EC) as the primary toxic compound existed in alcoholic beverages has attracted a great deal of attention than before. Until now, the tolerable levels of EC for different kinds of wines have been established by many countries.4,5 Assured conformance with the standards mentioned above demands the continuous monitoring of trace level of EC in alcoholic beverages.At present, the EC level in alcoholic beverages are usually determined by gas chromatography (GC)/mass spectroscopy (MS), GC/tandem MS, GC/high-resolution MS, high-performance liquid chromatography (HPLC) with fluorescence detection and enzyme-linked immunosorbent assay (ELISA).6,7 These methods are highly selective and very accurate. However, chromatographic methods are relatively sophisticated, expensive and tedious. In contrast, ELISA offers a rapid means of detection, but it is strongly dependent on the use of unstable enzymes and expensive test kits, which cannot meet the high-throughput detection required by the government and the beverage industry. Therefore, an alternative rapid and accurate approach is essentially required.
Spectroscopic methods are very promising and excellent tools for the determination of toxic or harmful substances in food because they are inherently rapid, specific, and possibly partially or completely computerized.8–10 Among spectroscopic methods, Raman spectroscopy is particularly valuable due to its great possibilities and advantages in detecting chemical compounds in liquid food (the interference from water in aqueous samples is very weak).11,12 However, compared to Rayleigh scattering, the cross-section of Raman scattering in normal Raman spectroscopy is weak, resulting in lack of sensitivity.13 Because of its significant enhancement of faint Raman scattering signals, surface-enhanced Raman spectroscopy (SERS) based on novel plasmonic nanoparticle substrates has overcome the technical barrier of normal Raman spectroscopy and become one of the most versatile quantitative approaches for detection down to the single-molecule level.14 SERS has been successfully used as a rapid and accurate detection method for determining many kinds of chemical compounds or bacteria in food during the past decade.15,16
However, SERS records vibrational signals associated with numerous functional groups in molecules, resulting in the fact that both target molecules and other constituents in matrices contribute to SERS spectra. This has become a major challenge to its analytical utility. For this reason, samples pretreatment is needed to eliminate the interferences to improve separation and detection and obtain satisfied results. Molecularly imprinting offers a new technique for efficient separation and enrichment of specific analytes from complicated matrices including fermented wines. It is a kind of biomimic technique to synthesize molecularly-imprinted polymers (MIPs) with artificially generated recognition sites able to specifically rebind a target molecule in preference to other closely-related compounds.17 MIPs synthesis is relatively cheap and easy, making it a great alternative to natural receptors. Because of these advantages, MIPs has attracted researchers' attention and been integrated with HPLC, quantum dots and graphene oxide for the determination of food chemical hazards in recent years.18,19 Lu et al. had successfully combined MIPs and SERS for accurate and high-throughput detection of melamine and α-tocopherol in milk and vegetable oils, respectively.20,21 To the best of our knowledge, no one has integrated MIPs and SERS to fabricate an innovative biosensing system for the separation and detection of EC in alcoholic beverages.
In this study, we tried to use MIPs as specific sorbent in solid phase extraction (SPE) to capture and separate EC in two kinds of alcoholic beverages, rice wine and fruit brandy, and then apply silver dendrite as SERS-active substrate to quantify the EC level in the eluent from SPE. The combination of the sensitivity of SERS and the selectivity of MIPs would immensely promote the development of sensitive and selective detection of chemical hazards in foods.
2. Materials and methods
2.1. Reagents and materials
Ethyl carbamate-d5 (EC-d5), ethylene glycol dimethacrylate (EGDMA), sodium chloride, α-methacrylic acid (MAA), 2,2′-azobis(isobutyronitrile) (AIBN), zinc plate (99.99% purity), ethanol and hydrochloric acid were purchased from Sigma-Aldrich (St. Louis, MO, USA). Silica, acetic acid, methylbenzene and methanol were obtained from Sinopharm Chemical Reagent Co., Ltd. (Shanghai, China). Silver nitrite and 3-(trimethoxysilyl)propyl methacrylate (γ-MPS) were purchased from Aladdin-Reagent Co., Ltd. (Shanghai, China). EC was provided by Jkchemical Inc. (Beijing, China). All reagents and solvents used were analytical or HPLC grade. The samples of rice wine and fruit brandy were obtained from two local supermarkets.2.2. Reference method for EC determination
EC in the samples of rice wine and fruit brandy was extracted using the classical liquid–liquid extract method reported by Valente et al. and Xu et al.22,23 with slight modifications. Briefly, 5 mL wine sample containing 50 μL of 400 μg L−1 EC-d5 (the internal standard) was added into a centrifuge tube, followed by adding 5 mL of dichloromethane and 1.8 g of NaCl. After that, the mixture was vortexed for 1 min, and centrifuged at 4000 × g for 3 min. The organic phase was obtained and the supernatant (wine sample) was suspended in 5 mL of dichloromethane followed by mixing and centrifugation (twice). The organic phase was merged and concentrated using vacuum distillation, dissolved in 2 mL of acetonitrile and purified using 2 mL 1-hexane (which was discarded after the mixed sample had been vortexed for 1 min and centrifuged at 4000g for 3 min). The residual solution was filtered through a Nylon filter (0.45 μm pore size) for the further analysis.For the detection of EC, a previous reported GC-MS method by our group was used.23 The GC-MS system used for detection was a SCION SQ 456-GC in combination with a PAL Combi-xt autosampler and mass selective detector (Bruker, USA). Data acquisition and analysis were conducted using standard software supplied by the manufacturer. Substances were separated on a DB-wax fused silica capillary column (30 m length, 0.25 mm i.d. and 0.25 μm film thickness, J&W Scientific, Folsom, CA, USA). The oven temperature was firstly held at 90 °C for 0.5 min, then the temperature was raised to 170 °C at a heating rate of 12 °C min−1. After that, the temperature was further raised to 230 °C at a heating rate of 25 °C min−1 and held at 230 °C for 4 min. The splitless injection mode and helium (carrier gas) with a constant flow rate of 1.0 mL min−1 were used. The mass spectrum in the electron impact mode was generated at 70 eV and the ion source temperature was 200 °C. The ions monitored were m/z 74 > 44 and 62 > 44 for EC, and m/z 64 > 44 for EC-d5. For quantification of the EC, the peak area ratios of the analytes to the EC-d5 (internal standard) were calculated.
2.3. Synthesis of MIPs
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2, v/v) for 24 h and 150 mL of methanol for another 24 h until no EC could be detected in the washing solutions. Finally, the composite particles were dried under vacuum (60 °C).
2, v/v) for 24 h and 150 mL of methanol for another 24 h until no EC could be detected in the washing solutions. Finally, the composite particles were dried under vacuum (60 °C).As a reference, nonimprinted polymer microspheres (NIPs) were also synthesized using the same synthesis protocol except for the omission of the template.
2.4. Rebinding assay
Static and kinetic adsorption tests were conducted individually to confirm the specific affinity of MIPs toward EC and the equilibration rates. In steady-state binding experiments, 10 mg of MIPs or NIPs was mixed with 2 mL of methanol solution of EC at various concentrations ranging from 10 mg L−1 to 120 mg L−1. The mixtures were shaken at 200 rpm for 120 min at 25 °C and then centrifuged at 4000 × g for 20 min. The supernatant was filtered through a nylon filter (0.45 μm pore size) and the concentration of the unbounded EC was measured by the GC-MS method abovementioned (Section 2.2.). The adsorption capacity (Q) was calculated by the following formula:
 | (1) |
In addition, the Scatchard, Freundlich, and Langmuir isotherm models were used to determine the affinity of MIPs and NIPs in the study. These three models were developed according to the equations shown in the ESI.†
For kinetic tests, 10 mg of either MIPs or NIPs was placed into centrifuge tubes and mixed 2 mL ethanol solution containing EC (60 mg L−1). The mixture was shaken for different time intervals ranging from 5 min to 240 min. After the centrifugation at 4000 × g for 20 min, the supernatant was treated and determined following the same procedures as for static adsorption.
The pseudo-second-order kinetic model was used to describe the adsorption process according to the formula:
 | (2) |
In order to evaluate the selective recognition ability, the analogues of EC, methyl carbamate, isoprocarb and meta-tolyl-N-methyl carbamate were selected as competitive agents to determine EC selectivity of the MIPs.
2.5. Molecularly imprinted solid phase extraction
500 mg of dried MIPs or NIPs particles were packed into a 6 mL empty SPE-cartridge with one PTFE frit (Agilent, Santa Clara, CA, USA) at each end. Prior to each extraction, the cartridge was preconditioned with 3 mL of methanol and water, respectively. A 3 mL of EC spiked rice wine and fruit brandy sample solution was loaded onto the cartridge at a flow rate of 2 mL min−1. After loading, the molecularly imprinted SPE column was washed with 3 mL of methanol–water (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 9, v/v) at 1.5 mL min−1, and finally, EC adsorbed on the sorbent was eluted with 3.0 mL of methanol/acetic acid (95
9, v/v) at 1.5 mL min−1, and finally, EC adsorbed on the sorbent was eluted with 3.0 mL of methanol/acetic acid (95![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 5, v/v) at 1.5 mL min−1. The eluent was collected in a test tube and condensed to dryness under a gentle flow of nitrogen at room temperature. The analytes were then redissolved in methanol solution. This solution was directly deposited onto the SERS-active nanosubstrate for spectral collection.
5, v/v) at 1.5 mL min−1. The eluent was collected in a test tube and condensed to dryness under a gentle flow of nitrogen at room temperature. The analytes were then redissolved in methanol solution. This solution was directly deposited onto the SERS-active nanosubstrate for spectral collection.
2.6. Synthesis of silver dendrite nanostructure
The silver dendrite was synthesized according to the following procedures: zinc plate (99.99%) was first treated by dilute hydrochloride acid (0.02 mol L−1) to remove surface contamination and was then rinsed with distilled water, followed by drying with cold air. The cleaned zinc plate was immersed into 150 mM AgNO3 solution and allowed to react at room temperature and ambient pressure for 1 min. The silver dendrite products were peeled carefully from the zinc plate with tweezers and put into a beaker. Then, the products were rinsed using distilled water and ethanol in sequence. Finally, the product was collected in glass vials. For use in a SERS experiment, the silver dendrite nanostructure was deposited onto a gold-coated glass slide (Thermo Electron, Waltham, MA, USA).2.7. Characterization
The surface groups of the polymer were measured using a FT-IR spectrometer (Nicolet iS10, Thermo Electron Corp., Madison, WI, USA) in the range of 500–4000 cm−1 with 256 scans at a resolution of 4 cm−1. The MIPs powders (5 mg) were ground with spectroscopic grade KBr powders (250 mg) and then pressed into 1 mm pellets.Thermogravimetric analysis (TGA) was carried out with a thermogravimetric analyzer (TGA-SDTA851e, Mettler Toledo Co., Ltd., Switzerland). Nitrogen gas was used as the carrier with flow rate of 50 mL min−1. Approximately 5.0 mg of each sample was used for analysis, and samples were heated from 30 °C to 600 °C at a heating rate of 10 °C min−1.
Morphologies of the silver dendrite nanostructure were characterized by a high-resolution transmission electron microscope (TEM, JEM-2100, JEOL, Japan) equipped with a charge-coupled device (CCD) camera operating at an acceleration voltage of 200 kV.
2.8. SERS spectra collection
A LabRAM HR Evolution system (HORIBA J-Y, France) coupled with an air-cooled He–Ne laser for 785 nm excitation, a motorized microscope and a CCD array detector with 1024 × 256 pixels was used to record the SERS spectra. The slit and pinhole were set at 100 and 400 μm, respectively, in the confocal configuration, with a holographic grating (600 g mm−1).After mounting the gold-coated microarray chip covered with multiple silver dendrite spots onto the standard stage of the microscope. The LabSpec 6 software (HORIBA Jobin Yvon) was utilized for spectral acquisition and instrument control. All experiments were carried out at least three times.
2.9. Statistical analysis
Raw SERS spectra usually contain noises (Fig. S1†). Thus, they were first baseline corrected and smoothed by the Savitzky–Golay algorithm to reduce spectral noise. Raman band at 1060 cm−1, which was attributed to NO3− on silver dendrite SERS substrate, and was used as an internal standard to normalize SERS spectra.Principal component analysis (PCA) was performed to examine any possible grouping of wine samples with different spiking levels of EC (0, 0.1, 0.2, 0.3, 0.4 and 0.5 mg L−1). The partial least-squares (PLS) regression model was established to correlate the actual EC concentrations and the predicted EC contents in wine samples. The correlation coefficient (R2 (cal)) and the root mean square error of calibration (RMSEC) and the correlation coefficient (R2 (pre)) and the root mean square error (RMSEP) of the prediction set were calculated to evaluate models. The calculations were performed by using the equations presented in the ESI.†
The spectral pretreatments were implemented in the commercial chemometric software The Unscrambler (v 10.2; CAMO Software AS, Oslo, Norway). PCA analysis and the construction of PLS models were carried out using Matlab R2010a (MathWorks, Natick, USA).
3. Results and discussions
The stepwise preparation process from activated SiO2 to EC-imprinted MIPs and the detailed fabrication procedures of the MIPs–SERS biosensing system for the determination of EC was schematically shown in Fig. 1. Firstly, a molecular memory was imprinted on the polymer, to make sure that the polymer could rebind the template selectively. After loading, washing, and eluting, the target molecular separated were finally deposited onto silver dendrite nanostructures for SERS signal collection. | ||
| Fig. 1 Schematic procedures for fabricating of MIPs–SERS biosensing system to detect EC in rice wine and fruit brandy. | ||
3.1. Characterization of MIPs and NIPs for EC
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O and saturated C–H, respectively.25 All the evidences demonstrated that the surface of SiO2 had been successfully modified by silane reagent. The vinyl groups introduced onto the surface of the activated silica by immobilization of a long chain group played a space-shield effect on the surrounding silanol groups, thus some silanol groups were still not bonded, which was confirmed by the existence of the peak at 3430 cm−1. Compared with the infrared data of silica, the intensities of absorption peaks of MIPs and NIPs at 1730 cm−1 and 2970 cm−1 were obviously enhanced while the peaks at around 800 cm−1 and 470 cm−1 weakened significantly. In addition, the peaks in the absorption band between 3200 cm−1 and 3800 cm−1 for the MIPs and NIPs shifted from 3430 cm−1 for activated and modified silica to 3460 cm−1, which might due to the interactions between the –COO or –NH2 group of the template molecule and the –NH2 or C
O and saturated C–H, respectively.25 All the evidences demonstrated that the surface of SiO2 had been successfully modified by silane reagent. The vinyl groups introduced onto the surface of the activated silica by immobilization of a long chain group played a space-shield effect on the surrounding silanol groups, thus some silanol groups were still not bonded, which was confirmed by the existence of the peak at 3430 cm−1. Compared with the infrared data of silica, the intensities of absorption peaks of MIPs and NIPs at 1730 cm−1 and 2970 cm−1 were obviously enhanced while the peaks at around 800 cm−1 and 470 cm−1 weakened significantly. In addition, the peaks in the absorption band between 3200 cm−1 and 3800 cm−1 for the MIPs and NIPs shifted from 3430 cm−1 for activated and modified silica to 3460 cm−1, which might due to the interactions between the –COO or –NH2 group of the template molecule and the –NH2 or C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O groups of MAA. The spectra for MIPs and NIPs had similar profiles (similar locations and appearances of the major bonds) indicated that they have same chemical nature. These results demonstrated that the polymerization on the surface of silica gel particles had been successfully carried out.
O groups of MAA. The spectra for MIPs and NIPs had similar profiles (similar locations and appearances of the major bonds) indicated that they have same chemical nature. These results demonstrated that the polymerization on the surface of silica gel particles had been successfully carried out.
3.1.3.1. Adsorption kinetics. The adsorption capacity of the synthesized MIPs was measured to evaluate its recognition and binding capacity of EC. The results of uptake kinetics of MIPs to EC were shown in Fig. 3. The profiles of MIPs and NIPs were similar. For both of them, the adsorption capacity gradually increased with incubation time and then became saturation with further increasing the time. The increase rate had been particularly high during the first 15 min. The equilibrium of MIPs reached within 30 min, much shorter than that of NIPs (∼50 min). It is worth noting that saturation time decreased with the reduction of the concentration of EC employed in the experiment. The binding capacity of NIPs was contributed by nonspecific binding that bonds analyte without recognizing its exact structure, while MIPs adsorb analyte by both specific and nonspecific bindings. As shown in Fig. 3, there was a significant difference between the values of QMIPs and QNIPs: the Q values of MIPs were constantly several times higher than that of NIPs along with the whole reaction process (5–240 min). These results reflected the fact that the nonspecific adsorption level of NIPs toward EC was very low. Thus, most of the binding sites in MIPs were specific ones and the number of nonspecific binding sites in MIPs were limited.
 | ||
| Fig. 3 Uptake kinetics study of MIPs and NIPs and pseudo-second-order kinetic model for the adsorption of EC by MIPs. | ||
As can be seen in Fig. 3, the adsorption reaction of MIPs towards EC can be well fitted by pseudo-second-order kinetic model. R2 calculated from the fitting curve (t/Qt versus t) was 0.9991, while the Qe value determined from the slope of the linear regression model was 3.91 mg g−1, agreed well with the experimental Qe value (3.84 mg g−1). Therefore, the adsorption process of MIPs towards EC was controlled by chemical adsorption mechanism through sharing electrons between them.26
3.1.3.2. Static binding characteristics. The static adsorption capacities of the MIPs and NIPs sorbents for EC were determined in the concentrations ranged from 10 mg L−1 to 120 mg L−1, and the relationship of their total adsorption capacities to the concentration of EC was presented in Fig. 4a.
It can be seen from the curves that the adsorption capacity of MIPs or NIPs prepared under optimum conditions increased and the increase rates of the adsorption capacity for both MIPs and NIPs gradually decreased with raising the initial concentration of EC. The growth rate of MIPs had been obviously smaller when the concentration was 120 mg L−1, by contrast, the adsorption capacity of NIPs had reached saturation at the same concentration. The performance of MIPs was always superior to that of NIPs (MIPs yielded higher Q values than NIPs) regardless of the initial ethyl carbamate concentration. Specifically, the Qe value of MIPs reached 5.32 mg g−1, which was about 2.8-fold that of NIPs (1.90 mg g−1) at the concentration of 120 mg L−1, verifying that high affinity binding cavities for EC had formed during the polymerization process. As a result, MIPs have higher specific recognition ability toward EC than NIPs and are suitable for further application of MIPs-SPE.
Scatchard relationship was determined to assess the affinity of MIPs toward EC. As shown in Fig. 4b, MIPs exhibited nonlinearity. Two intersecting straight lines with different slopes corresponding to the high- and low-affinity populations of binding sites were observed in the plot. The relatively low binding strength was nonspecific binding, which was hypothesized to be driven by the interruption of the hydrogen-bond interactions by the solvent media. The affinity of the imprinted receptor sites in the MIPs material for EC was ascribed to the host–guest shape and size recognition and the bond reaction between them.25 We are particularly interested in the high affinity sites as they contribute the most to the specific binding of the MIPs. The Kd and Qmax values of MIPs for EC were calculated from the slope and intercept of the Scatchard plot of the high-affinity sites and found to be 9.68 mg L−1 and 4.29 mg g−1, respectively. The isotherm absorption curves were also obtained according to Freundlich and Langmuir models and shown in Fig. 5. As could be seen in this figure, the binding process of EC onto the MIPs fitted well with both of these two isotherm models (R2 = 0.9538 and 0.9901 for Freundlich and Langmuir model, respectively), indicating that MIPs possessed a heterogeneous binding site distribution.26
3.1.3.3. Adsorption selectivity. The selectivity of MIPs and NIPs for EC and its structure analogues were investigated with 80 mg L−1 of standard solution and the molecular structures of the analytes are shown in Fig. 6. Fig. 4d illustrates the binding amounts of these targets on MIPs and NIPs. The adsorption amounts of EC in MIPs and NIPs were 4.48 and 1.85 mg g−1, which were much greater than that of methyl carbamate (2.49 and 1.38 mg g−1), isoprocarb (1.70 and 1.40 mg g−1) and meta-tolyl-N-methyl carbamate (2.00 and 1.59 mg g−1), suggesting satisfactory selectivity of the synthesized MIPs for EC.
In addition, the distribution coefficient (Kd), selectivity coefficient (k) and the relative selectivity coefficient (α) of the sorbent obtained in these comparative experiments were also used to evaluate the competitive selective capacity of MIPs and NIPs. There three parameters were calculated following the equations:
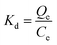 | (3) |
 | (4) |
 | (5) |
It is worth noting that among the three analogues of EC, methyl carbamate owns the highest Q value and Kd value (36.91 mL g−1) and the lowest values of k (2.11) and α (1.52) for MIPs, while isoprocarb has the lowest Q value and Kd value (23.81 mL g−1) and the highest values of k (3.27) and α (2.39). The differences between these two substances may be due to the larger structure differences between them. Among these three analogues, the degree of similarity between methyl carbamate and EC is the highest while the similarity degree between isoprocarb and EC was the lowest.
3.1.3.4. Stability and reusability of MIPs. To explore the reusability of the EC-imprinted MIPs synthesized in this study, ten successive adsorption–desorption cycles were carried out using the same MIPs sorbents. The adsorption capacity of the recycled EC-imprinted silica after each cycle is shown in Fig. 4c. It was found that the adsorption capacity of MIPs for EC remained essentially the same as cycle number increased from 1 to 6. After cycle number 6, the adsorption capacity of sorbents for EC gradually decreased. The MIPs for EC after ten cycles still remained 87.2% of the initial adsorption capacity, suggesting that MIPs employed in this experiment had good stability and reusability for EC adsorption.
3.2. MIPs-SPE for EC spiked wine samples
To apply MIPs-SPE for the extraction and enrichment of EC in an optimal condition, flow rate, extraction solvent and incubation time were firstly optimized. After the optimization of extraction conditions, the recovery of EC from rice wine and fruit brandy samples were tested. The recovery of EC in RW by MIPs-SPE and NIPs-SPE varied from 87% to 102% and 25% to 34%, respectively, while the recovery of EC in FB by MIPs-SPE and NIPs-SPE varied from 84% to 98% and 24% to 31%, respectively, revealing the existence of specific binding interaction between MIPs and target molecule.3.3. Determination of EC in rice wine and fruit brandy by SERS
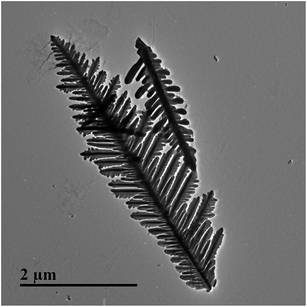
The silver dendrite nanostructures used in this study was synthesized based on the replacement reaction. The replacement reaction process is very simple and provides a straightforward and robust protocol for preparing nanostructures. In addition, this kind of nanostructure has high enhancement factor and good reproducibility. Thus, in our study, silver dendrite structure was employed as a substrate for collecting of SERS spectra. The size and morphology of silver nanostructure is mainly dependent upon the silver ion concentration and reaction time. It was found that 60 s of reaction between 150 mmol L−1 AgNO3 and zinc plate could produce the dendritic structure of silver with the best SERS effect. The silver dendrite structure can be clearly observed by TEM, as shown in Fig. 7. To prevent the generation of “coffee ring”, which may produce normal Raman signal,27 the solution of EC spotted on silver dendrite was blow-dried to shorten the drying time.Fig. 8 shows spectral features of EC determined by the normal Raman, SERS and MIPs–SERS spectral features of EC. Seven major bands at around 677 cm−1 (δ(OCO)), 846 cm−1 (ρ(NH2) + ρ(CH3)), 1257 cm−1 (ρ(CH3 + CH2)), 1382 cm−1 (νs(CC) + ν(CH3)), 1438 cm−1 (δ(CH)), 1648 cm−1 (β(NH2)) and 1671 cm−1 (ν(CO) + δ(CNH))28 were observed at all of three spectra, as shown in Fig. 8. In the SERS and MIPs–SERS spectra, some band shifts existed, which might be due to the SERS effect. When target molecules adsorbed onto the surface of substrate, some molecules interact with the noble-metallic nanostructures, resulting in the changes in dipole of the molecules and subsequent shifts in the location of SERS bands.29 The band at 1060 cm−1 is assigned to NO3−, which is derived from the synthesis of silver dendrite. In our study, this band is used as an internal standard. In addition, small bands at 641 cm−1, 1104 cm−1, 1162 cm−1, 1200 cm−1 and 1585 cm−1 appeared at similar locations in SERS spectrum and MIPs–SERS spectrum. Except bands assigned to EC, the limited interference bands (i.e., bands at 443 cm−1, 492 cm−1, 685 cm−1 and 1074 cm−1) appeared in MIPs–SERS might be related to wine residues. These results observed in Fig. 8 suggests MIPs could effectively adsorb EC while remove other interfering substances from rice wine and fruit brandy.
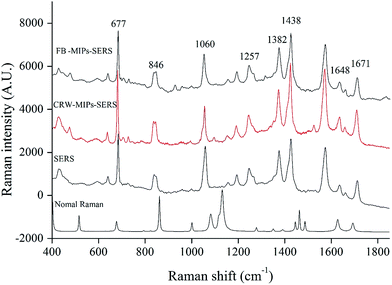 | ||
| Fig. 8 Representative SERS spectra of extract of EC-spiked rice wine (RW-MIPs-SERS) and fruit brandy (FB-MIPs-SERS) and standard EC solution (SERS), and normal Raman spectrum (normal Raman). | ||
PCA was performed on the full spectral region to explore the possibilities of differentiating wine samples spiked with different levels of EC. Fig. 9a shows the score plot of the first two PCs derived from the preprocessed SERS spectra, which account for 83% of the total variation. It was observed that all of the samples were clustered tightly and separated clearly according to different spiked levels, indicating PCA-based Raman spectroscopic analysis could successfully different various EC contents in wine samples.
Multivariate regression model were developed using PLS with leave-one-out full cross validation, since this method has been widely used. The optimum number of PLS factors used were selected according to the minimum root-mean-square error of cross-validation (RMSECV). In this study, the optimum number of factors for EC was seven for both of rice wine and fruit brandy, less than ten, indicating model overfitting did not occur. Satisfactory linear relationship was obtained from the constructed regression model in the current study. For rice wine, the R2 (cal) and R2 (pre) of this regression model was 0.9614 and 0.9456, respectively, while RMSEC and RMSEP was 0.0346 and 0.0399, respectively. For fruit brandy, the R2 (cal) and R2 (pre) of this regression model was 0.9559 and 0.9393, respectively, while RMSEC and RMSEP was 0.0369 and 0.0421, respectively. Generally, the higher the R2 (cal) and R2 (pre) value or the lower the RMESECV and RMSEP value is, the better performance the model has.30 The values of R2 (cal) and R2 (pre) obtained from our model were all higher than 0.90, indicating that excellent prediction precision was achieved using the established regression model. The RMESECV and RMSEP value was relatively low, and the difference between them was small, suggesting that the robustness of the models is also satisfactory. The prediction ability of the model also could be evaluated by the residual predictive deviation (RPD), which was calculated as the ratio of the standard deviation to RMSEP. The high value of the RPD indicated the great capacity of the model to predict the chemical composition accurately in samples outside the calibration set. It is generally acknowledged that, an RPD value higher than 3.0 considered to be indicative of excellent prediction, whereas values from 2.5 to 3.0 denote a good prediction. Approximate quantitative prediction is indicated by RPD values between 2.0 and 2.5. The possibility to distinguish between high and low values is revealed by values between 1.5 and 2.0. Unsuccessful prediction has RPD value lower than 1.5.31 In this study, RPD value was higher than 3.0 (4.29 and 4.06 for rice wine and fruit brandy, respectively), indicating the predictive ability of the models was excellent.
In order to further explain the regression model, the actual values versus SERS predicted values from the optimal models for EC contents in rice wine and fruit brandy samples were plotted in Fig. 9b and c, respectively. As shown in the figures, the correlations between the reference values and the values estimated by the SERS calibration for EC were very good, suggesting the model constructed here had satisfactory fitting results and predictive abilities.
The complex nature of rice wine and fruit brandy constitutes a real challenge to rapidly separate EC while maintain a relatively high recovery during sample pretreatment. Generally, conventional methods for extraction and recovery of EC from rice wine and fruit brandy samples includes complicated sample pretreatments (e.g., liquid–liquid extraction and rotary evaporation), which consumes lots of environmentally-unfriendly organic solvents and take long time. Additionally, these traditional pretreatment procedures always lead to partial loss of the target analyte. Due to the application of MIPs-SPE to separate EC from other components in wine samples, the time required was only ∼10 min in our study, significantly shorter than the time needed in traditional methods. Thus tremendous amount of time are saved. Even with MIPs as the pretreatment method, the chromatographic methods commonly used for the detection of ethyl carbamate in wines takes at least 10 min for one sample. On the contrary, the SERS spectral collection can be finished in 10 s. The overall analysis of wine sample can be finished in 15 min using our MIPs–SERS biosensor, including sample pre-treatment. In the food industry, great batch of samples need to be determined rapidly, and neither solvent extraction nor chromatographic methods can satisfy the requirement for high-throughput detection. The combination of MIPs and SERS technique can be more promising when the detection of large amount of food and agricultural products are required.
4. Conclusions
In this work, the applicability of MIPs combined with SERS for the prediction of the EC content of two kinds of alcoholic beverages was investigated. MIPs was synthesized and used as sorbent in SPE to separate EC from complicated wine samples, the eluted extract was subsequently placed onto silver dendrite nanostructure to detect EC concentrations based on SERS technology. The results demonstrated that the “two-step” MIPs–SERS biosensor can rapidly and accurately separate and enrich EC from rice wine and fruit brandy and performed well for quantification of EC. In summary, it is possible to specifically detect EC contents in rice wine and fruit brandy samples using the developed method in our study by combining MIPs with SERS. We believe the novel biosensor developed in this study has great potential for rapid determination of trace level of various chemical hazards in complex foods in future.Acknowledgements
This study was supported by the Nature Science Foundation of Jiangsu Province (No. BK20160168), the Nature Science Foundation of China (No. 31601413 and 31501418) and the graduate student innovation project of Jiangsu province (KYLX15_1134).References
- M. J. Dennis, N. Howarth, P. E. Key, M. Pointer and R. C. Massey, Food Addit. Contam., 1989, 6, 383–389 CrossRef CAS PubMed.
- F. A. Beland, R. W. Benson, P. W. Mellick, R. M. Kovatch, D. W. Roberts, J.-L. Fang and D. R. Doerge, Food Chem. Toxicol., 2005, 43, 1–19 CrossRef CAS PubMed.
- IARC, Alcoholic beverage consumption and ethyl carbamare (urethane), in IARC Monographs Working Group Experts Meeting, 2007, pp. 1–5 Search PubMed.
- R. R. Madrera and B. S. Valles, Food Control, 2009, 20, 139–143 CrossRef CAS.
- D. W. Lachenmeier, M. C. Lima, I. C. Nóbrega, J. A. Pereira, F. Kerr-Corrêa, F. Kanteres and J. Rehm, BMC Cancer, 2010, 10, 1 CrossRef PubMed.
- H.-S. Lim and K.-G. Lee, Food Chem., 2011, 126, 1373–1379 CrossRef CAS.
- D. W. Lachenmeier, W. Frank and T. Kuballa, Rapid Commun. Mass Spectrom., 2005, 19, 108–112 CrossRef CAS PubMed.
- H. He, D. Wu and D.-W. Sun, Food Chem., 2014, 156, 394–401 CrossRef CAS PubMed.
- J. Märk, M. Andre, M. Karner and C. W. Huck, Eur. J. Pharm. Biopharm., 2010, 76, 320–327 CrossRef PubMed.
- Z. Wu, E. Xu, J. Li, J. Long, A. Jiao, Z. Jin and X. Xu, Food Analytical Methods, 2016, 1–11, DOI:10.1007/s12161-016-0646-8.
- J. A. Iversen and B. K. Ahring, Bioresour. Technol., 2014, 172, 112–120 CrossRef CAS PubMed.
- Z. Wu, J. Long, E. Xu, F. Wang, X. Xu, Z. Jin and A. Jiao, Food Analytical Methods, 2016, 9, 1210–1219 CrossRef.
- K.-M. Lee, T. J. Herrman, Y. Bisrat and S. C. Murray, J. Agric. Food Chem., 2014, 62, 4466–4474 CrossRef CAS PubMed.
- J. Feng, L. Xu, G. Cui, X. Wu, W. Ma, H. Kuang and C. Xu, Biosens. Bioelectron., 2016, 81, 138–142 CrossRef CAS PubMed.
- B. Ankamwar, U. K. Sur and P. Das, Anal. Methods, 2016, 8, 2335–2340 RSC.
- K.-H. Chen, Y.-C. Pu, K.-D. Chang, Y.-F. Liang, C.-M. Liu, J.-W. Yeh, H.-C. Shih and Y.-J. Hsu, J. Phys. Chem. C, 2012, 116, 19039–19045 CAS.
- A. Martín-Esteban, TrAC, Trends Anal. Chem., 2013, 45, 169–181 CrossRef.
- M. L. Yola, T. Eren and N. Atar, Sens. Actuators, B, 2015, 210, 149–157 CrossRef CAS.
- S. E. Diltemiz, R. Say, S. Büyüktiryaki, D. Hür, A. Denizli and A. Ersöz, Talanta, 2008, 75, 890–896 CrossRef CAS PubMed.
- S. Feng, F. Gao, Z. Chen, E. Grant, D. D. Kitts, S. Wang and X. Lu, J. Agric. Food Chem., 2013, 61, 10467–10475 CrossRef CAS PubMed.
- Y. Hu, S. Feng, F. Gao, E. C. Y. Li-Chan, E. Grant and X. Lu, Food Chem., 2015, 176, 123–129 CrossRef CAS PubMed.
- I. M. Valente, M. R. Rui, L. M. Gonçalves and J. A. Rodrigues, Anal. Methods, 2014, 6, 9136–9141 RSC.
- E. Xu, Z. Wu, F. Wang, J. Long, X. Xu, Z. Jin and A. Jiao, J. Inst. Brew., 2016, 122, 55–62 CrossRef CAS.
- J. Hu, T. Feng, W.-L. Li, H. Zhai, Y. Liu, L.-Y. Wang, C.-L. Hu and M.-X. Xie, J. Chromatogr. A, 2014, 1330, 6–13 CrossRef CAS PubMed.
- B. Wei, X. Hu, H. Li, C. Wu, X. Xu, Z. Jin and Y. Tian, Food Hydrocolloids, 2014, 36, 369–373 CrossRef CAS.
- Y. Tang, J. Gao, X. Liu, J. Lan, X. Gao, Y. Ma, M. Li and J. Li, Food Chem., 2016, 201, 72–79 CrossRef CAS PubMed.
- F. Gao, S. Feng, Z. Chen, E. C. Li-Chan, E. Grant and X. Lu, J. Food Sci., 2014, 79, N2542–N2549 CrossRef CAS PubMed.
- D. Yang, N. E. Mircescu, H. Zhou, N. Leopold, V. Chiş, M. Oltean, Y. Ying and C. Haisch, J. Raman Spectrosc., 2013, 44, 1491–1496 CrossRef CAS.
- Y. Zhao, X. Liu, D. Y. Lei and Y. Chai, Nanoscale, 2014, 6, 1311–1317 RSC.
- Z. Wu, J. Long, E. Xu, F. Wang, X. Xu, Z. Jin and A. Jiao, Food Analytical Methods, 2015, 1–10 Search PubMed.
- F. Shen, X. Niu, D. Yang, Y. Ying, B. Li, G. Zhu and J. Wu, J. Agric. Food Chem., 2010, 58, 9809–9816 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c6ra23165a |
| This journal is © The Royal Society of Chemistry 2016 |