Synthesis and application of colloidal beta-cyclodextrin-decorated silver nanoparticles for rapid determination of malachite green in environmental water using surface-enhanced Raman spectroscopy
Fei Jia,
Xudong Yang and
Zhenyu Li*
School of Chemical Engineering, Changchun University of Technology, Changchun, 130012, China. E-mail: zhenyuli1974@163.com
First published on 23rd September 2016
Abstract
In this study, a green synthesis method of colloidal silver nanoparticles based on the traditional silver mirror reaction induced by adding beta-cyclodextrin (CD) to [Ag(NH3)2]+ solution was proposed. The beta-cyclodextrin-decorated silver nanoparticles (CD-AgNPs) were applied to the rapid determination of malachite green (MG) in environmental water using surface-enhanced Raman spectroscopy (SERS) with peaks at 1175, 1220, 1298, and 1618 cm−1. The effects of the synthesis parameters including the amount of [Ag(NH3)2]+ solution and CD, and the effects of analysis parameters including the amount and mixing time of CD-AgNPs and acidity regulator, were investigated. The analytical response to MG was linear in a concentration range of 0.010–0.15 mg L−1, with the correlation coefficients (R2) of 0.9916–0.9988 and a detection limit of 1 μg L−1. Recoveries obtained by analyzing the 6 spiked environmental water samples were between 85.3 and 103.2%.
1. Introduction
Malachite green (MG, Basic Green 4, C.I. 42000) is a kind of triphenylmethane dye, which has been extensively used in the production of silk, cotton, wool, jute, ceramics, leather and paper. Furthermore MG has been used worldwide as a fungicide or ectoparasiticide in fish culture since the 1930s.1 Nowadays because of potential animal carcinogenicity, mutagenicity and teratogenicity,2–5 MG has been prohibited for use in aquatic products for human consumption in many countries, including the United States, China and the European Union. For example, in Ireland the concentrations of malachite green in fish farm water effluent and water extracted for purposes of drinking water should not exceed 100 and 1.0 μg L−1, respectively.6 Conventional detection methods for MG include high performance liquid chromatography (HPLC),7–9 liquid chromatography/mass spectrometry (LC/MS)10–12 and enzyme-linked immunosorbent assay (ELISA).13,14 However, most of the methods require a time-consuming preconcentration step, and make the experiments difficult to conduct rapidly.The surface-enhanced Raman spectroscopy (SERS) is based on molecular adsorption on the nanostructure surface. Among metallic nanoparticles used for SERS, the silver nanoparticles are the best.15 The traditional silver nanoparticles colloid can be obtained easily by some simple methods.16,17 However the repeatability, stability and activity of the silver nanoparticles colloid are not satisfactory.18,19 To prepare stable nanoparticles, capping organic molecules are necessary for preventing the nanoparticles from irreversible aggregation and making the particles soluble in a given solvent. The modified cyclodextrin (CD) has already been applied in the preparation of nanometer materials,20–26 but the reports of preparing silver nanoparticles (AgNPs) using unmodified CD and the study of the interaction between CD and AgNPs are very rare. Based on the traditional silver mirror reaction by adding CD to [Ag(NH3)2]+ solution, the globular and water-soluble silver nanoparticles were greenly synthesized, and the influence of CD on AgNPs was discussed. The nanoparticles can be stored stably and applied to the detection for MG in four months with satisfactory results. This work is also helpful to facilitate the rapid and sensitive determination of trace MG in environmental water using SERS.
2. Experimental details
2.1 Chemicals and materials
Silver nitrate (AgNO3, 99.9%) and beta-cyclodextrin (CD, 99%) were purchased from J&K Chemical Company. Glucose, hydrochloric acid (HCl, 37% in water), malachite green (MG, 98%) were purchased from Beijing Chemical Reagent Company. All aqueous solution was prepared with deionized water prepared with Milli-Q water purification system (18.0 MΩ cm). All glassware was cleaned with freshly prepared aqua regia (HCl/HNO3, 3/1, v/v) and rinsed thoroughly with deionized water prior to use. Environmental water samples were collected from Changchun south lake (Jilin province, China). All other reagents were of analytical reagent grade and used without further purification or treatment.2.2 Instrument
The surface morphologies of the samples were measured on a JEOL JEM-1200EX transmission electron microscope (TEM, JEOL Ltd.). Absorption spectra were recorded on a TU-1810C UV-Vis spectrometer (Beijing Purkinje General Instrument Co., Ltd.) within the wavelength range of 250 to 650 nm. Raman spectra were obtained using a portable miniature Raman spectrometer (Changchun Jilin University Little Swan Instruments Co., Ltd., Changchun, China) with 1 cm quartz cells, a 785 nm diode laser and a fibre-optics probe. The instrument's physical dimensions are 20 cm (W) × 20 cm (L) × 10 cm (H) and it weighs about 2.5 kg. The laser power was chosen as 150 mW. The exposure time used for data collection was typically 10 s. The resolution of spectrum is 3 cm−1. The FT-IR spectrum was measured at wavenumbers ranging from 400 to 4000 cm−1 using a Nicolet Avatar 360 FT-IR spectrophotometer (Thermo Scientific). The samples for FT-IR were prepared by 250 μL colloidal silver glue 3 mL acetone followed by centrifuging at 12![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm, blowing to dry with N2 and tableting with KBr. Nanoparticle size measurements were performed using a Zetasizer NanoZS (Malvern Instruments).
000 rpm, blowing to dry with N2 and tableting with KBr. Nanoparticle size measurements were performed using a Zetasizer NanoZS (Malvern Instruments).
2.3 Synthesis of CD-AgNPs
In a typical synthesis, 4 mL of 0.1 mmol L−1 [Ag(NH3)2]+, 4 mL of 0.5 mmol L−1 glucose, 1.0 g of CD and 92 mL of water were added in a 150 mL conical flask, and refluxed with magnetic stirring in an oil bath for 30 min. After the flask was cooled down to room temperature, yellow-green and transparent colloidal CD-AgNPs solution was obtained. Different CD-AgNPs were synthesized by changing the ratio of [Ag(NH3)2]+ to CD.2.4 Analysis of MG
100–300 μL CD-AgNPs solution and 50–90 μL of 1 mol L−1 HCl were added into 1 mL sample solution. The solution was mixed for a few minutes in a 1 cm quartz cell and measured with a portable miniature Raman spectrometer. The quantitative analysis were performed based on the area of peaks at 1175, 1220, 1298 and 1618 cm−1 in SERS spectrum. All the experiments were performed in quintuplicate.3. Results and discussion
3.1 Characterization of CD-AgNPs
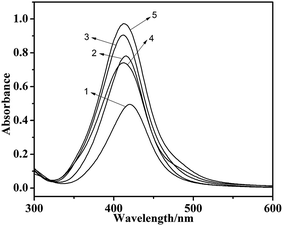
Spherical particles of the CD-AgNPs around with small irregular protuberances are observed by TEM (Fig. 1a). It can be found from the FT-IR spectrum (Fig. 2) that the red shift of νOH is 33 cm−1, due to the interaction between Ag and O of CDs. This result is the same as that reported in the literature.27 The existence of AgNPs did not affect the CDs frame vibration zone, so AgNPs should not enter into the hydrophobic cavity of CDs.28 According to the particle diameter distribution observed by TEM and ultraviolet spectrum of CD-AgNPs (Fig. 1b and 3), there were relationships between particle diameter and reactants. Some [Ag(NH3)2]+ could entered into the hydrophobic cavity of CDs and developed as silver particle seeds.29 Those free state [Ag(NH3)2]+ gathering at the surface of the silver particle seeds led to the increase of the seeds diameter. When the seeds grew up, they leave from the holes of CDs and formed AgNPs. The higher the concentration of [Ag(NH3)2]+, the more silver particle seeds would be formed, and the diameter of AgNPs obtained finally would be less than only a small amount of seeds. The amount of indispensable CD for the formation of colloid has little or no influence on the particle diameter of AgNPs. The formation process of AgNPs is shown in Fig. 4.3.2 Raman spectra of MG
As shown in Fig. 5, the dominating characteristic bands of the normal Raman and SERS spectra of MG ranging from 1000 to 1800 cm−1 were all observed clearly. According to previous studies,30,31 the predominant bands in the spectrum of MG located at 1172, 1218, 1294 1352, 1388, 1489, and 1609 cm−1. The in-plane aromatic C–H bending modes appeared as a single enhanced band in the SERS spectrum at 1175 cm−1 with a red shift of 3 cm−1 compared with normal Raman spectrum. The strong band in the solid Raman spectrum consisted of the in-plane C–H bending and symmetrical N–C-ring-C–C stretching appeared at 1352 and 1388 cm−1, respectively. The adsorbed bands were notably broad in the SERS spectra at 1368 and 1398 cm−1. The solid MG peak of the out-of-phase ring stretch (1609 cm−1) was split into two peaks appeared at 1588 and 1618 cm−1 in the SERS spectrum. In SERS spectrum, the benzene ring breathing vibration bands at 1218, 1294, 1489 cm−1 shifted to 1220, 1298, 1493 cm−1. Raman peaks at 1175 and 1618 cm−1 were barely able to observed in the Raman spectrum of 1000 mg L−1 MG solution, while strong Raman signals for 0.100 mg L−1 MG solution could be obtained using CD-AgNPs as the SERS substrate. The quantitative analysis of MG were performed based on the measured peak areas at 1175, 1220, 1298, 1618 cm−1 in SERS spectrum. In some of the discussions that follow, the SERS intensity was expressed as peak area. | ||
| Fig. 5 Raman spectrum of solid malachite green (a), SERS spectrum 0.100 mg L−1 malachite green solution (b) and Raman spectrum of 1000 mg L−1 malachite green solution (c). | ||
3.3 Calculation of enhancement factor
The SERS enhancement factor (EF) is an important parameter for demonstrating the SERS performance. We choose the strongest band at 1175 cm−1 of 1000 mg L−1 and 0.100 mg L−1 MG solution as the representation peak for EF calculation. Herein, the SERS EF is calculated from the standard equation32 defined as:| EF = ISERSCRaman/IRamanCSERS |
3.4 Optimization of conditions
Using 1 mL of 0.100 mg L−1 MG as probe, the following five major influencing parameters were optimized. Each experiment was conducted in quintuplicate. Once a parameter was optimized, it was set at its optimal value in subsequent experiments.3.5 Determination of MG
CD-AgNPs was prepared under the optimized conditions: 4 mL of 0.1 mmol L−1 [Ag(NH3)2]+, 4 mL of 0.5 mmol L−1 glucose and 1.0 g of CD. Determination of MG was under the optimized conditions: 1 mL of MG solution, 250 μL of colloidal CD-AgNPs solution and 70 μL of 1 mol L−1 HCl solution. The SERS spectra of the solutions containing the different concentrations of MG are shown in Fig. 7a. The calibration curve was plotted (Fig. 7b). The linear range was 0.010–0.150 mg L−1 and the limit of detection was 1.0 μg L−1, which meet the requirements of the preliminary screening of MG. The experimental results for the determination of MG obtained by using this present method and several other methods are listed in Table 1 for comparison.| Substrate | Linear range (mg L−1) | LOD (μg L−1) | Reference |
|---|---|---|---|
| β-Cyclodextrin-decorated silver nanoparticles | 0.010–0.150 | 1.0 | This work |
| Graphene oxide/gold nanocomposites | 0.91–36.5 | 912 | 36 |
| Microfluidic channel and aggregated silver colloids | 0.001–0.10 | 1–2 | 37 |
| Colloidal gold nanoparticles | 0.005–0.035 | 0.1 | 38 |
| Starch-coated silver nanoparticles | 0.005–0.035 | 0.08 | 39 |
| L-Cysteine decorated colloidal silver nanoparticles | 9.10–912 | 3.65 | 40 |
In order to validate the present method, the environmental water samples were pretreated under the optimized conditions. No MG was found in these real samples. The recovery test was carried out by sparking standard MG in water sample at levels from 0.010–0.100 mg L−1. The recoveries of MG are in the range of 85.3–103.2%, with RSDs between 1.40 and 5.20%. The results are illustrated in Table 2. These results in dictate that the present method is satisfactory to detect trace MG in environmental water samples.
| Added (mg L−1) | 1175 cm−1 | 1220 cm−1 | 1298 cm−1 | 1618 cm−1 | ||||
|---|---|---|---|---|---|---|---|---|
| Recovery% | RSD% | Recovery% | RSD% | Recovery% | RSD% | Recovery% | RSD% | |
| 0.01 | 92.4 | 3.50 | 90.1 | 4.20 | 85.3 | 5.20 | 93.2 | 2.70 |
| 0.015 | 93.6 | 2.10 | 91.7 | 3.70 | 86.1 | 4.50 | 94.5 | 2.60 |
| 0.02 | 95.5 | 1.90 | 94.7 | 3.20 | 90.0 | 3.80 | 95.6 | 2.20 |
| 0.03 | 97.6 | 1.50 | 93.5 | 4.00 | 91.2 | 3.60 | 98.9 | 1.50 |
| 0.05 | 99.5 | 1.60 | 95.5 | 2.90 | 93.5 | 2.70 | 102.6 | 1.60 |
| 0.10 | 101.2 | 1.70 | 98.6 | 3.10 | 93.9 | 2.70 | 103.2 | 1.40 |
4. Conclusions
In this work, a series of AgNP colloids were synthesized by silver mirror reaction in the presence of CDs. The influences of CD on the synthesis of AgNPs were discussed and a green and repeatable method for synthesis of AgNPs colloid was established. The effects of experimental parameters, including the volume of [Ag(NH3)2]+ solution, the amount of CD, the volume of silver colloid, the volume of HCl solution and the mixing time were investigated. The CD-AgNPs displayed a higher stability and sensitivity than the classic AgNPs. Combined with SERS, a rapid, convenient and efficient method for the determination of trace MG in environmental water samples was established successfully. The present method possesses the significant advantages of high reproducibility, simplicity and rapidness for the quantitative determination of MG. The approach may accelerate the application of rapid on-site detection of MG in environmental water.Acknowledgements
This work was supported by Jilin Province Youth Science Foundation (No. 20150520012JH).References
- D. J. Alderman, J. Fish Dis., 1985, 8, 289–298 CrossRef CAS.
- H. Yi, W. Qu and W. Huang, Microchim. Acta, 2008, 160, 291–296 CrossRef CAS.
- V. Fessard, T. Godard, S. Huet, A. Mourot and J. M. Poul, J. Appl. Toxicol., 1999, 19, 421–430 CrossRef CAS PubMed.
- M. A. Pierrard, P. Kestemont, E. Delaive, M. Dieu, M. Raes and F. Silvestre, Aquat. Toxicol., 2012, 114, 142–152 CrossRef PubMed.
- C. Long, Z. Mai, Y. Yang, B. Zhu, X. Xu, L. Lu and X. Zou, J. Chromatogr. A, 2009, 1216, 2275–2281 CrossRef CAS PubMed.
- K. Sagar, M. R. Smyth, J. G. Wilson and K. McLaughlin, J. Chromatogr. A, 1994, 659, 329–336 CrossRef CAS.
- G. Dowling, P. P. J. Mulder, C. Duffy, L. Regan and M. R. Smyth, Anal. Chim. Acta, 2007, 586, 411–419 CrossRef CAS PubMed.
- K. Mitrowska, A. Posyniak and J. Zmudzki, Anal. Chim. Acta, 2007, 586, 420–425 CrossRef CAS PubMed.
- G. Chen and S. Miao, J. Agric. Food Chem., 2010, 58, 7109–7114 CrossRef CAS PubMed.
- D. R. Doerge, M. I. Churchwell, T. A. Gehring, Y. M. Pu and S. M. Plakas, Rapid Commun. Mass Spectrom., 1998, 12, 1625–1634 CrossRef CAS PubMed.
- K. Halme, E. Lindfors and K. Peltonen, J. Chromatogr. B: Anal. Technol. Biomed. Life Sci., 2007, 845, 74–79 CrossRef CAS PubMed.
- K. Halme, E. Lindfors and K. Peltonen, J. Chromatogr. B: Anal. Technol. Biomed. Life Sci., 2007, 845, 74–79 CrossRef CAS PubMed.
- W. W. Xing, L. He, H. Yang, C. J. Sun, D. W. Li, X. M. Yang, Y. Li and A. P. Deng, J. Sci. Food Agric., 2009, 89, 2165–2173 CrossRef CAS.
- M. Yang, J. Fang, T. Kuo, D. Wang, Y. Huang, L. Liu, P. Chen and T. Chang, J. Agric. Food Chem., 2007, 55, 8851–8856 CrossRef CAS PubMed.
- W. Kiefer and S. Schlücker, Surface enhanced Raman spectroscopy: analytical, biophysical and life science applications, Wiley, 2011 Search PubMed.
- P. C. Lee and D. Meisel, J. Phys. Chem., 1982, 86, 3391–3395 CrossRef CAS.
- C. D. Tran, Anal. Chem., 1984, 56, 824–826 CrossRef CAS.
- J. Roh, H. Umh, J. Sim, S. Park, J. Yi and Y. Kim, Korean J. Chem. Eng., 2013, 30, 671–674 CrossRef CAS.
- R. Lu, J. Sha, W. Xia, Y. Fang, L. Gu and Y. Wang, CrystEngComm, 2013, 15, 6207–6212 RSC.
- J. Liu, J. Alvarez, W. Ong, E. Román and A. E. Kaifer, J. Am. Chem. Soc., 2001, 123, 11148–11154 CrossRef CAS PubMed.
- T. Carofiglio, R. Fornasier, L. Jicsinszky, U. Tonellato and C. Turco, Tetrahedron Lett., 2001, 42, 5241–5244 CrossRef CAS.
- Y. Xie, X. Wang, X. Han, X. Xue, W. Ji, Z. Qi, J. Liu, B. Zhao and Y. Ozaki, Analyst, 2010, 135, 1389–1394 RSC.
- P. Ma, F. Liang, Y. Sun, Y. Jin, Y. Chen, X. Wang, H. Zhang, D. Gao and D. Song, Microchim. Acta, 2013, 180, 1173–1180 CrossRef CAS.
- P. Ma, F. Liang, D. Wang, Q. Yang, B. Cao, D. Song, D. Gao and X. Wang, Microchim. Acta, 2015, 182, 167–174 CrossRef CAS.
- P. Ma, F. Liang, D. Wang, Q. Yang, Y. Ding, Y. Yu, D. Gao, D. Song and X. Wang, Microchim. Acta, 2015, 182, 863–869 CrossRef CAS.
- P. Ma, F. Liang, Q. Yang, D. Wang, Y. Sun, X. Wang, D. Gao and D. Song, Microchim. Acta, 2014, 181, 975–981 CrossRef CAS.
- J.-P. Sylvestre, A. V. Kabashin, E. Sacher, M. Meunier and J. H. T. Luong, J. Am. Chem. Soc., 2004, 126, 7176–7177 CrossRef CAS PubMed.
- Y.-Q. Ll, Y.-X. Zhu and Y. Zhang, Chem. Res. Chin. Univ., 2008, 29, 669–672 Search PubMed.
- J. Szejtli, Chem. Rev., 1998, 98, 1743–1754 CrossRef CAS PubMed.
- L. He, N. Kim, H. Li, Z. Hu and M. Lin, J. Agric. Food Chem., 2008, 56, 9843–9847 CrossRef CAS PubMed.
- M. Volny, A. Sengupta, C. B. Wilson, B. D. Swanson, E. J. Davis and F. Turecek, Anal. Chem., 2007, 79, 4543–4551 CrossRef CAS PubMed.
- J. Zhang, S. Qu, L. Zhang, A. Tang and Z. Wang, Spectrochim. Acta, Part A, 2011, 79, 625–630 CrossRef CAS PubMed.
- O. Siiman, L. A. Bumm, R. Callaghan, C. G. Blatchford and M. Kerker, J. Phys. Chem., 1983, 87, 1014–1023 CrossRef CAS.
- J. T. Krug, G. D. Wang, S. R. Emory and S. Nie, J. Am. Chem. Soc., 1999, 121, 9208–9214 CrossRef CAS.
- Y. Chen, L. Wu, Y. Chen, N. Bi, X. Zheng, H. Qi, M. Qin, X. Liao, H. Zhang and Y. Tian, Microchim. Acta, 2012, 177, 341–348 CrossRef CAS.
- W. L. Fu, S. J. Zhen and C. Z. Huang, Analyst, 2013, 138, 3075–3081 RSC.
- S. Lee, J. Choi, L. Chen, B. Park, J. B. Kyong, G. H. Seong, J. Choo, Y. Lee, K. H. Shin, E. K. Lee, S. W. Joo and K. H. Lee, Anal. Chim. Acta, 2007, 590, 139–144 CrossRef CAS PubMed.
- Y. Jin, P. Y. Ma, F. H. Liang, D. J. Gao and X. H. Wang, Anal. Methods, 2013, 5, 5609–5614 RSC.
- Y. Zhao, Y. Tian, P. Y. Ma, A. M. Yu, H. Q. Zhang and Y. H. Chen, Anal. Methods, 2015, 7, 8116–8122 RSC.
- G. N. Xiao and S. Q. Man, Spectrosc. Lett., 2013, 46, 577–582 CrossRef CAS.
| This journal is © The Royal Society of Chemistry 2016 |