New dihydropyrimidin-2(1H)-one based Hsp90 C-terminal inhibitors†
S. Terracciano‡
 a,
A. Foglia‡
a,
A. Foglia‡ a,
M. G. Chini
a,
M. G. Chini a,
M. C. Vaccaro
a,
M. C. Vaccaro a,
A. Russo
a,
A. Russo a,
F. Dal Piaz
a,
F. Dal Piaz ab,
C. Saturnino
ab,
C. Saturnino a,
R. Riccio
a,
R. Riccio a,
G. Bifulco
a,
G. Bifulco *a and
I. Bruno
*a and
I. Bruno *a
*a
aDepartment of Pharmacy, University of Salerno, via Giovanni Paolo II, 132, 84084, Fisciano, Italy. E-mail: bifulco@unisa.it; brunoin@unisa.it
bDepartment of Medicine and Surgery University of Salerno, Via Allende, 84081 Baronissi, SA, Italy
First published on 25th August 2016
Abstract
The inhibition of the C-terminal domain of heat shock protein 90 (Hsp90) is emerging as a novel strategy for cancer therapy, therefore the identification of a new class of C-terminal inhibitors is strongly required, also in consideration that to date only nature-inspired molecules have been largely expanded. Our recent discovery of potent antiproliferative dihydropyrimidone based C-terminal Hsp90 inhibitor (1, IC50 = 50.8 ± 0.2 μM and 20.8 ± 0.3 μM in A375 and Jurkat cell lines, respectively) drove us to further explore this very promising pharmacophoric core. In this study, we identified a new set of DHPM-derivatives that exhibited antiproliferative activity against two cancer lines by their modulation of Hsp90 C-terminus without inducing the undesired heat shock response. Our results strongly outline the high sensitivity of the Biginelli scaffold to structural decorations allowing us to point out also that small variations can deeply influence biological activity.
Introduction
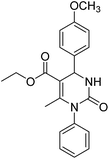
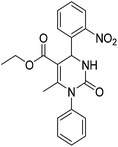
Heat shock protein 90 (Hsp90) is a molecular chaperone involved in the control of a wide range of physiological processes within cells, through directing the folding and the conformational maturation of many client proteins under both normal and stress conditions. Among Hsp90's clients, a surprising number is well recognized as target in oncology, including different oncoproteins (Her2, Bcr-Abl, Akt, etc.) that are linked to the six hallmarks of cancer.1–3 Moreover, in tumor cells, the dependency of oncoproteins on the chaperone function of Hsp90 is much higher than in normal cells. Cancer cells use Hsp90 chaperone machinery to protect mutated or over-expressed oncoproteins, which drive malignant progression.4,5 As a consequence, heat shock proteins are expressed at high levels in several tumors, including lung, breast, prostate and gastrointestinal cancers, and form a fostering environment that is essential for the disease development.6–9 The depletion of an array of clients oncoproteins and the simultaneous suppression of multiple oncogenic pathways, highlights the strategic approach of targeting Hsp90 machinery in cancer therapy.10 Therefore the inhibition of Hsp90 chaperone activity leads to proteasome-mediated degradation of deregulated oncogenic client proteins, which is essential in tumor genesis. Furthermore, Hsp90 inhibitors have been reported to target the chaperone within cancer cells with a higher selectivity compared to normal cells.11 For all these reasons Hsp90 has been widely studied as a promising druggable target to treat cancer and, in the last decades, many effective and selective inhibitors (Hsp90-I) have been identified.12 Among these, seventeen small molecules are under evaluation in over 50 clinical trials (http://ClinicalTrials.gov database) of several tumor types.13–16 To date, most Hsp90-I-clinical candidates included-, also known as “classical inhibitors”, modulate the N-terminus of the protein via interaction to its ATP-binding site.17,18 Despite their efficacy, these classical inhibitors have not yet achieved the expected success because they also stimulate a cytoprotective mechanism in cancer cells referred to as heat shock response (HSR), leading to an increase in the expression of heat shock proteins (mainly Hsp70 and Hsp27), which may limit their clinical potential.19,20 These Hsps support protein folding, prevent apoptosis, thus assisting tumor cell growth, survival and resistance to chemotherapy.21 Unlike N-terminal inhibitors, modulation of Hsp90 activity via the C-terminus, which does not trigger the pro-survival heat shock response, represents a convincing and intense area of research in an effort to develop more successful compounds for cancer treatment.22–28 Currently, high interest has been shown in the identification of C-terminal inhibitors that disrupt Hsp90 activity without inducing rescue mechanism and/or drug resistance. The earliest Hsp90 C-terminal inhibitor was the coumarin antibiotic novobiocin which weakly inhibits the molecular chaperone (IC50 = 700 μM in SKBr3 cells) and induces degradation of Hsp90 client proteins.29 Subsequent studies focused on structural modification of the natural compound and led to novobiocin analogues that exhibit greater potency and good anti-proliferative activity against multiple cancer cell lines.30–35 Other C-terminal inhibitors include clorobiocin, coumermycin A1, the macrocycles SM122, SM145, SM249, SM253 (SM series) and their structural analogues.22,25,36,37 However, crystal structures of Hsp90/C-terminal inhibitors have not yet been reported so far, consequently the absence of structural information is making the design of new modulators very challenging.38 Recently, in order to find out a novel class of non-natural inspired compounds targeting Hsp90 C-terminus, we disclosed a 3,4-dihydropyrimidin-2(1H)-one (DHPM) based compound 1, which showed antiproliferative effect in two different cancer cell lines (IC50 = 50.8 ± 0.2 μM and 20.8 ± 0.3 μM in A375 and Jurkat cell lines, respectively), without any apparent cytotoxic activity in non-tumor cells.39 The effect of compound 1 on Hsp90 client proteins levels confirmed that the antitumor activity was carried out through Hsp90 oncoproteins degradation. Interestingly, the levels of Hsp90 and Hsp70 did not increase in both cell lines, suggesting that the undesired heat shock response was not induced. Finally, limited proteolysis experiments, oligomerization assays and molecular docking studies allowed us to identify the binding of compound 1 with the C-terminal domain of Hsp90.39 Therefore, in an ongoing effort to develop new C-terminal Hsp90 modulators we decided to explore a new small set of easily accessible (synthesized) derivatives, by using compound 1 as lead for punctual structural modifications in order to produce a second generation of DHPM based molecules 2–13 (Fig. 1).Result and discussion
Design and synthesis of novel 3,4-dihydropyrimidin-2(1H)-ones
As above mentioned, despite the great interest reawakened by the C-terminal Hsp90 inhibitors as potential therapeutic agents, the lack of a co-crystal structure of the chaperone C-terminus bound to inhibitors severely limits the discovery and structural optimization of this class of compounds. In our previous study, in order to explore the influence of chemical functionalization of the six positions around the DHPM core and to provide valuable insight into structure–activity relationship, we generated a first collection of molecules in which different building blocks (urea, aromatic aldehyde and β-ketoester) were employed.39 In the present study, from a design perspective, our efforts are focused on the exploration of the 3,4-dihydropyrimidin-2(1H)-one scaffold at C4 position. In more details diverse commercially available aromatic aldehydes, differently substituted in the o-, m-, p-, and o,m-positions, with either electron-withdrawing or -donating groups were chosen, with the aim of shedding more light on the influence of chemical diversity on this position around the scaffold. The chemical structures of this second generation of DHPM analogs are reported in Fig. 2.The synthesis of compounds 2–13 was efficiently realized by one-pot Biginelli microwave-assisted reaction, following the optimized experimental conditions previously reported by us. Briefly, the synthesis was accomplished using trimethylsilyl chloride (TMSCl), as an efficient reaction mediator for the Biginelli condensation, especially for the synthesis of N1-substituted DHPMs more difficult to obtain, using microwave heating (Scheme 1).40–43 Following this synthetic procedure we prepared compounds 2–13 in short reaction times and high yields (see Experimental).
Evaluation of Hsp90 binding by SPR approach
In order to assess the effect on Hsp90, as a first step, we evaluated the ability of compound 2–13 to interact with Hsp90α by a surface plasmon resonance (SPR) based binding assay, which was recently successfully applied to investigate the interaction of small molecules to Hsp90.39,44–48 The lead compound 1 and the known Hsp90 inhibitor 17-N-allylamino-17-demethoxygeldanamycin (17-AAG)49 were used as positive controls. This assay allowed us to establish the affinity of our synthesized compounds toward the investigated protein (see Table 1). In more details, eight out of the twelve tested compounds efficiently interacted with the immobilized protein, as demonstrated by the concentration dependent responses, and by the clearly discernible exponential curves, during both the association and dissociation phases. Each constant was calculated fitting at least 12 curves, obtained by injecting three times the investigated compounds 2–13 at four different concentrations, ranging from 0.020 μM to 1 μM. On the basis of the KD values measured for the interaction with Hsp90α, compound 2–6, 8, and 11 showed high affinity toward the molecular chaperone with low KD values in the nM range (ranging from 2.9 to 76.2 nM). These preliminary screening allowed us to confirm the ability of this second generation of DHPM derivatives to bind the immobilized protein. In particular, by analyzing the data obtained, all the compounds carrying a p-substituted aromatic aldehyde at C4 of DHPM ring, were able to interact with the molecular chaperone, showing (except for 2) an higher affinity than 1 toward HSP90 (see Table 1). The presence of a NO2 group at ortho position, seems to improve the interaction with the protein (8 versus 2, KD 2.9 ± 0.8 nM and 76.2 ± 1.9 nM, respectively), while a CF3 moiety at the same position was not tolerated (compound 7). Moreover, the use of m-substituted benzaldehydes, 9 and 10, dropped the affinity to the protein. Finally, compounds with ortho–meta disubstituted aromatic ring at C4 position of the DHPM scaffold resulted in a non-homogeneous behaviour; indeed, compound 11 showed to bind the immobilized protein with nanomolar affinity (KD of 23.6 ± 0.7 nM), while 12 and 13 showed no activity.| Compound | KD (nM) | Compound | KD (nM) |
|---|---|---|---|
| a NB: no binding observed in the SPR experiment for this compound.b Data previously reported.c 17-N-Allylamino-17-demethoxygeldanamycin. | |||
| 1b | 76 ± 7 | 8 | 2.9 ± 0.8 |
| 2 | 76.2 ± 1.9 | 9 | NB |
| 3 | 17.6 ± 4.9 | 10 | NB |
| 4 | 13.0 ± 4.9 | 11 | 23.6 ± 0.7 |
| 5 | 12.0 ± 1.9 | 12 | NB |
| 6 | 3.7 ± 0.9 | 13 | NB |
| 7 | NBa | 17-AAGb,c | 388 ± 89 |
Evaluation of anti-proliferative activity of compounds 2–13
After measuring the affinity for Hsp90α by SPR assay, (in order to correlate the observed molecular chaperone interaction of the tested compounds with cytotoxic effect), the eight selected DHPM binders, listed in Table 1, were further investigated for their anti-proliferative activity against two cancer cell lines: A375 (human melanoma) and Jurkat (human leukemic). The IC50 values are reported in Tables 2 and 3, respectively. The C-terminal inhibitor novobiocin was also used as reference compound in our assay. The anti-proliferative activity manifested by this second generation of 3,4-dihydropyrimidinones, allowed us to identify three new compounds 4, 5 and 11, endowed with moderate cytotoxic effects at micromolar concentration in both cancer cell lines (see Tables 2 and 3). Moreover, all these tested compounds exhibited a better anti-proliferative profile than the novobiocin and quite comparable to that observed for the lead compound 1 (IC50 values of 50.8 ± 0.2 and 20.8 ± 0.3 μM in A375 and Jurkat, respectively). Interestingly, referred to human leukemic cell line, the highest IC50 value was obtained for 11 (21.3 ± 0.9 μM after 24 h-treatment; novobiocin 170.6 ± 1.1 μM under the same experimental condition). Compounds 4 and 5 displayed the same potency with an IC50 = ∼50 μM, 3 showed only some effect in Jurkat cell after 48 h of treatment, while 2, 6 and 8 have not affected cell viability. In addition, it should be emphasized also that no negative effect was observed in PHA-stimulated proliferating PBMC, used as control non-tumor cell line, for which the percentage of non-viable cells after 24 h of treatment was similar to the value observed in DMSO treated control cells. The anti-proliferative trend observed for these new designed molecules highlights the role of DHPM ring as a suitable scaffold for the identification of new C-terminal Hsp90 inhibitors.| Compound | IC50 (μM) 24 h | IC50 (μM) 48 h |
|---|---|---|
| 2 | — | — |
| 3 | — | — |
| 4 | — | — |
| 5 | 85.1 ± 0.8 | 74.2 ± 1.1 |
| 6 | — | — |
| 8 | — | — |
| 11 | 81.0 ± 1.2 | 70.5 ± 1.4 |
| Novobiocin | 550.3 ± 1.3 | 460.5 ± 0.9 |
| Compound | IC50 (μM) 24 h | IC50 (μM) 48 h |
|---|---|---|
| 2 | — | — |
| 3 | — | 86.1 ± 0.9 |
| 4 | 51.2 ± 0.8 | 40.3 ± 0.6 |
| 5 | 55.0 ± 0.6 | 43.5 ± 1.0 |
| 6 | — | — |
| 8 | — | — |
| 11 | 21.3 ± 0.9 | 15.2 ± 1.1 |
| Novobiocin | 170.6 ± 1.1 | 150.5 ± 0.7 |
Western blot analyses of Hsp90 client proteins
In order to offer further evidence that links the anti-proliferative effects showed by compounds 4, 5 and 11 to Hsp90 inhibition, we verified by western blot analyses the Hsp90 client protein expression in the same treated and untreated cell lines used in the antiproliferative assay. The A375 and Jurkat cells were incubated for 24 h with DMSO or 4, 5 and 11 used at the concentrations corresponding to the IC50 values. Actin was used as control, since this protein is not an Hsp90-dependent substrate. As shown in Fig. 3, all the tested compounds induced a strong down-regulation (80–60%) of Hsp-90-depent clients Raf-1 and p-Akt, arising as new potent Hsp90 modulators. As above mentioned, the cytoprotective heat shock response (HRS) triggered by N-terminal inhibitors induces a large increase in HSPs levels, whereas C-terminal modulators did not cause high levels of heat shock proteins. Several studies report that over-expression of Hsp70 and Hsp27 increases tumorigenicity and protects malignant cells against apoptosis through several mechanisms.50,51 Notably, our data showed that exposure of compounds 5 and 11 in A375 and 4, 5 and 11 in Jurkat cell lines did not induce any significant increase in Hsp90, Hsp70 and Hsp27 protein levels,50,51 thus revealing that they did not trigger a HSR, inhibiting Hsp90 by modulation of the C-terminus. | ||
| Fig. 3 Western blot analysis of the Hsp90 and client proteins expression in lysate of A375 and Jurkat cells treated for 24 h with compounds 4, 5 and 11. β-Actin was used as loading control. | ||
Furthermore, the treatment with 5 and 11 in A375 for 24 h, has caused a decrease in Hsp27 protein levels (Fig. 3). Jurkat cell line was completely Hsp27 negative (according Sedlackova, et al.).50 In contrast, the incubation with 17-AAG led to a reduction in protein Raf-1 and p-Akt, however it induced a considerable upregulation of Hsp90, Hsp70 and Hsp27 expression levels (Fig. 3).
Analyzing the computational results of compound 11, the presence of disubstituted ring at C-4 directs efficiently the binding mode in the C terminal domain (Fig. 4). The bromine atom, in fact, forms a crucial halogen bond with the side chain of Arg591Chain B and the –OH establishes a hydrogen bonds with backbone of Ala597Chain A; the dihydropyrimidin-2(1H)-one ring, moreover, creates a π–π interaction with Arg591Chain B and two hydrogen bonds with Ser657Chain A and Lys594Chain A. The key role of bromine atom on the aromatic ring at C-4, is also confirmed by the biological profile exhibited for 5, in which, it forms a halogen bond with Arg591Chain B (Fig. 4).
 | ||
| Fig. 4 Three dimensional model of 11 (cyan sticks) and 5 (light brown sticks) in the Hsp90 C-terminal domain. | ||
On the other hand, even if the bromine atom of 13 forms a halogen bond with Ala597Chain A, the CF3 group decrease the molecule-target complex affinity creating an ugly interaction with the key amino acid Arg591Chain B. The same trend is also observed for 12, where the CF3 group negatively affects its activity forming a bad interaction with the Lys423Chain B. The high influence of the substitution of the aromatic ring at C-4 is also corroborated by the complete biological inactivity of 7, in which the CF3 group at ortho position forms a bad interaction with key Arg591Chain B, drafting this group as not favorable in the design of new Hsp90 C-terminal inhibitors. Considering the molecules belonging to the second subset, with para substitution (2–4 and 6) (Fig. 5), they show a good calculated interactions between the key aminoacids Arg591, Lys594 and the ring at C-4. These results outline the fundamental role of the decoration of the phenyl moiety that strongly affects the modulation of Hsp90 activity, as showed from a not homogeneous (4, 5 vs. 2, 3 and 6) profile in antiproliferative assay.
Conclusions
Anticancer drug development strategies critically involve the identification of novel molecular targets which are crucial for tumorigenesis and metastasis. In this context, Hsp90 and especially its C-terminal domain has gained great interest as a promising anticancer drug target. The main advantage of C-terminal modulators is their ability to inhibit numerous oncogenic pathways regulated by Hsp90 without inducing heat shock response. In this field the identification of new class of no-nature expired C-terminal inhibitors represents a valid and alternative chemotherapeutic approach. We have already disclosed the DHPM scaffold as an efficacious C-terminal modulator, which is easily accessible by a fast high-yielding multicomponent reaction. Starting from some key interactions previously identified by us, here we reported a new set of closely related DHPM derivatives endowed with a comparable biological profile of lead compound and better with respect to novobiocin. In this study, our structural results disclose the halogen bonding between bromine atom and the side chain of Arg591Chain B as a new key interaction suitable for the design of novel DHPM Hsp90. Moreover, these findings outline that only punctual changes at specific position of the aromatic ring at C-4 of Biginelli scaffold are tolerated. These data, in fact, reflect a high sensitive steric environment within the C-terminal domain, and prompted us to further optimize the Biginelli core in order to identify more powerful Hsp90 inhibitors.Experimental
General information
All commercially available starting materials were purchased from Sigma-Aldrich and were used as received. All solvents used for the synthesis were of HPLC grade; they were purchased from Sigma-Aldrich and Carlo Erba Reagenti. NMR spectra (1H, HMBC, HSQC) were recorded on a Bruker 400 MHz instrument. All compounds were dissolved in 0.5 mL of 99.95% CDCl3 (Carlo Erba, 99.95 atom% D). Coupling constants (J) are reported in hertz, and chemical shifts are expressed in parts per million (ppm) on the delta (δ) scale relative to CHCl3 (7.26 ppm for 1H and 77.2 ppm for 13C) as internal reference. Electrospray mass spectrometry (ESI-MS) was performed on a LCQ DECA TermoQuest (San Josè, California, USA) mass spectrometer.Reactions were monitored on silica gel 60 F254 plates (Merck) and the spots were visualized under UV light. Analytical and semi-preparative reversed-phase HPLC was performed on Agilent Technologies 1200 Series high performance liquid chromatography using a Jupiter Proteo C18 reversed-phase column (250 × 4.60 mm, 4μ, 90 Å, flow rate = 1 mL min−1; 250 × 10.00 mm, 10μ, 90 Å, flow rate = 4 mL min−1 respectively, Phenomenex®). The binary solvent system (A/B) was as follows: 0.1% TFA in water (A) and 0.1% TFA in CH3CN (B). The absorbance was detected at 280 nm. The purity of all tested compound (>97%) was determined by HPLC analysis. All microwave irradiation experiments were carried out in a dedicated CEM-Discover® Focused Microwave Synthesis apparatus, operating with continuous irradiation power from 0 to 300 W utilizing the standard absorbance level of 300 W maximum power. The reactions were carried out in 10 mL sealed microwave glass vials. The Discover™ system also offers controllable ramp time, hold time (reaction time) and uniform stirring. The temperature was monitored using the CEM-Discover built-in-vertically-focused IR temperature sensor. After the irradiation period, the reaction vessel was cooled rapidly (60–120 s) to ambient temperature by air jet cooling.
Synthesis
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) (3 × 3 mL), to give the desired product in good yields (68–90%). HPLC purification was performed by semi-preparative reversed-phase HPLC (on a Jupiter Proteo C18 column: 250 × 10.00 mm, 10μ, 90 Å, flow rate = 4 mL min−1) using the gradient conditions reported below for each compound. The final products were obtained with high purity (>95%) as detected by HPLC analysis and were fully characterized by ESMS and NMR spectra.
1) (3 × 3 mL), to give the desired product in good yields (68–90%). HPLC purification was performed by semi-preparative reversed-phase HPLC (on a Jupiter Proteo C18 column: 250 × 10.00 mm, 10μ, 90 Å, flow rate = 4 mL min−1) using the gradient conditions reported below for each compound. The final products were obtained with high purity (>95%) as detected by HPLC analysis and were fully characterized by ESMS and NMR spectra.Compound 2 was obtained by following the general procedure as a yellow solid (200.0 mg, 66% yield). RP-HPLC tR = 33.5 min, gradient condition: from 5% B to 100% B in 50 min, flow rate of 4 mL min−1, λ = 280 nm. 1H NMR (400 MHz, CDCl3): δ = 1.21 (t, J = 7.1 Hz, 3H), 2.13 (s, 3H), 4.16 (q, J = 7.0 Hz, 2H), 5.61 (s, 1H), 7.14 (br s, 2H), 7.41–7.50 (m, 3H), 7.55–7.60 (d, J = 8.5 Hz, 2H), 8.22 (d, J = 8.4 Hz, 2H); 13C NMR (100 MHz, CDCl3): δ = 15.2, 19.6, 54.5, 61.0, 105.6, 122.6, 125.0, 128.3, 128.9, 130.6, 133.5, 138.3, 143.4, 148.1, 166.5. ESMS, calcd for C20H19N3O5 381.1; found m/z = 382.5 [M + H]+.
Compound 3 was obtained by following the general procedure as a yellow gelatinous solid (210.0 mg, 60% yield). RP-HPLC tR = 33.2 min, gradient condition: from 5% B to 100% B in 50 min, flow rate of 4 mL min−1, λ = 280 nm. 1H NMR (400 MHz, CDCl3): δ = 1.21 (t, J = 7.1 Hz, 3H), 2.13 (s, 3H), 3.80 (s, 3H), 4.16 (q, J = 7.0 Hz, 2H), 5.60 (s, 1H), 6.82–6.97 (m, 2H), 7.12–7.24 (m, 4H), 7.28–7.36 (m, 3H), 8.20–8.25 (d, 2H); 13C NMR (100 MHz, CDCl3): δ = 14.6, 18.9, 55.0, 55.8, 60.1, 105.6, 113.8, 127.5, 128.4, 128.9, 132.8, 138.6, 150.3, 158.7, 167.5. ESMS, calcd for C21H22N2O4 366.4; found m/z = 367.3 [M + H]+.
Compound 4 was obtained by following the general procedure as a yellow solid (200.0 mg, 75% yield). RP-HPLC tR = 33.9 min, gradient condition: from 5% B to 100% B in 50 min, flow rate of 4 mL min−1, λ = 280 nm. 1H NMR (400 MHz, CDCl3): δ = 1.24 (t, J = 7.1 Hz, 3H), 2.13 (s, 3H), 2.40 (s, 3H), 4.16 (q, J = 7.1 Hz, 2H), 5.50 (s, 1H), 7.18–7.22 (d, J = 7.8 Hz, 2H), 7.24 (s, 1H), 7.29–7.33 (m, 3H), 7.42–7.50 (m, 3H); 13C NMR (100 MHz, CDCl3): δ = 15.1, 19.6, 20.7, 55.5, 61.1, 104.9, 125.6, 127.9, 128.4, 128.9, 129.4, 136.5, 140.2, 147.8, 152.9, 166.7 ESMS, calcd for C21H22N2O3 350.4; found m/z = 351.4 [M + H]+.
Compound 5 was obtained by following the general procedure as a yellow solid (190.0 mg, 65% yield). RP-HPLC tR = 34.8 min, gradient condition: from 5% B to 100% B in 50 min, flow rate of 4 mL min−1, λ = 280 nm. 1H NMR (400 MHz, CDCl3): δ = 1.24 (t, J = 7.1 Hz, 3H), 2.14 (s, 3H), 4.16 (q, J = 7.0 Hz, 2H), 5.50 (s, 1H), 7.20 (s, 2H), 7.28–7.31 (d, J = 8.5 Hz, 2H), 7.42–7.53 (m, 5H); 13C NMR (100 MHz, CDCl3): δ = 14.3, 19.5, 55.0, 61.3, 105.6, 122.8, 123.4, 128.9, 129.1, 132.8, 133.5, 138.6, 143.4, 149.7, 167.3. ESMS, calcd for C20H19BrN2O3 415.3; found m/z = 415.5–417.5 [M + H]+.
Compound 6 was obtained by following the general procedure as a yellow solid (200.0 mg, 75% yield). RP-HPLC tR = 29.4 min, gradient condition: from 5% B to 100% B in 50 min, flow rate of 4 mL min−1, λ = 280 nm. 1H NMR (400 MHz, CDCl3): δ = 1.27 (t, J = 7.1 Hz, 3H), 2.40 (s, 3H), 4.16 (q, J = 7.0 Hz, 2H), 5.50 (s, 1H), 6.76 (d, J = 8.4 Hz, 2H), 7.07 (d, J = 8.3 Hz, 2H), 7.18 (t, J = 9.4 Hz, 3H), 7.28 (dd, J = 8.5 Hz, 2H); 13C NMR (100 MHz, CDCl3): δ = 15.3, 16.8, 59.1, 61.0, 114.5, 124.6, 126.3, 128.1, 129.6, 130.1, 140.2, 148.7, 115.4, 167.2. ESMS, calcd for C20H20BrN2O4 352.4; found m/z = 353.4 [M + H]+.
Compound 7 was obtained by following the general procedure as a white solid (190.0 mg, 55% yield). RP-HPLC tR = 36.6 min, gradient condition: from 5% B to 100% B in 50 min, flow rate of 4 mL min−1, λ = 280 nm. 1H NMR (400 MHz, CDCl3): δ = 1.04 (t, J = 7.1 Hz, 3H), 2.28 (s, 3H), 4.02 (q, J = 7.0 Hz, 2H), 5.62 (s, 1H), 7.29 (s, 1H), 7.45–7.52 (m, 5H), 7.66–7.69 (d, J = 6.6 Hz, 2H), 7.73–7.76 (d, J = 7.7 Hz, 1H); 13C NMR (100 MHz, CDCl3): δ = 14.8, 19.7, 51.2, 61.5, 103.5, 115.1, 123.2, 127.2, 128.9, 129.3, 129.6, 130.1, 133.6, 161.9. ESMS, calcd for C21H19F3N2O3 404.4; found m/z = 405.5 [M + H]+.
Compound 8 was obtained by following the general procedure as a white solid (200.0 mg, 70% yield). RP-HPLC tR = 34.7 min, gradient condition: from 5% B to 100% B in 50 min, flow rate of 4 mL min−1, λ = 280 nm. 1H NMR (400 MHz, CDCl3): δ = 0.98 (t, J = 7.1 Hz, 3H), 2.27 (s, 3H), 3.98 (q, J = 7.1 Hz, 2H), 5.92 (s, 1H), 7.21–7.25 (s, 2H), 7.42–7.54 (m, 4H), 7.65–7.74 (m, 2H), 7.98 (dd, J = 8.1, 1.3 Hz, 1H); 13C NMR (100 MHz, CDCl3): δ = 14.8, 19.4, 51.3, 61.1, 101.6, 123.9, 128.2, 128.5, 133.0, 135.8, 136.2, 147.3, 148.1, 150.9, 152.6, 164.2. ESMS, calcd for C20H19N3O5 381.4; found m/z = 382.3 [M + H]+.
Compound 9 was obtained by following the general procedure as a colorless gelatinous solid (170.0 mg, 70% yield). RP-HPLC tR = 35.4 min, gradient condition: from 5% B to 100% B in 50 min, flow rate of 4 mL min−1, λ = 280 nm. 1H NMR (400 MHz, CDCl3): δ = 1.23 (t, J = 7.1 Hz, 3H), 2.14 (s, 3H), 4.16 (q, J = 7.0 Hz, 2H), 5.47 (s, 1H), 7.25 (s, 2H), 7.36 (t, J = 7.2 Hz, 1H), 7.44–7.53 (m, 5H), 7.61 (s, 1H); 13C NMR (100 MHz, CDCl3): δ = 15.3, 19.5, 55.1, 61.5, 105.4, 124.2, 125.7, 128.0, 128.9, 130.5, 132.4, 147.2, 151.2, 154.9, 166.4. ESMS, calcd for C20H19BrN2O3 415.3; found m/z = 415.5–417.5 [M + H]+.
Compound 10 was obtained by following the general procedure as a white solid (200.0 mg, 70% yield). RP-HPLC tR = 34.2 min, gradient condition: from 5% B to 100% B in 50 min, flow rate of 4 mL min−1, λ = 280 nm. 1H NMR (400 MHz, CDCl3): δ = 1.26 (t, J = 7.1 Hz, 3H), 2.20 (s, 3H), 4.17 (q, J = 7.0 Hz, 2H), 5.67 (s, 1H), 6.91 (s, 1H), 7.30 (s, 1H), 7.47–7.53 (m, 3H), 7.63 (t, J = 8.1 Hz, 1H), 7.78 (d, J = 7.9 Hz, 1H), 8.24 (d, J = 8.4 Hz, 1H), 8.36 (s, 1H); 13C NMR (100 MHz, CDCl3): δ = 13.9, 18.4, 53.7, 60.4, 104.0, 121.5, 122.8, 128.5, 129.5, 130.0, 132.2, 137.0, 145.2, 148.2, 149.6, 153.1, 165.1. ESMS, calcd for C20H19BrN3O5 381.4; found m/z = 382.4 [M + H]+.
Compound 11 was obtained by following the general procedure as a pale yellow gelatinous solid (170.0 mg, 74% yield). RP-HPLC tR = 38.7 min, gradient condition: from 5% B to 100% B in 50 min, flow rate of 4 mL min−1, λ = 280 nm. 1H NMR (400 MHz, CDCl3): δ = 1.10 (t, J = 7.1 Hz, 3H), 2.21 (s, 3H), 4.16 (q, J = 7.1 Hz, 2H), 5.83 (s, 1H), 6.64 (dd, J = 8.6, 2.9 Hz, 1H), 6.89 (br s, 1H), 7.17–7.24 (m, 2H), 7.39–745 (m, 4H); 13C NMR (100 MHz, CDCl3): δ = 13.9, 19.4, 53.4, 60.2, 114.4, 115.0, 116.9, 128.9, 129.2, 129.5, 133.5, 134.4, 138.6, 143.4, 149.7, 166.3. ESMS, calcd for C20H19BrN2O4 430.1; found m/z = 431.0 [M + H]+.
Compound 12 was obtained by following the general procedure as a white solid (150.0 mg, 70% yield). RP-HPLC tR = 38.2 min, gradient condition: from 5% B to 100% B in 50 min, flow rate of 4 mL min−1, λ = 280 nm. 1H NMR (400 MHz, CDCl3): δ = 1.13 (t, J = 7.1 Hz, 3H), 2.27 (s, 3H), 4.13 (q, J = 7.0 Hz, 2H), 5.98 (s, 1H), 7.15–7.24 (m, 2H), 7.43–7.50 (m, 3H), 7.57 (s, 2H), 7.66 (s, 1H); 13C NMR (100 MHz, CDCl3): δ = 15.0, 19.6, 53.9, 61.3, 105.6, 121.9, 125.6, 127.1, 128.9, 129.8, 130.2, 129.2, 141.9, 149.7, 167.3. ESMS, calcd for C21H18BrF3N2O3 483.3; found m/z = 483.2–485.2 [M + H]+.
Compound 13 was obtained by following the general procedure as a colorless gelatinous solid (100.0 mg, 80% yield). RP-HPLC tR = 39.2 min, gradient condition: from 5% B to 100% B in 50 min, flow rate of 4 mL min−1, λ = 280 nm. 1H NMR (400 MHz, CDCl3): δ = 1.14 (t, J = 7.1 Hz, 3H), 2.27 (s, 3H), 4.13 (q, J = 7.0 Hz, 2H), 5.97 (s, 1H), 7.18–7.24 (m, 2H), 7.43–7.50 (m, 4H), 7.66 (s, 1H), 7.78 (d, J = 8.1 Hz, 1H); 13C NMR (100 MHz, CDCl3): δ = 14.9, 19.2, 54.9, 61.9, 105.6, 121.9, 125.6, 127.1, 128.9, 129.8, 130.1, 129.2, 141.9, 149.7, 167.3. ESMS, calcd for C21H18ClF3N2O3 438.8; found m/z = 439.0 [M + H]+.
Biological methods
SPR analyses were carried out according to our previously published procedures.39,44–48 Briefly, SPR analyses were performed using a Biacore 3000 optical biosensor equipped with research-grade CM5 sensor chips (GE Healthcare). Using this platform, two separate recombinant Hsp90α surfaces, a BSA surface and one unmodified reference surface were prepared for simultaneous analyses. Proteins (100 μg mL−1 in 10 mM CH3COONa, pH 5.0) were immobilized on individual sensor chip surfaces at a flow rate of 5 μL min−1 using standard amine-coupling protocols to obtain densities of 8–12 kRU.
Compounds 2–13, as well as 17-AAG, were dissolved in 100% DMSO to obtain 4 mM solutions, and diluted 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 200 (v/v) in PBS (10 mM NaH2PO4, 150 mM NaCl, pH 7.4) to a final DMSO concentration of 0.5%. Compounds concentration series were prepared as twofold dilutions into running buffer: for each sample, the complete binding study was performed using a six-point concentration series, typically spanning 0.025–1 μM, and triplicate aliquots of each compound concentration were dispensed into disposable vials. Binding experiments were performed at 25 °C, using a flow rate of 50 μL min−1, with 60 s monitoring of association and 300 s monitoring of dissociation (Table 1).
200 (v/v) in PBS (10 mM NaH2PO4, 150 mM NaCl, pH 7.4) to a final DMSO concentration of 0.5%. Compounds concentration series were prepared as twofold dilutions into running buffer: for each sample, the complete binding study was performed using a six-point concentration series, typically spanning 0.025–1 μM, and triplicate aliquots of each compound concentration were dispensed into disposable vials. Binding experiments were performed at 25 °C, using a flow rate of 50 μL min−1, with 60 s monitoring of association and 300 s monitoring of dissociation (Table 1).
Simple interactions were suitably fitted to a single-site bimolecular interaction model (A + B = AB), yielding a single KD. Sensorgram elaborations were performed using the BIAevaluation software provided by GE Healthcare.
A375 (1 × 104 per well) and Jurkat (2 × 104 per well) cells were seeded in triplicate in 96 well-plates and incubated with increasing concentrations of compounds 2–6, 8, 11 (concentration between 10 μM to 100 μM) or novobiocin (50 μM to 800 μM) or DMSO 0.15% (v/v) for the 24 h and 48 h. The number of viable cells was determined by using a [3-4,5-dimethyldiazol-2-yl]-2,5-diphenyl tetrazolium bromide (MTT, Sigma-Aldrich) conversion assay, according to the method described by Mosmann. Briefly, following the treatment, 25 μL of MTT (5 mg mL−1 in PBS) was added and the cells were incubated for additional 3 h at 37 °C. Thereafter, cells were lysed and suspended with 100 μL of buffer containing 50% (v/v) N,N-dimethylformamide, 20% SDS (pH 4.5). The absorbance was measured with a microplate reader (Titertek multiskan MCC7340, LabSystems, Vienna, VA, USA) equipped with a 620 nm filter. The cell population growth inhibition was also tested by cytometric counting (trypan blue exclusion). IC50 values were calculated from cell viability dose–response curves and defined as the concentration resulting in 50% inhibition of cell survival, compared to control cells treated with DMSO. PBMC were treated with 4, 5 and 11 used at the concentrations proximity to the IC50 values of A375 and Jurkat cells (85 μM, 50 μM or 20 μM, respectively) to evaluate their effects on non-cancer cells viability.
After washing, the membranes were incubated at room temperature (1 h), with an appropriate peroxidase-conjugate secondary antibodies and were detected with Pierce ECL Western Blotting Substrate (Pierce ECL, Thermo Scientific, Rockford, IL, USA) according the manufacturer's instructions. Quantitative densitometry analyses were performed using a ImageQuant LAS 4000 system (GE Healthcare Life Sciences, NY, USA).
Acknowledgements
Financial support from Ministero Istruzione e Ricerca (MIUR), Italy, under Grant ORSA151559 is gratefully acknowledged.Notes and references
- J. J. Barrott and T. A. Haystead, FEBS J., 2013, 280, 1381–1396 CrossRef CAS PubMed.
- J. A. Hall, L. K. Forsberg and B. S. Blagg, Future Med. Chem., 2014, 6, 1587–1605 CrossRef CAS PubMed.
- D. Hanahan and R. A. Weinberg, Cell, 2011, 144, 646–674 CrossRef CAS PubMed.
- G. Chiosis, H. Huezo, N. Rosen, E. Mimnaugh, L. Whitesell and L. Neckers, Mol. Cancer Ther., 2003, 2, 123–129 CAS.
- L. Whitesell and S. L. Lindquist, Nat. Rev. Cancer, 2005, 5, 761–772 CrossRef CAS PubMed.
- Y. Miyata, H. Nakamoto and L. Neckers, Curr. Pharm. Des., 2013, 19, 347–365 CrossRef CAS PubMed.
- T. Stivarou, D. Stellas, G. Vartzi, D. Thomaidou and E. Patsavoudi, Cancer Biol. Ther., 2016, 799–812 CrossRef CAS PubMed.
- M. I. Gallegos Ruiz, K. Floor, P. Roepman, J. A. Rodriguez, G. A. Meijer, W. J. Mooi, E. Jassem, J. Niklinski, T. Muley, N. van Zandwijk, E. F. Smit, K. Beebe, L. Neckers, B. Ylstra and G. Giaccone, PLoS One, 2008, 3, e0001722 Search PubMed.
- D. B. Solit, A. D. Basso, A. B. Olshen, H. I. Scher and N. Rosen, Cancer Res., 2003, 63, 2139–2144 CAS.
- G. Jego, A. Hazoume, R. Seigneuric and C. Garrido, Cancer Lett., 2013, 332, 275–285 CrossRef CAS PubMed.
- A. Kamal, L. Thao, J. Sensintaffar, L. Zhang, M. F. Boehm, L. C. Fritz and F. J. Burrows, Nature, 2003, 425, 407–410 CrossRef CAS PubMed.
- K. Moulick, J. H. Ahn, H. Zong, A. Rodina, L. Cerchietti, E. M. Gomes DaGama, E. Caldas-Lopes, K. Beebe, F. Perna, K. Hatzi, L. P. Vu, X. Zhao, D. Zatorska, T. Taldone, P. Smith-Jones, M. Alpaugh, S. S. Gross, N. Pillarsetty, T. Ku, J. S. Lewis, S. M. Larson, R. Levine, H. Erdjument-Bromage, M. L. Guzman, S. D. Nimer, A. Melnick, L. Neckers and G. Chiosis, Nat. Chem. Biol., 2011, 7, 818–826 CrossRef CAS PubMed.
- S. Modi, C. Saura, C. Henderson, N. U. Lin, R. Mahtani, J. Goddard, E. Rodenas, C. Hudis, J. O'Shaughnessy and J. Baselga, Breast Cancer Res. Treat., 2013, 139, 107–113 CrossRef CAS PubMed.
- N. Gandhi, A. T. Wild, S. T. Chettiar, K. Aziz, Y. Kato, R. P. Gajula, R. D. Williams, J. A. Cades, A. Annadanam, D. Song, Y. Zhang, R. K. Hales, J. M. Herman, E. Armour, T. L. DeWeese, E. M. Schaeffer and P. T. Tran, Cancer Biol. Ther., 2013, 14, 347–356 CrossRef CAS PubMed.
- E. A. Ronnen, G. V. Kondagunta, N. Ishill, S. M. Sweeney, J. K. Deluca, L. Schwartz, J. Bacik and R. J. Motzer, Invest. New Drugs, 2006, 24, 543–546 CrossRef CAS PubMed.
- S. Pacey, M. Gore, D. Chao, U. Banerji, J. Larkin, S. Sarker, K. Owen, Y. Asad, F. Raynaud, M. Walton, I. Judson, P. Workman and T. Eisen, Invest. New Drugs, 2012, 30, 341–349 CrossRef CAS PubMed.
- A. Khandelwal, V. M. Crowley and B. S. Blagg, Med. Res. Rev., 2016, 36, 92–118 CrossRef CAS PubMed.
- L. Neckers and P. Workman, Clin. Cancer Res., 2012, 18, 64–76 CrossRef CAS PubMed.
- A. K. McCollum, C. J. Teneyck, B. M. Sauer, D. O. Toft and C. Erlichman, Cancer Res., 2006, 66, 10967–10975 CrossRef CAS PubMed.
- R. Bagatell, G. D. Paine-Murrieta, C. W. Taylor, E. J. Pulcini, S. Akinaga, I. J. Benjamin and L. Whitesell, Clin. Cancer Res., 2000, 6, 3312–3318 CAS.
- X. Wang, M. Chen, J. Zhou and X. Zhang, Int. J. Oncol., 2014, 45, 18–30 Search PubMed.
- A. Donnelly and B. S. Blagg, Curr. Med. Chem., 2008, 15, 2702–2717 CrossRef CAS PubMed.
- Y. C. Koay, J. R. McConnell, Y. Wang and S. R. McAlpine, RSC Adv., 2015, 5, 59003–59013 RSC.
- J. R. McConnell, L. A. Alexander and S. R. McAlpine, Bioorg. Med. Chem. Lett., 2014, 24, 661–666 CrossRef CAS PubMed.
- Y. C. Koay, J. R. McConnell, Y. Wang, S. J. Kim, L. K. Buckton, F. Mansour and S. R. McAlpine, ACS Med. Chem. Lett., 2014, 5, 771–776 CrossRef CAS PubMed.
- J. Gavenonis, N. E. Jonas and J. A. Kritzer, Bioorg. Med. Chem., 2014, 22, 3989–3993 CrossRef CAS PubMed.
- Y. Wang and S. R. McAlpine, Org. Biomol. Chem., 2015, 13, 4627–4631 CAS.
- H. Zhao, G. Garg, J. Zhao, E. Moroni, A. Girgis, L. S. Franco, S. Singh, G. Colombo and B. S. Blagg, Eur. J. Med. Chem., 2015, 89, 442–466 CrossRef CAS PubMed.
- M. G. Marcu, A. Chadli, I. Bouhouche, M. Catelli and L. M. Neckers, J. Biol. Chem., 2000, 275, 37181–37186 CrossRef CAS PubMed.
- A. C. Donnelly, H. P. Zhao, B. R. Kusuma and B. S. J. Blagg, MedChemComm, 2010, 1, 165–170 RSC.
- H. Zhao, M. Anyika, A. Girgis and B. S. Blagg, Bioorg. Med. Chem. Lett., 2014, 24, 3633–3637 CrossRef CAS PubMed.
- B. R. Kusuma, A. Khandelwal, W. Gu, D. Brown, W. Liu, G. Vielhauer, J. Holzbeierlein and B. S. Blagg, Bioorg. Med. Chem., 2014, 22, 1441–1449 CrossRef CAS PubMed.
- M. Anyika, M. McMullen, L. K. Forsberg, R. T. Dobrowsky and B. S. Blagg, ACS Med. Chem. Lett., 2016, 7, 67–71 CrossRef CAS PubMed.
- K. M. Byrd, C. Subramanian, J. Sanchez, H. F. Motiwala, W. Liu, M. S. Cohen, J. Holzbeierlein and B. S. Blagg, Chem.–Eur. J., 2016, 22, 6921–6931 CrossRef CAS PubMed.
- J. B. Zhao, H. P. Zhao, J. A. Hall, D. Brown, E. Brandes, J. Bazzill, P. T. Grogan, C. Subramanian, G. Vielhauer, M. S. Cohen and B. S. J. Blagg, Medchemcomm, 2014, 5, 1317–1323 RSC.
- R. P. Sellers, L. D. Alexander, V. A. Johnson, C. C. Lin, J. Savage, R. Corral, J. Moss, T. S. Slugocki, E. K. Singh, M. R. Davis, S. Ravula, J. E. Spicer, J. L. Oelrich, A. Thornquist, C. M. Pan and S. R. McAlpine, Bioorg. Med. Chem., 2010, 18, 6822–6856 CrossRef CAS PubMed.
- J. A. Burlison and B. S. Blagg, Org. Lett., 2006, 8, 4855–4858 CrossRef CAS PubMed.
- E. Moroni, H. Zhao, B. S. Blagg and G. Colombo, J. Chem. Inf. Model., 2014, 54, 195–208 CrossRef CAS PubMed.
- M. Strocchia, S. Terracciano, M. G. Chini, A. Vassallo, M. C. Vaccaro, F. Dal Piaz, A. Leone, R. Riccio, I. Bruno and G. Bifulco, Chem. Commun., 2015, 51, 3850–3853 RSC.
- L. Pisani, H. Prokopcova, J. M. Kremsner and C. O. Kappe, J. Comb. Chem., 2007, 9, 415–421 CrossRef CAS PubMed.
- C. O. Kappe, Eur. J. Med. Chem., 2000, 35, 1043–1052 CrossRef CAS PubMed.
- A. Stadler and C. O. Kappe, J. Comb. Chem., 2001, 3, 624–630 CrossRef CAS PubMed.
- S. Terracciano, G. Lauro, M. Strocchia, K. Fischer, O. Werz, R. Riccio, I. Bruno and G. Bifulco, ACS Med. Chem. Lett., 2015, 6, 187–191 CrossRef CAS PubMed.
- F. Dal Piaz, A. Vassallo, A. Temraz, R. Cotugno, M. A. Belisario, G. Bifulco, M. G. Chini, C. Pisano, N. De Tommasi and A. Braca, J. Med. Chem., 2013, 56, 1583–1595 CrossRef PubMed.
- F. Dal Piaz, A. Vassallo, M. G. Chini, F. M. Cordero, F. Cardona, C. Pisano, G. Bifulco, N. De Tommasi and A. Brandi, PLoS One, 2012, 7, e43316 CAS.
- C. Giommarelli, V. Zuco, E. Favini, C. Pisano, F. Dal Piaz, N. De Tommasi and F. Zunino, Cell. Mol. Life Sci., 2010, 67, 995–1004 CrossRef CAS PubMed.
- M. A. Cooper, Anal. Bioanal. Chem., 2003, 377, 834–842 CrossRef CAS PubMed.
- A. Vassallo, M. C. Vaccaro, N. De Tommasi, F. Dal Piaz and A. Leone, PLoS One, 2013, 8, e74266 CAS.
- W. Guo, P. Reigan, D. Siegel, J. Zirrolli, D. Gustafson and D. Ross, Cancer Res., 2005, 65, 10006–10015 CrossRef CAS PubMed.
- L. Sedlackova, M. Spacek, E. Holler, Z. Imryskova and I. Hromadnikova, Tumor Biol., 2011, 32, 33–44 CrossRef CAS PubMed.
- A. Aghdassi, P. Phillips, V. Dudeja, D. Dhaulakhandi, R. Sharif, R. Dawra, M. M. Lerch and A. Saluja, Cancer Res., 2007, 67, 616–625 CrossRef CAS PubMed.
- Induced Fit Docking, protocol 2015-2, Glide version 6.4, Prime version 3.7, Schrödinger, LLC, New York, NY, 2015 Search PubMed.
- S. Picaud, M. Strocchia, S. Terracciano, G. Lauro, J. Mendez, D. L. Daniels, R. Riccio, G. Bifulco, I. Bruno and P. Filippakopoulos, J. Med. Chem., 2015, 58, 2718–2736 CrossRef CAS PubMed.
- B. Renga, C. Festa, S. De Marino, S. Di Micco, M. V. D'Auria, G. Bifulco, S. Fiorucci and A. Zampella, Steroids, 2015, 96, 121–131 CrossRef CAS PubMed.
- L. d'Aquino, M. C. de Pinto, L. Nardi, M. Morgana and F. Tommasi, Chemosphere, 2009, 75, 900–905 CrossRef PubMed.
- W. Sherman, T. Day, M. P. Jacobson, R. A. Friesner and R. Farid, J. Med. Chem., 2006, 49, 534–553 CrossRef CAS PubMed.
- W. Sherman, H. S. Beard and R. Farid, Chem. Biol. Drug Des., 2006, 67, 83–84 CAS.
- M. M. U. Ali, S. M. Roe, C. K. Vaughan, P. Meyer, B. Panaretou, P. W. Piper, C. Prodromou and L. H. Pearl, Nature, 2006, 440, 1013–1017 CrossRef CAS PubMed.
- C. C. Lee, T. W. Lin, T. P. Ko and A. H. Wang, PLoS One, 2011, 6, e19961 CAS.
- G. M. Sastry, M. Adzhigirey, T. Day, R. Annabhimoju and W. Sherman, J. Comput.-Aided Mol. Des., 2013, 27, 221–234 CrossRef PubMed.
- Maestro, version 10.2, Schrödinger, LLC, New York, 2015 Search PubMed.
- LigPrep, version 3.4, Schrödinger, LLC, New York, 2015 Search PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c6ra17235k |
| ‡ These authors contributed equally to this work. |
| This journal is © The Royal Society of Chemistry 2016 |