Number-controlled spatial arrangement of gold nanoparticles with DNA dendrimers†
Ping Chen*a,
Tao Zhang‡
b,
Tao Zhoub and
Dongsheng Liub
aJiangsu Key Laboratory for Chemistry of Low-Dimensional Materials, School of Chemistry and Chemical Engineering, Huaiyin Normal University, Huaian 223300, P. R. China. E-mail: chenping@iccas.ac.cn
bKey Laboratory of Organic Optoelectronics & Molecular Engineering of the Ministry of Education, Department of Chemistry, Tsinghua University, Beijing 100084, P. R. China
First published on 20th July 2016
Abstract
In this study we described a controlled self-assembly of gold nanoparticles mediated by DNA dendrimers. Stepwise titration of three-arm Y-shaped DNA monomers through a divergent route can assemble different generations of DNA dendrimers with discrete DNA sticky ends on the surface. Hybridizing gold nanoparticles bearing one single-stranded DNA to different generations of DNA dendrimers thereby results in assembly of number-controlled discrete gold nanoparticle groupings. The conjugation strategy used herein provides a novel model for spatial positioning of functionalized nanoparticles and precise construction of multivalent nanomaterials.
Gold nanoparticles (AuNPs) are being actively developed as building blocks for nanophotonic and nanoelectronic materials.1,2 Many of their properties, such as surface plasmon resonance, depend critically on the interaction to their neighbouring particles.3,4 Bottom-up self-assembly provides a decent way to organize nanoparticles over different distances and geometries precisely.5 Among various self-assembly methods, DNA is now widely employed as a template for nanoparticle assembly6–17 because of its remarkable molecular recognition properties and structural features.18 However, number-controlled spatial arrangement of nanoparticles groupings still remains as a challenging goal. Meanwhile, dendrimers,19 a class of highly branched macromolecules with uniform size, receive increasing attention for their multivalent and nanosized structures providing great potential to program nanoparticle assemblies. But most of dendrimers involve multi-step synthesis and purification processes, as well as complex procedures to incorporate more branches. In contrast to conventional wisdom, DNA dendrimers based solely on DNA self-assembly could be obtained in high yields without any purification owing to the precise recognition of DNA hybridization.20–22 Herein we report a straightforward method to control the assembly of discrete structures of AuNPs using DNA dendrimers. The outermost sticky ends of the DNA dendrimers could be evenly distributed in three dimensions as their globular structures, which make them extremely suitable for being used as scaffolds to place nanoparticles in precise spatial positions.
Our strategy is described in Scheme 1. DNA dendrimers were prepared using step-by-step assembly of Y-shaped DNA (Y-DNA). As the structure of Y-DNA is approximately planar,23 a three-dimensional scaffold can be accessed by choosing the lengths of DNA duplex between each layer to be approximately 3.75 turns (39 basepairs). In particular, to eliminate the possible steric issues caused by multiple branched sides, 60 basepairs (∼5.75 turns) spacer were introduced between the first layer and the core. This design can not only delay the saturation occurring at a certain layer, but also improve assembly efficiency of the sticky ends. As each Y-DNA bearing three arms, the number of outermost branches could be controlled to certain value. DNA mono-modified AuNPs were then assembled via hybridization to the sticky ends on DNA dendrimers surface, which were used as scaffolds to control programmed spatial arrangement of nanoparticles.
 | ||
| Scheme 1 Strategies employed in controlled spatial arrangement of gold nanoparticles using DNA dendrimers as scaffolds. | ||
To obtain different generations of DNA dendrimers, equal moles of oligonucleotides (for example, Yna, Ynb and Ync) were mixed together to form dendrimer monomers (Yn, the nth-generation Y-DNA). The 13-base or 23-base sticky ends on the Y-DNA arms (Scheme 1) construct stable self-assembled structures at room temperature.24 The nth-generation DNA dendrimer was named Gn, and stepwise titration of Yn to Gn−1 (n ≥ 1), DNA dendrimers of different generations can be synthesized via the hybridization of sticky ends (Fig. 1a). Here every new layers of Yn that were added on Gn−1 are different and we assemble the G series dendrimer in a manner of A-B-C-D-E. As we had found in the previous report,21 such assembly strategy yielded the best quality of DNA dendrimers. Therefore the sticky ends on surface of each generation Gn are different, either 13 or 23 bases in our experiments, which can avoid crosstalk between the different layers. The self-assembled DNA dendrimers were then characterized by agarose gel electrophoresis and dynamic light scattering (DLS). As shown in Fig. 1b, DNA dendrimers of different generations appeared in agarose gel as a single band with a high yield in the absence of purification. DNA dendrimers of higher generations exhibited reduced mobility since the larger sizes, the slower the migrations. Here the size effect (but not the charges) dominates in the gel electrophoresis. Thus the mobility of Gn+1 was slower than that of Gn as shown in the gel. DLS measurement (Fig. 1c) showed, hydrodynamic radius increased from ∼12 nm for G1 to ∼28 nm for G4, which is consistent to the gel electrophoresis result. The structure of DNA dendrimer (G4) was further confirmed by atomic force microscopy (AFM). The G4 samples were deposited on mica and imaged by tapping mode under buffer, which revealed highly branched dendritic nanostructure (Fig. 1d).
 | ||
| Fig. 1 (a) The assembly strategies to construct DNA dendrimers; (b) agarose gel electrophoresis of G0–G4; (c) size distribution by DLS of G1–G4; (d) AFM images of G4. | ||
Such nanoscale, size-controllable DNA dendritic structures with known surface sticky ends can be used as scaffolds for nanoparticles arrangement. Here we assemble AuNPs which bear only one single-stranded DNA (in terms of DNA monofunctionalized AuNPs), because such monovalent AuNPs can preclude crosstalk during assembly and significantly improve the assembly accuracy. The synthesis of DNA monofunctionalized AuNPs was carried out by incubation 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 molar ratio of BSPP (bis(p-sulfonatophenyl)phenylphosphine dehydrate dipotassium salt) capped Au particles with lipoic acid modified DNA (lipoic-DNA) and purification of the desired adducts from agarose gels (Fig. 2) (the detailed protocol can also be found in the ref. 25). Since agarose gel electrophoresis-based separation of monofunctionalized AuNPs to two-, three- or even more DNA functionalized AuNPs requires a significantly long single-stranded DNA (usually >50 bases for 5 nm AuNPs) for a better resolution, lipoic-DNA (13 bases or 23 bases) used to hybridize on DNA dendrimers surface were obviously too short. To solve this problem, we hybridized a longer strand (EXT-Yn) as a helper to ‘extend’ the lipoic-DNA strand (LA-cYn).9 The resulting AuNP–DNA conjugates were then separated by gel electrophoresis as shown in Fig. 2a. After extracting DNA monofunctionalized nanoparticles from the corresponding band, EXT-Yn was removed upon hybridization to a fully complementary DNA strand (cEXT-Yn) (Fig. 2b) and the desired short lipoic-DNA modified AuNPs were obtained.
1 molar ratio of BSPP (bis(p-sulfonatophenyl)phenylphosphine dehydrate dipotassium salt) capped Au particles with lipoic acid modified DNA (lipoic-DNA) and purification of the desired adducts from agarose gels (Fig. 2) (the detailed protocol can also be found in the ref. 25). Since agarose gel electrophoresis-based separation of monofunctionalized AuNPs to two-, three- or even more DNA functionalized AuNPs requires a significantly long single-stranded DNA (usually >50 bases for 5 nm AuNPs) for a better resolution, lipoic-DNA (13 bases or 23 bases) used to hybridize on DNA dendrimers surface were obviously too short. To solve this problem, we hybridized a longer strand (EXT-Yn) as a helper to ‘extend’ the lipoic-DNA strand (LA-cYn).9 The resulting AuNP–DNA conjugates were then separated by gel electrophoresis as shown in Fig. 2a. After extracting DNA monofunctionalized nanoparticles from the corresponding band, EXT-Yn was removed upon hybridization to a fully complementary DNA strand (cEXT-Yn) (Fig. 2b) and the desired short lipoic-DNA modified AuNPs were obtained.
With DNA dendrimer scaffolds and DNA mono-modified AuNPs in hand, we proceeded to test the ability of these scaffolds to yield nanoparticle groupings with spatial control. When G0, which has three branched sticky ends, was incubated with certain amount of corresponding complementary DNA monofunctionalized AuNPs, Au trimers were obtained as transmission electron microscopy (TEM) images showed in Fig. 3a. Due to the conformational freedom of Y-DNA, triangles of gold nanoparticles have angular flexibility and not showed as perfect triangle-like arrangements. Likewise, well-defined gold particle hexamers could be obtained by the hybridization of G1 with six complementary gold monoconjugates (Fig. 3b), and hybridization of G2 and nanoparticle–DNA conjugates results in the synthesis of complexes containing 12-nanoparticle (Fig. 3c). To our knowledge, the precise control over the spatial arrangement of 12 discrete nanoparticles in such a straightforward manner has not been reported, although this strategy becomes less feasible as the generations of the dendrimers increase. As we found that, the desired assemblies were not formed by incubation of G3 and G4 with corresponding AuNP monoconjugates, respectively. TEM images revealed that 22 or less nanoparticles groupings were yielded (Fig. 3d, S6 and S7†). The most likely explanation is that the higher generation dendrimers, the more floppy the DNA structures are, and the more crowded sticky ends on surface. Steric effect and electrostatic repulsion may cause serious obstacle for site-specific assembly.
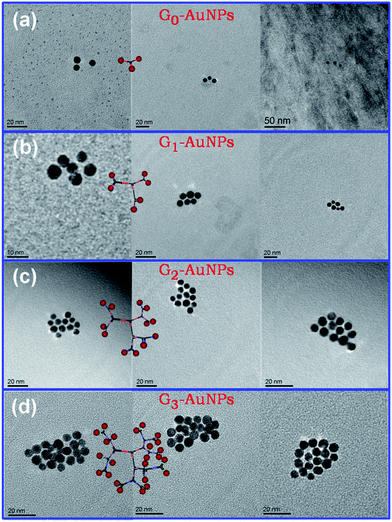 | ||
| Fig. 3 TEM images of DNA monofunctionalized AuNPs incubated with (a) G0, (b) G1, (c) G2 and (d) G3, respectively. | ||
To sum up, we reported the use of DNA dendrimers as scaffolds to direct the assembly of discrete gold nanoparticle groupings. DNA dendrimers can be constructed in a straightforward way with high yields. Different numbers of AuNPs clusters were formed according to TEM images. The capability of using DNA dendrimer-based assembly allowed one to use several components at same time and only the right component with right sequence can assemble at the specific position. The arrangement of nanoparticles on DNA dendrimers provides a new strategy for multiple components such as sliver nanoparticles,26 quantum dots,27 nanodiamonds28 etc. for nanophotonic and sensoring applications.
Acknowledgements
The authors appreciate financial support from the National Natural Science Foundation of China (No. 51403073).Notes and references
- M.-C. Daniel and D. Astruc, Chem. Rev., 2004, 104, 293–346 CrossRef CAS PubMed.
- Y. Ofir, B. Samanta and V. M. Rotello, Chem. Soc. Rev., 2008, 37, 1814–1825 RSC.
- S. Eustis and M. A. El-Sayed, Chem. Soc. Rev., 2006, 35, 209–217 RSC.
- S. K. Ghosh and T. Pal, Chem. Rev., 2007, 107, 4797–4862 CrossRef CAS PubMed.
- G. M. Whitesides and B. Grzybowski, Science, 2002, 295, 2418–2421 CrossRef CAS PubMed.
- C. A. Mirkin, R. L. Letsinger, R. C. Mucic and J. J. Storhoff, Nature, 1996, 382, 607–609 CrossRef CAS PubMed.
- A. P. Alivisatos, K. P. Johnsson, X. Peng, T. E. Wilson, C. J. Loweth, M. P. Bruchez and P. G. Schultz, Nature, 1996, 382, 609–611 CrossRef CAS PubMed.
- S. A. Claridge, S. L. Goh, J. M. J. Fréchet, S. C. Williams, C. M. Micheel and A. P. Alivisatos, Chem. Mater., 2005, 17, 1628–1635 CrossRef CAS.
- F. A. Aldaye and H. F. Sleiman, J. Am. Chem. Soc., 2007, 129, 4130–4131 CrossRef CAS PubMed.
- J. Sharma, R. Chhabra, C. S. Andersen, K. V. Gothelf, H. Yan and Y. Liu, J. Am. Chem. Soc., 2008, 130, 7820–7821 CrossRef CAS PubMed.
- S. J. Tan, M. J. Campolongo, D. Luo and W. Cheng, Nat. Nanotechnol., 2011, 6, 268–276 CrossRef CAS PubMed.
- L. H. Tan, H. Xing and Y. Lu, Acc. Chem. Res., 2014, 47, 1881–1890 CrossRef CAS PubMed.
- R. Schreiber, J. Do, E.-M. Roller, T. Zhang, V. J. Schuller, P. C. Nickels, J. Feldmann and T. Liedl, Nat. Nanotechnol., 2014, 9, 74–78 CrossRef CAS PubMed.
- A. Kuzyk, R. Schreiber, H. Zhang, A. O. Govorov, T. Liedl and N. Liu, Nat. Mater., 2014, 13, 862–866 CrossRef CAS PubMed.
- G. Yao, H. Pei, J. Li, Y. Zhao, D. Zhu, Y. Zhang, Y. Lin, Q. Huang and C. Fan, NPG Asia Mater., 2015, 7, e159, DOI:110.1038/am.2014.1131.
- T. G. W. Edwardson, K. L. Lau, D. Bousmail, C. J. Serpell and H. F. Sleiman, Nat. Chem., 2016, 8, 162–170 CAS.
- Y. Zhang, J. Chao, H. Liu, F. Wang, S. Su, B. Liu, L. Zhang, J. Shi, L. Wang, W. Huang, L. Wang and C. Fan, Angew. Chem., Int. Ed., 2016, 55, 8036–8040 CrossRef CAS PubMed.
- N. C. Seeman, Nature, 2003, 421, 427–431 CrossRef PubMed.
- D. Astruc, E. Boisselier and C. Ornelas, Chem. Rev., 2010, 110, 1857–1959 CrossRef CAS PubMed.
- Y. Li, Y. D. Tseng, S. Y. Kwon, L. d'Espaux, J. S. Bunch, P. L. McEuen and D. Luo, Nat. Mater., 2004, 3, 38–42 CrossRef CAS PubMed.
- T. Zhou, P. Chen, L. Niu, J. Jin, D. Liang, Z. Li, Z. Yang and D. Liu, Angew. Chem., Int. Ed., 2012, 51, 11271–11274 CrossRef CAS PubMed.
- H.-M. Meng, X. Zhang, Y. Lv, Z. Zhao, N.-N. Wang, T. Fu, H. Fan, H. Liang, L. Qiu, G. Zhu and W. Tan, ACS Nano, 2014, 8, 6171–6181 CrossRef CAS PubMed.
- S. Chatterjee, J. B. Lee, N. V. Valappil, D. Luo and V. M. Menon, Nanoscale, 2012, 4, 1568–1571 RSC.
- Y. Xing, E. Cheng, Y. Yang, P. Chen, T. Zhang, Y. Sun, Z. Yang and D. Liu, Adv. Mater., 2011, 23, 1117–1121 CrossRef CAS PubMed.
- T. Zhang, P. Chen, Y. Sun, Y. Xing, Y. Yang, Y. Dong, L. Xu, Z. Yang and D. Liu, Chem. Commun., 2011, 47, 5774–5776 RSC.
- D. Zhu, J. Chao, H. Pei, X. Zuo, Q. Huang, L. Wang, W. Huang and C. Fan, ACS Appl. Mater. Interfaces, 2015, 7, 11047–11052 CAS.
- G. Tikhomirov, S. Hoogland, P. E. Lee, A. Fischer, E. H. Sargent and S. O. Kelley, Nat. Nanotechnol., 2011, 6, 485–490 CrossRef CAS PubMed.
- T. Zhang, A. Neumann, J. Lindlau, Y. Wu, G. Pramanik, B. Naydenov, F. Jelezko, F. Schüder, S. Huber, M. Huber, F. Stehr, A. Högele, T. Weil and T. Liedl, J. Am. Chem. Soc., 2015, 137, 9776–9779 CrossRef CAS PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available: Materials and methods, DNA sequences, experimental details, melting curves, and additional TEM images. See DOI: 10.1039/c6ra16653a |
| ‡ Current address: PhD student at Fakultät für Physik-Lehrstuhl Prof. Rädler, Ludwig-Maximilians-Universität München, Munich 80539, Germany. |
| This journal is © The Royal Society of Chemistry 2016 |

