Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Cellular uptake studies of antisense oligonucleotides using G-quadruplex-nanostructures. The effect of cationic residue on the biophysical and biological properties†
Santiago
Grijalvo
 *ab,
Adele
Alagia
a,
Raimundo
Gargallo
c and
Ramon
Eritja
*ab
*ab,
Adele
Alagia
a,
Raimundo
Gargallo
c and
Ramon
Eritja
*ab
aInstitute of Advanced Chemistry of Catalonia (IQAC-CSIC), Jordi Girona 18-26, E-08034 Barcelona, Spain. E-mail: recgma@cid.csic.es; santiago.grijalvo@iqac.csic.es
bNetworking Centre on Bioengineering, Biomaterials and Nanomedicine (CIBER-BBN), Spain
cUniversity of Barcelona, Department of Chemical Engineering and Analytical Chemistry, Martí i Franquès 1-11, E-08028 Barcelona, Spain
First published on 4th August 2016
Abstract
Oligonucleotides carrying cationic peptides have been used to improve hybridization and cellular uptake of oligonucleotides. In this study, a series of four cationic amino acid derivatives were covalently linked at the 3′-termini with the objective of modifying the Tetrahymena telomeric repeat sequence d(TGGGGT). This led to the synthesis of a small cationic G-quadruplex-forming oligonucleotide series containing lysine, ornithine, homo-arginine and arginine moieties. The preparation of the appropriate solid-supports afforded the synthesis of 3′-lysine and 3′-ornithine oligonucleotides which were converted to homo-arginine and arginine conjugates through post-synthetic modifications. Circular dichroism and thermal denaturation experiments confirmed that the presence of the four cationic residues did not affect the formation of stable parallel G-quadruplex structures. Afterwards, a phosphorothioate oligonucleotide targeting Renilla luciferase mRNA was prepared at the 5′-termini of this telomeric sequence which afforded a series of phosphorothioate oligonucleotide/[d(TG4T)]4 hybrid constructs modified with lysine, ornithine, homo-arginine and arginine units after incubation. In addition, the phosphodiester/phosphorothioate mixed backbone contributed to the degradation of the G-quadruplex moiety by exonucleases liberating the antisense sequence. Cell culture analysis of gene expression showed that the formation of self-assembled G-quadruplex nanostructures did not disrupt the antisense mechanism and therefore were able to induce luciferase gene inhibition in mammalian cells without using cationic lipids. Flow cytometry analyses confirmed that fluorescently labelled antisense G4-quadruplex nanostructures were efficiently taken up by HeLa cells. These results suggest that G-quadruplex nanostructures may be used to improve cellular uptake of therapeutic oligonucleotides.
Introduction
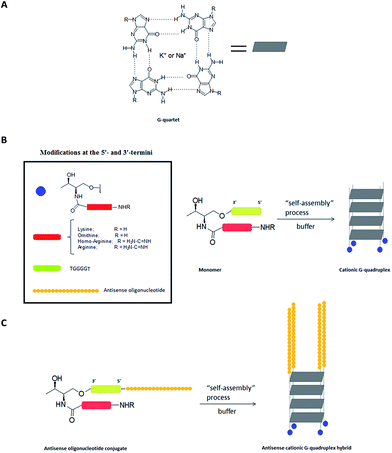
The G-quadruplex is a stable secondary structure that is formed in guanine-rich nucleic sequences. This structure is based on the π–π stacking of two or more planar arrangements of four guanine bases associated through Hoogsteen base-pairings. These structures are usually stabilized by monovalent cations that are placed in the central cavity of the G-quartets, forming cation–dipole interactions with eight guanines.1,2 This stabilization has been evaluated in the presence of several cations, with Na+ and K+ being the most studied.G-Quadruplex structures may show different topologies, depending on strand number, their orientation (parallel or anti-parallel), the oligonucleotide sequence, the guanine base conformations around the glycosidic bond (syn or anti) and others.3 G-Quadruplexes have received remarkable attention in areas ranging from supramolecular chemistry and nanobiotechnology to medicinal chemistry. Additionally, several examples can be found in the use of extended nanoscale G-wires,4 chip-based biosensors5 and DNA nanodevices.6
Hurley and Neidle described the use of G-quadruplex-binding small molecules to target and inhibit the enzyme telomerase in cancer cells for the first time.7 Since then, the biological application of G-quadruplexes has received a lot of attention in the search for novel cancer therapeutics8 and in the field of structure-based drug design.9 These studies have shown that planar aromatic ligands, molecules based on macrocyclic or helically folded oligomers, porphyrin-like units10,11 and some natural products like telomestatin displayed a clear preference for recognizing, binding and stabilizing G-quadruplexes.12–15
Besides their importance in gene expression regulation,16 some G-quadruplexes have been also described as potential anti-HIV drugs due to their capacity to interact with proteins present in HIV-1 infections.17 Based on this, extensive work has been carried out in the search of antiviral compounds with the synthesis of novel G-quadruplex-forming oligonucleotide derivatives. Examples can be found in the case of T30177, a 17-mer G-rich sequence that inhibits HIV replication cycles,18 PS-ISIS5320 (TTGGGGTT)19 and the Hotoda's 6-mer sequence d(TGGGAG).20 Further modifications and biophysical studies with carbohydrates,21 cholesterol22 and large aromatic groups at the 5′-termini of the Hotoda's sequence23 have helped understand the behavior of these modified G-quadruplexes and thus obtain series of conjugates with improved stabilities and inhibitory activities than unmodified sequences. Furthermore, it is worth mentioning that a good number of aptamers with biological activity can also contain G-quadruplex structures, indicating that G-quadruplex can be considered a privileged structure since it is able to interact and recognize proteins and more specifically, membrane proteins.24
The structure and kinetics of the tetramolecular G-quadruplex structure of the [d(TG4T)]4 have been well-studied and characterized.25 This Tetrahymena 6-mer telomeric repeat sequence only contains guanine (G) and thymine (T) bases which tend to form stable parallel G-quadruplexes with all guanine residue in the anti-conformation in the presence of Na+ and K+ cations in solution. The use of solid-phase synthesis,26 phosphoramidite chemistry27 or molecular self-assembly28 strategies have offered accessible and robust approaches for modifying this tetramer either at their ends (3′- or 5′-termini) or at internal positions by introducing modified guanine derivatives.29 Additionally, the effect of branched oligonucleotides,26,27 dendrimers with a wide range of functionalities28 and 8-aminoguanine derivatives29 are some examples that have been studied to evaluate the tetrameric structure stability, kinetics and mechanism of the [d(TG4T)]4 G-quadruplex formation.
Cationic peptides have been used to improve both biophysical and cellular uptake properties when combined with nucleic acids (e.g. antisense or siRNA oligonucleotides) through covalent or electrostatic approaches.30,31 For example, cell-penetrating peptides like penetratin, Tat peptide or other delivery systems containing arginine derivatives are representative examples recently reported in literature.32,33 However, there are few precedents in the literature to date in which G-quadruplexes have been combined or covalently linked with cationic peptides. One of the most representative studies was carried out by Chatterjee et al. who described the effect promoted by the human cathelicidin cationic peptide LL37 on the stabilization of telomeric G-quadruplex and its evaluation of the enzyme telomerase inhibition activity in cancer cells.34
In light of the important properties that cationic peptides can exert on nucleic acids at both biophysical and biologically levels, novel hybrid systems based on the linkage of cationic amino acids with G-quadruplexes offer not only new challenges from a synthetic point of view, but also valuable information about these systems from a biological point of view. There are a good number of cationic amino acids that have been used to enhance both cellular uptake processes and the distribution of nucleic acids. Some examples can be found in the case of lysine (Lys), ornithine (Orn) together with their guanidinium counterparts, which have been introduced in several cell-penetrating peptide sequences to facilitate cellular internalization and thus deliver-therapeutic nucleic acids to their respective targets.35–37
Taking into account the ability of short guanine-rich oligonucleotide to self-assemble and generate high-order structures, the presence of Lys, Orn, homo-arginine (HArg) and arginine (Arg) residue covalently linked at the 3′-termini of the telomeric repeat sequence d(TGGGGT) could lead to interesting cationic structures that would endow the G-quadruplex with additional properties not only on a biophysical level, but also on a biological level (Fig. 1A).
Toward this goal, we herein describe a synthetic strategy to introduce these four amino acids (Lys, Orn, HArg and Arg) together with biophysical characterization of the resulting cationic G-quadruplex-forming oligonucleotide conjugates. Additionally, the synthesis of a small series of a 17-mer antisense oligonucleotide (ASO)/[d(TG4T)]4 hybrids containing Lys, Orn, HArg, and Arg residue and their ability to silence Renilla luciferase using antisense technology in the absence of cationic lipids was also described (Fig. 1C). The results shown here open the possibility of using G-quadruplex nanostructures to enhance cell delivery of antisense oligonucleotide derivatives.
Experimental
General
All sensitive reactions to air were carried out under nitrogen atmosphere in flame-dried using dry solvents and were carried out under glassware with magnetic stirring. All solvents and other reagents were purchased from Sigma-Aldrich (St. Louis, Missouri, USA) and were used as supplied. All the standard phosphoramidites (A, C, G and T) together with ancillary reagents used for oligonucleotide synthesis were purchased from Link Technologies (Lanarkshire, Scotland). Analytical thin-layer chromatography (TLC) was performed using TLC sheets ALUGRAM SIL G/UV254, 0.20 mm. Purification of reaction crudes was carried out by flash chromatography using silica gel 60A (230–400 mesh). Visualization of the TLC sheets was accomplished with UV light and phosphomolybdic stain.1H (400 MHz) and 13C NMR (proton-decoupled 13C, 125 MHz) spectra were recorded on a Varian Inova 400 (400 MHz) spectrometer (RMN core facility, IQAC-CSIC). Chemical shifts were reported in ppm on the δ scale relative to either CDCl3 (δ = 7.26 for 1H NMR and δ = 77.0 for 13C NMR) or CD3OD (δ = 3.49 for 1H NMR and δ = 49.0 for 13C NMR) as internal references. Data are reported as follows: s = singlet, d = doublet, t = triplet, m = multiplet, br = broad and coupling constant(s) in Hz for the peaks integration.
HPLC analysis and separations were performed using a Waters 2695 Separations Module equipped with a Waters 2998 Photodiode Array Detector. The used HPLC solvents were: solvent (A) 5% ACN in 100 mM triethylammonium acetate (TEAA) (pH = 7) and solvent (B) 70% ACN in 100 mM TEAA (pH = 7).
Molecular absorption-monitored melting experiments were performed using a Jasco V-650 instrument equipped with a thermo-regulated cell holder. Circular Dichroism (CD) spectra were recorded on a JASCO J-810 equipped with a thermo-regulated cell holder, as well. Melting temperatures (Tm) were calculated from melting experiments as described elsewhere,38 using MATLAB routines (R2009b version; Math-Works, Natick, MA, USA).
Electrospray ionization mass spectra (ESI-MS) were recorded on a Micromass ZQ instrument with single quadrupole detector coupled to an HPLC, and high-resolution (HR) ESI-MS on an Agilent 1100 LC/MS-TOF instrument (Servei d'Espectrometría de Masses, Universitat de Barcelona). Matrix-assisted laser desorption ionization time-of-flight (MALDI-TOF) mass spectra were recorded on a Voyager-DE™RP spectrometer (Applied Biosystems) in negative mode (2,4,6-trihydroxyacetophenone matrix with ammonium citrate as an additive) (Servei d'Espectrometría de Masses, Universitat de Barcelona).
An antisense phosphorothioate oligonucleotide (sequence 5′-CGT TTC CTT TGT TCT GGA-3′) complementary to the mRNA of the Renilla luciferase gene which target to a predominant accessible site between 20 and 40 nt, was purchased from Sigma-Aldrich. A 18-mer scrambled antisense oligonucleotide (sequence 5′-CTG TCT GAC GTT CTT TGT-3′) was synthesized in-house and purified according to DMToff-based protocols. Lipofectamine 2000 was purchased from Invitrogen. Dulbecco's Modified Eagle's Medium (DMEM) was supplemented with 10% heat-inactive fetal serum bovine (FBS). DMEM, PBS buffer and distilled water (DNAse/RNAse free) were purchased from Gibco (Waltham, Massachusetts, USA). Nuclease-free water was prepared by using 0.1% of diethylpyrocarbonate (DEPC) to ensure the removal of RNAse contamination, autoclaved and filtered before using. Luciferase assay kits were purchased from Promega (Madison, Wisconsin, USA). Qiagen Giga plasmid purification kit was purchased from Qiagen. Luminescence was measured in a Promega Glomax Multidetection System instrument. Flow cytometer analyses were carried out in a Guava® easyCyte 8HT instrument (Millipore).
Protocol for G-quadruplex formation
G-Quadruplex complexes were formed by dissolving both lyophilized conjugates 3′-(7–10) and the unmodified oligonucleotide d(TG4T) in 10 mM lithium cacodylate buffer (pH 7.2) supplemented with 0.11 M NaCl. Also, phosphorothioate oligonucleotide conjugates Luc-(11–14) were dissolved in a buffer solution containing 100 mM PBS (pH 7.4). Both solutions were annealed by heating at 93 °C for 2 minutes and slowly cooling down to room temperature. The resulting oligonucleotide solutions (G4-7, G4-8, G4-9, G4-10, Luc-G4-11, Luc-G4-12, Luc-G4-13 and Luc-G4-14, respectively) and the unmodified tetramer [d(TG4T)]4 were stored at 4 °C for one week before recording CD spectra, melting profiles and carrying out transfection studies.CD spectroscopy and thermal denaturation experiments
CD spectra of the G-quadruplex conjugates [G4-(7–10)] and the unmodified G-quadruplex [d(TG4T)]4 were registered between 220 and 320 nm at 20 °C in the appropriate buffer. CD thermal denaturation experiments of G-quadruplex-forming oligonucleotides (G4-7, G4-8, G4-9 and G4-10) (5–10 μM) were performed in a range of temperature between 20 and 85 °C using a heating rate of 0.8 °C min−1 and monitoring the CD values at 263 nm.Electrophoretic mobility shift assay (EMSA)
Phosphorothioate conjugates Luc-(11–15) (0.5 μM) were analyzed by electrophoresis on 20% polyacrylamide gel under native conditions. 1× TBE were used as a running buffer. Aliquots were dissolved in a mixture containing 1× TBE and 50% glycerol (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) and samples were run at 150 V for at least 8 hours at 25 °C. Oligonucleotide bands were stained with SYBR green according to the manufacturer's instructions.
1) and samples were run at 150 V for at least 8 hours at 25 °C. Oligonucleotide bands were stained with SYBR green according to the manufacturer's instructions.
3′-Exonuclease digestions
MTT-based cytotoxicity assay
HeLa cells were regularly passaged to maintain exponential growth. Cells were seeded (5 × 103 cells per well) into each well of a 96-well cell culture plate and were incubated overnight at 37 °C and 5% CO2 in DMEM supplemented with 10% FBS without antibiotics. Cells were incubated for 8 hours with the oligonucleotide conjugate monomers Luc-(11–14) and Luc-G4-(11–14) with increasing concentrations (60, 120 and 300 nM) in 200 μL of DMEM (10% FBS). The unmodified tetramer Luc-G4-15 was used as a control. Growth medium was removed and cells were washed with PBS (200 μL). Fresh DMEM (10% FBS) was added (200 μL) and cells were incubated again for 12 hours at 37 °C. Then, MTT dye solution (25 μL; 5 mg mL−1) was added per well and cells were incubated for two additional hours. Medium was removed and DMSO (100 μL) was added to dissolve formazan crystals. The plate was read at 570 nm.In vitro antisense oligonucleotide conjugates transfection assay in the presence of Lipofectamine
HeLa cells were regularly passaged to maintain exponential growth. Cells were seeded (105 cells per well) into each well of a 24-well plate and were incubated overnight at 37 °C and 5% CO2 in growth medium (60% confluence). This medium was composed of DMEM supplemented with 10% FBS, 100 units per mL penicillin and 100 μg mL−1 streptomycin. Cells were transfected with a mixture containing luciferase plasmids and antisense oligonucleotides. Two luciferase plasmids – Renilla luciferase (pRL-TK) and Firefly luciferase (pGL4) – were used as reporter and control, respectively. Plasmids pGL4-control (1.0 μg) and pRL-TK (0.1 μg) together with antisense oligonucleotides Luc-(11–14) at 60 nM and Lipofectamine 2000 (1.3 μL) were formulated into liposomes and added to each well with a final volume of 600 μL. After 18 hours of incubation at 37 °C, medium was discharged and cells were thoroughly washed with PBS. The unmodified oligonucleotide (Luc) and the unmodified Luc-TG4T monomer (Luc-15) were used as controls at the same concentration. Transfections were performed in triplicate. Finally, lysates were analyzed for luciferase expression and Firefly and Renilla luciferase expression were measured according to manufacturer's protocol.In vitro G-quadruplex-forming antisense oligonucleotide conjugates transfection assay in the absence of Lipofectamine
HeLa cells were regularly passaged to maintain exponential growth. Cells were seeded (105 cells per well) into each well of a 24-well cell culture plate and were incubated overnight at 37 °C and 5% CO2 in growth medium (60% confluence). This medium was composed of DMEM supplemented with 10% FBS, 100 units per mL penicillin and 100 μg mL−1 streptomycin. Cells were transfected with Renilla luciferase (pRL-TK; 0.1 μg) and Firefly luciferase (pGL4; 1.0 μg) plasmids in combination with Lipofectamine 2000 in OptiMEM, according to manufacturer's protocol. Before transfection, DMEM was removed from the cells and wells were washed with PBS (500 μL). The mixtures were incubated 6 hours at 37 °C to allow the plasmid uptake by cells in DMEM supplemented with 10% FBS without antibiotics. The growth medium was removed from the cells that were thoroughly washed with PBS (2 × 400 μL). Finally, fresh DMEM supplemented with 10% FBS was added (500 μL). 100 μL of (Luc-G4-11, Luc-G4-12, Luc-G4-13 and Luc-G4-14) in 100 mM PBS with increasing concentrations (60 nM, 120 nM and 300 nM) were added with a final volume of 600 μL. The unmodified tetramer (Luc-G4-15) and the antisense oligonucleotide (Luc) were used as controls at the same concentrations. Transfections were performed in triplicate. Cells were allowed to grow for 24 hours at 37 °C. Medium was discharged and cells were washed with PBS (400 μL). Finally, 1× lysis buffer was added (15 min of stirring at room temperature) and the resultant lysates were analyzed for luciferase expression. Firefly and Renilla luciferase expression were measured according to manufacturer's protocol.Flow cytometry
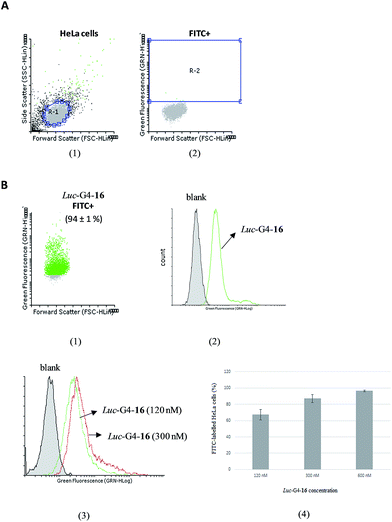
HeLa cells (105 cells per well) were seeded on 24-well cell culture plate following the same transfection procedure described before. Cells were analyzed after 24 hours incubation. Following fluorescently labelled Luc-G4-16 incubation, cells were washed with PBS (3 × 500 μL) and harvested with trypsin–EDTA (200 μL) at 37 °C for 5 minutes. DMEM (800 μL) was added and cells were centrifuged ((3.0 rcf × 3 min), 8 min). Cells were washed with PBS again (3 × 500 μL) and the anticipated pellets were analyzed by a flow cytometer instrument. For each sample, 104 events were collected in a selected gate (R1) that corresponds to the HeLa cell population. Flowing Software 2.5.1 (University of Turku, Finland) was used to analyze the relationship between fluorescently labelled and untreated cell populations.Statistical analysis
All data were shown as mean ± standard deviation (SD) and are the result of an average of three to six replicates. Statistical differences (*p < 0.05, and **p < 0.01) were determined using a regular two-way ANOVA analysis of variance combined with Bonferroni post-test which was used for multiple comparisons.Results and discussion
Synthesis of cationic G-quadruplex-forming oligonucleotide conjugates containing lysine, ornithine, homo-arginine and arginine
A short guanine-rich oligonucleotide of sequence d(TGGGGT) was covalently modified with four types of cationic residue which were incorporated at the 3′-termini. We used threoninol as a starting material to introduce modifications because of the presence of two alcohol groups with different reactivity among each other. This selectivity makes threoninol compatible with solid-phase chemistries, in particular with DNA and RNA syntheses.39–41 Furthermore, the presence of an amino group in the threoninol molecule provides an additional degree of diversity besides the possibility of reacting with other functional electrophile groups like alkyl halides, aldehydes, ketones or carboxylic acids among others. This high degree of versatility allows for DNA and RNA molecules to be achieved with additional modifications and therefore endow them with an increased complexity.42Lys and Orn are two amino acids that have been used for modifying nucleic acids43–45 and have provided important properties to oligonucleotides in terms of stability and cell entry.46,47 The synthesis of oligonucleotide conjugates containing Lys and Orn is displayed in Scheme 1. We decided that the fluorenylmethyloxycarbonyl group (Fmoc) was best suited for protecting the site chain of Lys and Orn. The activation of the (Fmoc)2Lys-OH and (Fmoc)2Orn-OH carboxylic acids with N-hydroxysuccinimide allowed for their selective incorporation into the threoninol molecule affording the lysine (3) and ornithine (4) derivatives with good yields (67% and 87%, respectively). The selective protection of the primary alcohol was carried out using dimethoxytrityl chloride (DMTrCl) which generated the formation of the DMTr-threoninol derivatives (5) and (6) with good to moderate yields (92% and 42%, respectively). Finally, the functionalization of the secondary alcohol with succinic anhydride afforded the corresponding carboxylic acids which were rapidly incorporated into a Controlled-Pore Glass (CPG) solid support yielding the modified resins CPG-1 (L = 31.2 μmol g−1) and CPG-2 (20.7 μmol g−1).
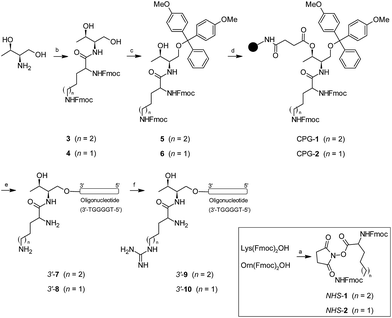 | ||
| Scheme 1 Synthesis of oligonucleotide conjugates containing lysine (Lys), ornithine (Orn), homo-arginine (HArg) and arginine (Arg) and the telomeric sequence d(TGGGGT) as a model at the 3′-termini. | ||
The assembly of oligonucleotide conjugates 3′-(7) and 3′-(8) containing the telomeric sequence d(TGGGGT) as a model was obtained using an automated DNA synthesizer and standard phosphoramidite chemistry as well as DMToff-based protocols. We purified detached oligonucleotide conjugates 3′-(7) and 3′-(8) by using a semi-preparative HPLC and verified it using MALDI-TOF mass spectrometry (Table 1). The final step was the incorporation of the guanidinium moiety according to post-synthetic approaches by adding a O-methylisourea solution.47,48 Although conjugates 3′-(7) and 3′-(8) have two amino groups susceptible to modification, we only observed the mono-guanidinylated conjugates containing homo-arginine 3′-(9) and arginine 3′-(10) cationic residue, respectively. After desalting, the aforementioned oligonucleotide conjugates were isolated without further purification, analyzed by analytical HPLC and finally confirmed by using MALDI-TOF mass spectrometry (Table 1). For further experimental details regarding synthesis of both threoninol derivatives and oligonucleotide conjugates modified at 3′-termini, see ESI.†
| Sequencea | Backboneb | 3′-Modification | 5′-Modification | MS [calcd] | MS [found] | T 1/2 c (°C) K+ | T 1/2 d (°C) Na+ | ΔT (°C) |
|---|---|---|---|---|---|---|---|---|
a TG4T = G4 = d(5′-![[T with combining low line]](https://www.rsc.org/images/entities/char_0054_0332.gif) ![[G with combining low line]](https://www.rsc.org/images/entities/char_0047_0332.gif) ![[G with combining low line]](https://www.rsc.org/images/entities/char_0047_0332.gif) ![[G with combining low line]](https://www.rsc.org/images/entities/char_0047_0332.gif) ![[G with combining low line]](https://www.rsc.org/images/entities/char_0047_0332.gif) ![[T with combining low line]](https://www.rsc.org/images/entities/char_0054_0332.gif) -3′); Luc: d(5′- -3′); Luc: d(5′-![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) ![[G with combining low line]](https://www.rsc.org/images/entities/char_0047_0332.gif) ![[T with combining low line]](https://www.rsc.org/images/entities/char_0054_0332.gif) ![[T with combining low line]](https://www.rsc.org/images/entities/char_0054_0332.gif) ![[T with combining low line]](https://www.rsc.org/images/entities/char_0054_0332.gif) ![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) ![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) ![[T with combining low line]](https://www.rsc.org/images/entities/char_0054_0332.gif) ![[T with combining low line]](https://www.rsc.org/images/entities/char_0054_0332.gif) ![[T with combining low line]](https://www.rsc.org/images/entities/char_0054_0332.gif) ![[G with combining low line]](https://www.rsc.org/images/entities/char_0047_0332.gif) ![[T with combining low line]](https://www.rsc.org/images/entities/char_0054_0332.gif) ![[T with combining low line]](https://www.rsc.org/images/entities/char_0054_0332.gif) ![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) ![[T with combining low line]](https://www.rsc.org/images/entities/char_0054_0332.gif) ![[G with combining low line]](https://www.rsc.org/images/entities/char_0047_0332.gif) ![[G with combining low line]](https://www.rsc.org/images/entities/char_0047_0332.gif) ![[A with combining low line]](https://www.rsc.org/images/entities/char_0041_0332.gif) TGGGGT-3′); scrambled = Scr: d(5′- TGGGGT-3′); scrambled = Scr: d(5′-![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) ![[T with combining low line]](https://www.rsc.org/images/entities/char_0054_0332.gif) ![[G with combining low line]](https://www.rsc.org/images/entities/char_0047_0332.gif) ![[T with combining low line]](https://www.rsc.org/images/entities/char_0054_0332.gif) ![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) ![[T with combining low line]](https://www.rsc.org/images/entities/char_0054_0332.gif) ![[G with combining low line]](https://www.rsc.org/images/entities/char_0047_0332.gif) ![[A with combining low line]](https://www.rsc.org/images/entities/char_0041_0332.gif) ![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) ![[G with combining low line]](https://www.rsc.org/images/entities/char_0047_0332.gif) ![[T with combining low line]](https://www.rsc.org/images/entities/char_0054_0332.gif) ![[T with combining low line]](https://www.rsc.org/images/entities/char_0054_0332.gif) ![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) ![[T with combining low line]](https://www.rsc.org/images/entities/char_0054_0332.gif) ![[T with combining low line]](https://www.rsc.org/images/entities/char_0054_0332.gif) ![[T with combining low line]](https://www.rsc.org/images/entities/char_0054_0332.gif) ![[G with combining low line]](https://www.rsc.org/images/entities/char_0047_0332.gif) ![[T with combining low line]](https://www.rsc.org/images/entities/char_0054_0332.gif) -3′).
b PO: phosphodiester; PS: phosphorothioate (underlined).
c (Non-equilibrium) melting temperature of the preformed G-quadruplex in 10 mM potassium phosphate buffer, 200 mM KCl, 0.1 mM EDTA (pH 7.0) with a temperature gradient of 0.8 °C min−1. Uncertainty: ±0.5 °C.
d (Non-equilibrium) melting temperature of the preformed G-quadruplex in 10 mM lithium cacodylate buffer with 0.11 M NaCl (pH 7.2) with a temperature gradient of 0.8 °C min−1.
e ΔT: difference between guanosine-rich oligonucleotide conjugate T1/2 and unmodified guanosine-rich oligonucleotide T1/2.
f [M + Na + K]+. To confirm the purity of the conjugate, a polyacrylamide gel electrophoresis (PAGE) was carried out (see ESI for further details).
g The [GGGGT-Luc]+ fragment is observed. To confirm the purity of the conjugate, a polyacrylamide gel electrophoresis (PAGE) was carried out (see ESI for further details).
h nd: not determined. -3′).
b PO: phosphodiester; PS: phosphorothioate (underlined).
c (Non-equilibrium) melting temperature of the preformed G-quadruplex in 10 mM potassium phosphate buffer, 200 mM KCl, 0.1 mM EDTA (pH 7.0) with a temperature gradient of 0.8 °C min−1. Uncertainty: ±0.5 °C.
d (Non-equilibrium) melting temperature of the preformed G-quadruplex in 10 mM lithium cacodylate buffer with 0.11 M NaCl (pH 7.2) with a temperature gradient of 0.8 °C min−1.
e ΔT: difference between guanosine-rich oligonucleotide conjugate T1/2 and unmodified guanosine-rich oligonucleotide T1/2.
f [M + Na + K]+. To confirm the purity of the conjugate, a polyacrylamide gel electrophoresis (PAGE) was carried out (see ESI for further details).
g The [GGGGT-Luc]+ fragment is observed. To confirm the purity of the conjugate, a polyacrylamide gel electrophoresis (PAGE) was carried out (see ESI for further details).
h nd: not determined.
|
||||||||
| G4-7 | PO | Lysine | None | 2158 | 2153 | >80 | 62.2 | 3.0 |
| G4-8 | PO | Ornithine | None | 2144 | 2138 | >80 | 65.6 | 6.4 |
| G4-9 | PO | Homoarginine | None | 2201 | 2195 | >80 | 64.8 | 5.6 |
| G4-10 | PO | Arginine | None | 2186 | 2180 | >80 | 63.0 | 3.8 |
| Luc-11 | PO/PS | Lysine | None | 7968 | 7963 | nd | nd | — |
| Luc-12 | PO/PS | Ornithine | None | 7954 | 7959 | nd | nd | — |
| Luc-13 | PO/PS | Homoarginine | None | 8010 | 8009 | nd | nd | — |
| Luc-14 | PO/PS | Arginine | None | 7996 | 8053f | nd | nd | — |
| Luc-15 | PO/PS | None | None | 7672 | 7365g | nd | nd | — |
| Luc-16 | PO/PS | None | Fluorescein | 8226 | 8216 | nd | nd | — |
| TG4T | PO | None | None | 1862 | 1857 | >80 | 59.2 | 0 |
| Scr | PS | None | None | nd | nd | nd | nd | — |
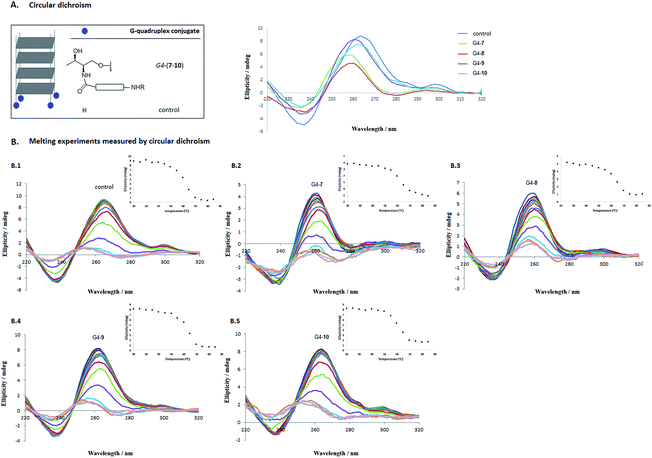
With oligonucleotide conjugates 3′-(7–10) and the unmodified d(TGGGGT) oligonucleotide in our hands, self-assembling processes took place when subjected to the presence of either a 10 mM potassium phosphate buffer, 200 mM KCl, 0.1 mM EDTA (pH 7.0) or a 10 mM lithium cacodylate buffer supplemented with 0.11 M NaCl (pH 7.2). Circular dichroism confirmed the presence of the parallel-stranded tetramolecular G-quadruplex structures at 20 °C for the conjugates (G4-7, G4-8, G4-9 and G4-10, respectively) displaying a typical positive band at 264 nm and a negative band around 240 nm. Fig. 2A shows the circular dichroism spectra for G4-(7–10) conjugates together with the unmodified tetramer [d(TG4T)]4 in cacodylate buffer.
CD melting analysis
The effect of cationic modifications on the G-quadruplex stabilization G4-(7–10) compared to the unmodified [d(TG4T)]4 tetramer was studied by thermal denaturation using CD spectroscopy. Two buffers were considered. In the presence of 10 mM potassium phosphate buffer, all G-quadruplex sequences were stable at temperatures up to 80 °C (Table 1). However, when potassium ions were replaced by sodium (10 mM cacodylate buffer with 0.11 M NaCl) the melting of the quadruplexes G4-(7–10) took place, as we had expected. These results agree with the literature25 due to the preference of G-quadruplex central cavities for potassium over sodium ions.49As displayed on Fig. 2B.1–B.5, as the temperature increased, the positive and the negative bands at 264 and 240 nm reduced in magnitude and shifted to lower wavelengths. Additionally, the apparent melting temperature (T1/2) of the cationic G4-quadruplexes G4-(7–10) was also determined considering these tetrameric G-quadruplex structures were not in thermodynamic equilibrium during the unfolding process.25,50 Interestingly, the introduction of the cationic residue (Lys, Orn, HArg and Arg) clearly stabilized the G-quadruplex structure (62.2, 65.6, 64.8 and 63.0 °C for G4-7, G4-8, G4-9 and G4-10, respectively) compared to the apparent melting temperature of the unmodified [d(TG4T)]4 tetramer (control) (59.2 °C) (see Table 1 and ESI† for further details).48,51
In vitro transfection studies
After confirming that these cationic moieties introduced at the ends of the tetrameric sequence were able to stabilize the G-quadruplex structure, we decided to move forward and study new hybrid constructs. In addition to having the same telomeric unit previously described, they also contained additional oligonucleotide sequences with biological relevance in their structures.We selected a phosphorothioate oligonucleotide of sequence d(5′-CGT TTC CTT TGT TCT GGA-3′)52 which was conveniently assembled at the 5′-termini of the telomeric sequence in order to obtain the mixed phosphodiester/phosphorothioate oligonucleotides Luc-(11–14) containing lysine (Luc-11), ornithine (Luc-12), homo-arginine (Luc-13) and arginine (Luc-14) conjugates, respectively. We also prepared the phosphodiester/phosphorothioate oligonucleotide Luc-TG4T (Luc-15) not containing a cationic amino acid residue as a control. According to DMTon-based protocols, all conjugates were purified by semi-preparative HPLC except for Luc-15, which was obtained directly without further purification (DMToff protocol). All conjugates were characterized by MALDI-TOF mass spectrometry and verified using native PAGE gel (Table 1 and ESI, Fig. S4†). Afterwards, G-quadruplex-forming oligonucleotide hybrids Luc-G4-(11–14) along with the unmodified tetramer (Luc-G4-15) were formed in 100 mM PBS buffer (pH 7.2) according to the same methodology previously described (CD spectra of Luc-G4-11, Luc-G4-12 and Luc-G4-15 are displayed in the ESI, Fig. S5† as representative examples).
Before starting the transfection experiments, the cytotoxicity of the phosphorothioate monomers [Luc-(11–14)] and G-quadruplex constructs [Luc-G4-(11–14)] were first carried out by examining the cell viability for HeLa cells using the MTT colorimetric assay (Fig. S6†).53 As displayed on Fig. S6A (see ESI†), none of phosphorothioate [Luc-(11–14)] antisense monomers tested nor cationic G-quadruplex conjugates [Luc-G4-(11–14)] were detrimental in HeLa cell viability compared to the untreated cells (blank) at several concentrations (60, 120 and 300 nM). This indicates the lack of toxicity of both amine and guanidine derivatives (Fig. S6B†).
Degradation by exonucleases
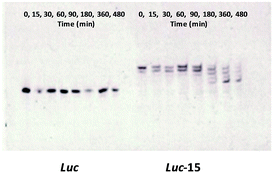
An important issue related to the cellular delivery of therapeutic oligonucleotides is lysosomal escape in order to reach the target mRNA. For this reason, we considered keeping the d(TGGGGT) oligonucleotide sequence in its phosphodiester form, so lysosomal degradation of the antisense sequence would help liberate the antisense oligonucleotide sequence intact due to the presence of the protected phosphorothioate bonds.The liberation of the antisense Luc oligonucleotide from the TG4T building block was studied using degradation experiments. Stability experiments of the phosphorothioate conjugate (Luc-15) in the presence of snake venom phosphodiesterase revealed phosphate bond degradation within a short period of time starting from the 3′-termini until reaching the first phosphorothioate bond which remained constant during the degradation experiment (Fig. 3). As a consequence, this liberation from the d(TGGGGT) oligonucleotide would allow the Luc antisense oligonucleotide to reach the mRNA molecule and then block the luciferase protein. Taking into account our PAGE denaturizing gels, we noticed that Luc oligonucleotide (Fig. 3-left, control) migrated in a different way than the phosphorothioate oligonucleotide liberated from Luc-15 (Fig. 3-right). This effect could be attributed to the presence of the first phosphorothioate bond that joins the 5′-termini of d(3′-TGGGGT-5′) and the antisense Luc residue together. Consequently, the degradation process produced by snake venom phosphodiesterase would generate the liberation of T-Luc residue instead of the Luc oligonucleotide alone.
Transfection experiments in the presence of Lipofectamine
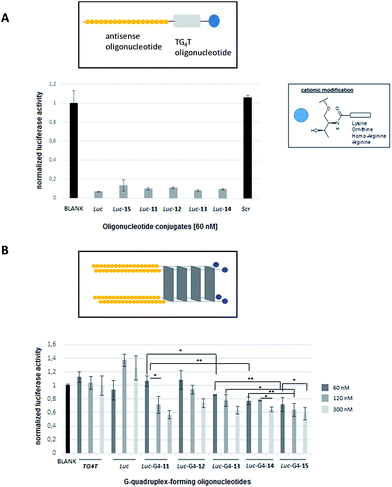
The efficiency of phosphorothioate conjugates [Luc-(11–15)] to knockdown Renilla luciferase mRNA was evaluated in the presence of Lipofectamine 2000 at 60 nM. Fig. 4A shows the inhibition activities generated by Luc-(11–15) conjugates taking into account the normalized ratio between the reporter Renilla and the control (Firefly) luciferase genes, respectively.As displayed on Fig. 4A, the inhibition profiles of antisense oligonucleotide conjugates showed comparable silencing activities to the unmodified phosphorothioate (Luc) oligonucleotide which was used as a positive control (around 90% inhibition). Additionally, a scrambled sequence (Scr) was also tested indicating the selectivity of the transfection experiment. This result suggested that the presence of both cationic residue – lysine, ornithine, homo-arginine and arginine – along with the additional guanine-rich oligonucleotide did not have a negative effect on the antisense activity and indicated that such modifications did not disrupt the antisense mechanism.
Transfection experiments in the absence of Lipofectamine
The ability of the preformed cationic G-quadruplex conjugates [Luc-G4-(11–15)] to silence Renilla luciferase mRNA was also evaluated in the absence of commercially available cationic lipids (Fig. 4B). Three concentrations of G-quadruplex-forming oligonucleotide conjugates were tested (60, 120 and 300 nM). As displayed on Fig. 4B, all antisense G-quadruplex hybrid conjugates were able to inhibit Renilla luciferase in a dose–response manner thereby affording moderate inhibitory activities in the case of G-quadruplex conjugates at 300 nM (around 40% inhibition).These results showed us the presence of the guanidinium group at the ends of conjugates (Luc-G4-13 and Luc-G4-14) slightly improved the efficiency in luciferase inhibition at 60 nM (14% and 23%, respectively) compared to the G-quadruplex conjugates containing amine groups (Luc-G4-11 and Luc-G4-12) which did not show any evidence in the transfection process at the same concentration (*p < 0.05 in the case of Luc-G4-11versus Luc-G4-13 and **p < 0.01 for Luc-G4-11versus Luc-G4-14 at 60 nM).
Antisense G-quadruplex conjugates (Luc-G4-11, Luc-G4-13 and Luc-G4-14) displayed similar silencing at both 120 nM (29%, 23% and 23%) and 300 nM (43%, 37% and 35%) respectively, as the G-quadruplex construct concentration was increased. These results represented a significant reduction in the Renilla luciferase when the Luc-G4-11 concentration (from 60 to 120 nM; *p < 0.05) and Luc-G4-14 (from 120 to 300 nM; *p < 0.05) were increased. These inhibition results, though modest, showed that both cationic modifications and G4-quadruplex nanostructures did not have a negative impact on the antisense activity, as confirmed by the transfection assay carried out with Lipofectamine. Curiously, the unmodified tetramer (Luc-G4-15) was also able to efficiently silence Renilla luciferase mRNA with comparable activities just like modified G-quadruplex conjugates at the three concentrations tested (28%, 35% and 41% for 60, 120 and 300 nM, respectively). This dose–response profile significantly demonstrated an inhibition potency at high concentration (300 nM) when compared to silencing activities at 60 nM (*p < 0.05). The antisense Luc-G4-15 construct also displayed significant luciferase inhibition values in comparison with Luc-G4-13 at 60 and 120 nM (**p < 0.01 and *p < 0.05, respectively) and Luc-G4-14 at 120 nM (**p < 0.01).
To confirm that the antisense effect promoted by the cationic G4-quadruplex conjugates was only gene-specific, a d[(TG4T)]4 construct not containing the antisense oligonucleotide (Luc) was also tested. As expected, the G-quadruplex alone did not show either luciferase inhibition activity or anti-proliferative activity on HeLa cervical carcinoma cells. The lack of the d[(TG4T)]4 cytotoxicity in HeLa cells is in accordance with the results obtained by Dapic et al.54 It has recently been described that the anti-proliferative activity of several guanine-rich oligonucleotides like AS1411 among others may come from the metabolites generation of the enzymatic digestion G-quadruplexes such as G monophosphate.55 However, the concentration used in these experiments was quite high compared to the concentrations used in this study on antisense therapy.
While the presence of the lysine protonable ε-amino group and the positively charged guanidine groups may have a positive impact on transfection due to the presence of cationic residue,56–58 additional factors could be also hypothesized in accordance with the results obtained in Fig. 4B. Alternatively, the ability of the antisense G-quadruplex constructs (Luc-G4-11, Luc-G4-13, Luc-G4-14 and Luc-G4-15) to silence Renilla luciferase gene was studied with serum-free medium conditions.59 As displayed in Fig. S7 (see ESI†), inhibition was practically abolished at 120 nM in serum-free media. These preliminary results suggest that transfection of G-quadruplex structures could be mediated by factors present either in serum60 or in the cell membrane.
Interestingly, some studies have suggested nucleolin (NCL)-mediated endocytosis as the main cellular uptake mechanism for G-quadruplex structures.61 NCL is a phosphoprotein highly expressed at the proliferating surface cells.62 According to Maizels findings,63 NCL is able to bind G–G paired structures as natural binding targets with a high affinity and has become an important target for anti-cancer therapies among others.64,65 It might be conceivable that the presence of electrostatic interactions between cationic Luc-G4-(11–14) conjugates and the anionic NCL at the cell surface could contribute to better G-quadruplex absorption into cells.66 Interestingly, this enhanced efficiency has been observed with G-quadruplexes modified with positively lysine-rich (Luc-G4-11), homo-arginine (Luc-G4-13) and arginine (Luc-G4-14) cationic residue. However, we cannot assert in a peremptory fashion that NCL is the only mechanism involved in the G-quadruplex cellular uptake since other mechanisms like micropinocytosis have been also proposed for the internalization of these kinds of nanostructures into cells.55,67 To solve these inconclusive results, further studies would need to be carried out in order to have a deeper understanding of the cellular uptake mechanism as well as its gene silencing activities.
To evaluate the cellular uptake, flow cytometry was used to study the amount of fluorescently labelled cell populations by analyzing changes in fluorescence between both untreated and treated cells when transfected with FITC-Luc-G4-16 in the absence of Lipofectamine (Luc-G4-16) after 24 hours of incubation at several concentrations (120, 300 and 600 nM) (Fig. 5). We first analyzed fluorescently labelled and unlabelled cell populations by selecting a normal R1 gate for HeLa cells based on their light scattering properties (Fig. 5A). An increase in the G-quadruplex conjugate (Luc-G4-16) concentration up to 600 nM produced a gradual internalization of the nanostructure inside cells, as illustrated in Fig. 5B. This produced a greater number of transfected cells containing the FITC dye and displayed a remarkable histogram shift in comparison with untreated cell populations (Fig. 5B.2). This effect was clearly visible inside HeLa cells after 24 hours of incubation (Fig. S8†). Finally, a relationship between G-quadruplex concentration and the amount of fluorescently labelled cells was observed (67%, 86% and 96% for 120, 300 and 600 nM, respectively) (Fig. 5B.3 and B.4).
As control experiments, the fluorescently labelled Luc-G4-16 was formulated into liposomes by adding Lipofectamine 2000 in order to transfect HeLa cells (Fig. S9†). As displayed in Fig. S9A,† a clear distinction between treated and untreated cell populations with regard to the size was observed which afforded more than 90% of the cells labelled with FITC dye.
A similar amount of transfected cells was obtained when the fluorescently labelled monomer (Luc-16) was used at the same concentration (60 nM) with Lipofectamine 2000. This transfection efficiency mediated by the G-quadruplex nanostructure (Luc-G4-16) and the antisense oligonucleotide (Luc-16) produced comparable shifts in the histogram for fluorescently labelled cell populations with FITC dye (Fig. S9B†).
Conclusions
A series of oligonucleotides having a telomeric G-quadruplex forming sequence modified with amino and guanidinium residue at the 3′-end were synthesized using solid-phase and post-synthetic protocols and threoninol linker and Fmoc-amino acids. The presence of the amino and guanidinium groups did not alter the formation of a parallel G-quadruplex and stabilized the anticipated nanostructures compared to the unmodified oligonucleotide tetramer. Additionally, a phosphorothioate antisense sequence was introduced at the 5′-end of the telomeric sequence containing the same cationic residue which resulted in the formation of the expected cationic antisense G-quadruplex hybrid constructs. The ability of G-rich antisense oligonucleotides to hybridize with a complementary RNA sequence had previously been shown from PAGE analysis by Wolfe.68 Luciferase gene inhibition studies in the presence of Lipofectamine clearly showed that both cationic modifications and the additional TGGGGT residue did not disrupt the antisense mechanism and obtained similar inhibition activities like the unmodified antisense oligonucleotide (Luc). Furthermore, the efficiency of these antisense G-quadruplex constructs for inhibiting the Renilla luciferase mRNA was also evaluated in the absence of Lipofectamine. The results showed moderate silencing activities for the G-quadruplex structures modified with lysine (Luc-G4-11), homo-arginine (Luc-G4-13) and arginine (Luc-G4-14) residue in the presence of 10% serum. However, in the absence of serum, the gene expression inhibition was practically abolished. Although there is little mechanistic understanding of G-quadruplex-mediated delivery, these results suggest that NCL might mediate gene transfection as well as act as a cell surface receptor59 and thus facilitate the cellular internalization of these kinds of nanostructures. However, the effect of the serum content and alternative cellular uptake mechanisms should also be taken into account. Finally, the transfection efficiency of the G-quadruplex-forming oligonucleotide (Luc-G4-16) was analyzed using flow cytometry with several concentrations. This promoted a shifted histogram and clearly distinguished fluorescently labelled cell populations with FITC dye and non-treated cells. Further insight into cellular uptake mechanisms, cell trafficking as well as introducing alternative labile linkers for improving lysosomal release may help to understand the proper role played by these G-quadruplex nanostructures in gene delivery.Acknowledgements
This work was supported by the Ministerio de Economía y Competitividad (CTQ2014-52588-R and CTQ2012-38616-C02-02 grants) and the Generalitat de Catalunya (2014-SGR-1106 and 2014-SGR-187 grants). CIBER-BBN is an initiative funded by the VI National R&D&I Plan 2008–2011, Iniciativa Ingenio 2010, Consolider Program, CIBER Actions and financed by the Instituto de Salud Carlos III with assistance from the European Regional Development Fund. We would like to thank Dr Gemma Fabriàs and Mireia Casasampere for their assistance and access to the flow cytometer instrument and Dr Cristina Garrido for her help and advice on cytometer data processing.Notes and references
- D. J. Patel, A. T. Phan and V. Kuryavyi, Nucleic Acids Res., 2007, 35, 7429–7455 CrossRef CAS PubMed.
- S. Balasubramanian, L. H. Hurley and S. Neidle, Nat. Rev. Drug Discovery, 2011, 10, 261–275 CrossRef CAS PubMed.
- J. L. Hupper, Chem. Soc. Rev., 2008, 37, 1375–1384 RSC.
- T. Ilc, P. Sket, J. Plavec, M. Webba da Silva, I. Drevensek-Olenik and L. Spindler, J. Phys. Chem. C, 2013, 117, 23208–23215 CAS.
- C. Teller and I. Willner, Curr. Opin. Biotechnol., 2010, 21, 376–391 CrossRef CAS PubMed.
- L. A. Yatsunyk, O. Mendoza and J.-L. Mergny, Acc. Chem. Res., 2014, 47, 1836–1844 CrossRef CAS PubMed.
- D. Sun, B. Thompson, B. E. Cathers, M. Salazar, S. M. Kerwin, J. O. Trent, T. C. Jenkins, S. Neidle and L. H. Hurley, J. Med. Chem., 1997, 40, 2113–2116 CrossRef CAS PubMed.
- S. Balasubramanian and S. Neidle, Curr. Opin. Biotechnol., 2009, 13, 345–353 CrossRef CAS PubMed.
- A. R. O. Cousins, D. Ritson, P. Sharma, M. F. G. Stevens, J. E. Moses and M. S. Searle, Chem. Commun., 2014, 50, 15202–15205 RSC.
- M. Franceschin, D. Nocioni, A. Biroccio, E. Micheli, S. Cacchione, C. Cingolani, A. Venditti, P. Zizza, A. Bianco and A. Altieri, Org. Biomol. Chem., 2014, 12, 9572–9582 CAS.
- X.-X. Huang, L.-N. Zhu, B. Wu, Y.-F. Huo, N.-N. Duan and D.-M. Kong, Nucleic Acids Res., 2014, 42, 8719–8731 CrossRef CAS PubMed.
- L.-N. Zhu, B. Wu and D.-M. Kong, Nucleic Acids Res., 2013, 41, 4324–4335 CrossRef CAS PubMed.
- S. Müller, K. Laxmi-Reddy, P. V. Jena, B. Baptiste, Z. Dong, F. Godde, T. Ha, R. Rodríguez, S. Balasubramanian and I. Huc, ChemBioChem, 2014, 15, 2563–2570 CrossRef PubMed.
- A. Laguerre, N. Desbois, L. Stefan, P. Richard, C. P. Gros and D. Monchaud, ChemMedChem, 2014, 9, 2035–2039 CrossRef CAS PubMed.
- T. Tauchi, K. Shin-ya, G. Sashida, M. Sumi, S. Okabe, J. H. Ohyashiki and K. Ohyashiki, Oncogene, 2006, 25, 5719–5725 CrossRef CAS PubMed.
- M. Wieland and J. S. Hartig, Chem. Biol., 2007, 14, 757–763 CrossRef CAS PubMed.
- M. Métifiot, S. Amrane, S. Litvak and M.-L. Andreola, Nucleic Acids Res., 2014, 42, 12352–12366 CrossRef PubMed.
- V. T. Mukundan, N. Q. Do and A. T. Phan, Nucleic Acids Res., 2011, 39, 8984–8991 CrossRef CAS PubMed.
- J. R. Wyatt, T. A. Vicker, J. L. Roberson, J. R. W. Buckheit, T. Klimkait, E. DeBaets, P. W. Davis, B. Rayner, J. L. Imbach and D. J. Ecker, Proc. Natl. Acad. Sci. U. S. A., 1994, 91, 1356–1360 CrossRef CAS.
- H. Hotoda, M. Koizumi, R. Koga, M. Kaneko, K. Momota, T. Ohmine, H. Furukawa, T. Agatsuma, T. Nishigaki and J. Sone, J. Med. Chem., 1998, 41, 3655–3663 CrossRef CAS PubMed.
- J. D'Onofrio, L. Petraccone, L. Martino, G. Di Fabio, A. Iadonisi, J. Balzarini, C. Giancola and D. Montesarchio, Bioconjugate Chem., 2008, 19, 607–616 CrossRef PubMed.
- D. Musumeci and D. Montesarchio, Molecules, 2012, 17, 12378–12392 CrossRef CAS PubMed.
- G. Di Fabio, J. D'Onofrio, M. Chiapparelli, B. Hoorelbeke, D. Montesarchio, J. Balzarini and L. De Napoli, Chem. Commun., 2011, 47, 2363–2365 RSC.
- R. Shadid, A. Bugaut and S. Balasubramanian, Biochemistry, 2010, 49, 8300–8306 CrossRef PubMed.
- J.-L. Mergny, A. De Cian, A. Ghelab, B. Saccà and L. Lacroix, Nucleic Acids Res., 2005, 33, 81–94 CrossRef CAS PubMed.
- G. Oliviero, N. Borbone, A. Galeone, M. Varra, G. Piccialli and L. Mayol, Tetrahedron Lett., 2004, 45, 4869–4872 CrossRef CAS.
- R. Ferreira, M. Alvira, A. Aviñó, I. Gómez-Pinto, C. González, V. Gabelica and R. Eritja, ChemistryOpen, 2012, 1, 106–114 CrossRef CAS PubMed.
- P. S. Ghosh and A. D. Hamilton, Chem.–Eur. J., 2012, 18, 2361–2365 CrossRef CAS PubMed.
- J. Gros, A. Aviñó, J. López de La Osa, C. González, L. Lacroix, A. Pérez, M. Orozco, R. Eritja and J.-L. Mergny, Chem. Commun., 2008, 25, 2926–2928 RSC.
- M. E. Favretto and R. Brock, Small, 2014, 11, 1414–1417 CrossRef PubMed.
- A. Astriab-Fisher, D. Sergueev, M. Fisher, B. Ramsay Shaw and R. L. Juliano, Pharm. Res., 2002, 19, 744–754 CrossRef CAS.
- T. Endoh and T. Ohtsuki, Adv. Drug Delivery Rev., 2009, 61, 704–709 CrossRef CAS PubMed.
- S. Futaki, Adv. Drug Delivery Rev., 2005, 57, 547–558 CrossRef CAS PubMed.
- J. Jana, R. K. Kar, A. Ghosh, A. Biswas, S. Ghosh, A. Bhunia and S. Chatterjee, Mol. BioSyst., 2013, 9, 1833–1836 RSC.
- H. Margus, K. Padari and M. Pooga, Mol. Ther., 2012, 20, 525–533 CrossRef CAS PubMed.
- K. Ezzat, S. EL Andaloussi, E. M. Zaghloul, T. Lehto, S. Lindberg, P. M. D. Moreno, J. R. Viola, T. Magdy, R. Abdo, P. Guterstam, R. Sillard, S. H. Hammon, M. J. A. Wood, A. A. Arzumanov, M. J. Gait, C. I. Edvard Smith, M. Hällbrink and U. Langel, Nucleic Acids Res., 2011, 39, 5284–5298 CrossRef CAS PubMed.
- R. J. Naik, P. Chandra, A. Mann and M. Ganguli, J. Biol. Chem., 2011, 286, 18982–18993 CrossRef CAS PubMed.
- J. D. Puglisi and I. Tinoco, Methods Enzymol., 1989, 180, 304–325 CAS.
- K. Murayama, Y. Tanaka, T. Toda, H. Kashida and H. Asanuma, Chem.–Eur. J., 2013, 19, 14151–14158 CrossRef CAS PubMed.
- H. Kashida, F. Hiromu and H. Asanuma, Org. Biomol. Chem., 2008, 6, 2892–2899 CAS.
- A. K. Sharma, P. Kumar and K. C. Kailash, Helv. Chim. Acta, 2001, 84, 3643–3649 CrossRef CAS.
- S. Pérez-Rentero, S. Grijalvo, G. Peñuelas, C. Fabrega and R. Eritja, Molecules, 2014, 19, 10495–10523 CrossRef PubMed.
- E. A. Englund and D. H. Apella, Angew. Chem., Int. Ed., 2007, 46, 1414–1418 CrossRef CAS PubMed.
- N. V. Sumbatyan, E. A. Artsatbanova, O. V. Baeva, K.-C. Hyun, S. A. Kuznetsova, M. B. Gottikh and G. A. Korshunova, Nucleosides Nucleotides, 1999, 18, 1489–1490 CAS.
- J. G. Harrison and S. Balasubramanian, Nucleic Acids Res., 1998, 26, 3136–3145 CrossRef CAS PubMed.
- T. Shiraishi, R. Hamzavi and P. E. Nielsen, Nucleic Acids Res., 2008, 36, 4424–4432 CrossRef CAS PubMed.
- S. Grijalvo and R. Eritja, Mol. Diversity, 2012, 16, 307–317 CrossRef CAS PubMed.
- G. Deglane, S. Abes, T. Michel, P. Prévot, E. Vives, F. Debart, I. Barvik, B. Lebleu and J. J. Vasseur, ChemBioChem, 2006, 7, 684–692 CrossRef CAS PubMed.
- P. Alberti, A. Bourdoncle, B. Saccà, L. Lacroix and J.-L. Mergny, Org. Biomol. Chem., 2006, 4, 3383–3391 CAS.
- L. Petraccone, E. Erra, V. Esposito, A. Randazzo, A. Galeone, G. Barone and C. Giancola, Biopolymers, 2005, 77, 75–85 CrossRef CAS PubMed.
- R. Noir, M. Kotera, B. Pons, J.-S. Remy and J.-P. Behr, J. Am. Chem. Soc., 2008, 130, 13500–13505 CrossRef CAS PubMed.
- H.-Y. Zhang, J. Mao, D. Zhou, Y. Xu, H. Thonberg, Z. Liang and C. Wahlestedt, Nucleic Acids Res., 2003, 31, e72 CrossRef PubMed.
- T. Mosmann, J. Immunol. Methods, 1983, 65, 55–63 CrossRef CAS PubMed.
- V. Dapic, V. Abdomerovic, R. Marrington, J. Peberdy, A. Rodger, J. O. Trent and P. J. Bates, Nucleic Acids Res., 2003, 31, 2097–2107 CrossRef CAS PubMed.
- N. Zhang, T. Bing, X. Liu, C. Qi, L. Shen, L. Wang and D. Shangguan, Chem. Sci., 2015, 6, 3831–3838 RSC.
- M. L. Jain, P. Y. Bruice, I. E. Szabó and T. C. Bruice, Chem. Rev., 2012, 112, 1284–1309 CrossRef CAS PubMed.
- P. Zhou, M. Wang, L. Du, G. W. Fisher, A. Waggoner and D. H. Ly, J. Am. Chem. Soc., 2003, 125, 6878–6879 CrossRef CAS PubMed.
- C. Plank, M. X. Tang, A. R. Wolfe and F. C. Szoka, Hum. Gene Ther., 1999, 10, 319–332 CrossRef CAS PubMed.
- X. Chen, D. M. Kube, M. J. Cooper and P. B. Davis, Mol. Ther., 2007, 16, 333–342 CrossRef PubMed.
- A. E. Felber, N. Bayo-Puxan, G. F. Deleavey, B. Castagner, M. J. Damha and J. C. Leroux, Biomaterials, 2012, 33, 5955–5965 CrossRef CAS PubMed.
- P. J. Bates, J. B. Kahlon, S. D. Thomas, J. O. Trent and D. M. Miller, J. Biol. Chem., 1999, 274, 26369–26377 CrossRef CAS PubMed.
- A. G. Hovanessian, C. Soundaramourty, D. El Khoury, I. Nondier, J. Svab and B. Krust, PLoS One, 2010, 5, e15787 CAS.
- L. A. Hanakahi, H. Sun and N. Maizels, J. Biol. Chem., 1999, 274, 15908–15912 CrossRef CAS PubMed.
- V. Brázda, L. Hároníková, J. C. C. Liao and M. Fojta, Int. J. Mol. Sci., 2014, 15, 17493–17517 CrossRef PubMed.
- M. Sader, J. Courty and D. Destouches, J. Nanomed. Nanotechnol., 2015, 6, 1–9 Search PubMed.
- D. Destouches, N. Page, Y. Hamma-Kourbali, V. Machi, O. Chaloin, S. Frechault, C. Birmpas, P. Katsoris, J. Beyrath, P. Albanese, M. Maurer, G. Carpentier, J.-M. Strub, A. van Dorsselaer, S. Muller, D. Bagnard, J. P. Briand and J. Courty, Cancer Res., 2011, 71, 3296–3305 CrossRef CAS PubMed.
- E. M. Reyes-Reyes, Y. Teng and P. J. Bates, Cancer Res., 2010, 70, 8617–8629 CrossRef CAS PubMed.
- J. L. Wolfe and J. Goodchild, J. Am. Chem. Soc., 1996, 118, 6301–6302 CrossRef CAS.
Footnote |
| † Electronic supplementary information (ESI) available: Experimental procedures, 1H-NMR and 13C-NMR spectra, HPLC profiles and MALDI-TOF mass spectra, CD thermal denaturation experiments, native PAGE gel, CD spectra of selected conjugates, MTT colorimetric assay, transfection efficiencies in serum-free conditions, flow cytometry analyses. See DOI: 10.1039/c6ra15336d |
| This journal is © The Royal Society of Chemistry 2016 |