Open Access Article
Open Access ArticleNanotemplate-directed DNA segmental thermal motion†
E. V.
Dubrovin
*ab,
M.
Schächtele
b and
T. E.
Schäffer
b
aLomonosov Moscow State University, Faculty of Physics, Leninskie Gory, 1/2, 119991 Moscow, Russian Federation. E-mail: dubrovin@polly.phys.msu.ru
bUniversity of Tübingen, Institute of Applied Physics, Auf der Morgenstelle 10, 72076 Tübingen, Germany
First published on 17th August 2016
Abstract
DNA adsorption on molecular nanotemplates represents a promising system for use both in fundamental studies and biotechnological applications. Using atomic force microscopy (AFM) operated in a liquid, we have investigated the conformation and real time thermal motion of phage T7 DNA adsorbed on stearylamine and stearic acid nanotemplates on highly oriented pyrolytic graphite (HOPG) in aqueous solutions. Though local linear straightening of adsorbed DNA segments along the lamellas of nanotemplates was always observed, DNA adsorbed on a stearic acid nanotemplate revealed a larger surface coverage and higher mobility, indicating a weaker DNA–surface interaction than for a stearylamine nanotemplate. The DNA adopted a nearly two-dimensional conformation on the molecular nanotemplates, which can be described by a two-dimensional compact globule state for stearylamine and by a model of a self-avoiding walk for stearic acid. Unlike previously reported thermal motion of single DNA molecules, DNA segmental thermal motion on the nanotemplates mostly proceeded along preferred directions, defined by the lamellas of the nanotemplates. However, DNA hopping to adjacent lamellas and out-of-plane movement (movement through temporary desorption of DNA segments) were also observed. Owing to their peculiar properties, molecular nanotemplates on graphite are ideal candidates for utilization as model substrates for AFM studies of two-dimensional conformations of DNA at the nanoscale, for exploring conformational relaxation times of DNA on a single molecule level and for investigation of the dynamics of DNA–protein interaction.
Introduction
Controlled adsorption and assembly of DNA molecules on solid supports, especially conductive ones, are in great demand for use both in fundamental studies and biotechnological applications. Directed DNA adsorption on solid substrates is essential for high precision DNA mapping and for investigation of conductive and mechanical properties of single DNA molecules.1–4 Ordered DNA structures on a surface are used for the development of biosensors and molecular electronics devices.5 For example, the development of DNA immobilization methodologies that strongly stabilize DNA on an electrode surface is thought to be one of the key factors in DNA biosensor design, and the degree of DNA surface coverage is the critical issue in the development of a DNA electrochemical biosensor for rapid detection of DNA interaction and damage by hazardous compounds.6The use of the “bottom-up” approach is attractive for the development of controlled DNA architectures on the surface. Unlike widely developed systems based on DNA-origami,7,8 nanotemplate-driven ordering of DNA molecules does not depend on the specific DNA sequence (and normally does not utilize base pairing of complementary strands), does not require computational design and further synthesis of each molecule included in the structure. Nanotemplate-mediated DNA assembly can be used for any natural or artificial dsDNA molecules irrespective of their nucleotide sequence and length.
Nanotemplates on crystal surfaces such as highly oriented pyrolytic graphite (HOPG) can be formed by long chain alkanes and their derivatives.9–14 Normally, hydrophobic carbon tails lay parallel to each other reflecting the hexagonal structure of HOPG (C–C bonds in alkanes and HOPG have similar length), while functional groups (if any) separate into straight lamellas.15 These lamellas can serve as nanoscopic “rails” which, depending on the charge of the headgroups and the lengths of the alkyl chains, can differently orient single polymer molecules, including DNA.16–18 The most popular method for studying molecular nanotemplates is scanning probe microscopy, mainly atomic force microscopy (AFM).19 These methods allow direct visualization of nanotemplates with molecular or even atomic resolution (e.g., ref. 9).
Directed assembly of DNA molecules was for the first time reported on a dodecylamine layer on HOPG:20 DNA molecules were oriented along the directions of three-fold symmetry of the underlying pattern at room temperature. Similar effect of DNA assembly on a stearylamine layer was investigated in ref. 21 and 22. In the latter work, different scenarios of DNA adsorption on modified HOPG were reported for three types of nanotemplates differing only by a functional group (amine, acid and alcohol).
Despite some progress in this area our knowledge about nanotemplate-driven DNA adsorption remains incomplete, since it was acquired in static experiments in air. For deeper insight a detailed study of the dynamics of this process and its dependence on physicochemical factors such as ionic composition is required. Movement of a single polymeric molecule on the surface is connected with different surface phenomena, such as adhesion, wetting and friction.23 A dynamical study of single DNA molecules adsorbed on molecular nanotemplates will provide novel information on DNA thermal motion confined by a surface with periodic structure. Observing the dynamical behavior of single DNA molecules on the surface in real time will allow analyzing their diffusion and give better understanding of DNA interaction with nanotemplates. AFM operated in liquid is an effective tool for studying single DNA molecules adsorbed on the substrate24 including DNA conformation.25 One of the main advantages of this method is the possibility to study the dynamics of adsorbed DNA molecules in solutions.26–29
The purpose of this work was to achieve deeper understanding of controlled DNA immobilization, assembly and organization on molecular nanotemplates of stearylamine and stearic acid on HOPG using an AFM dynamical study in liquid. We have characterized DNA conformation on these nanotemplates in solution and explored DNA segmental movement in real time during several tens of minutes. The obtained results reveal peculiarities in thermal motion of DNA molecules adsorbed on the nanotemplates and allow discriminating DNA conformation and mobility on different nanotemplates. DNA adsorbed on stearylamine and stearic acid nanotemplates represents good model systems of two-dimensional compact globules and self-avoiding walk. Also, these nanotemplates are ideal candidates for utilization as model substrates for investigation of the dynamics of DNA–protein interaction using AFM in solution.
Experimental
For preparation of molecular nanotemplates on graphite, 20 μl of 100 μg ml−1 stearic acid or stearylamine (Sigma-Aldrich) solution in propanol-2 (p.a.) were deposited onto a rotated surface (75 rotations per s) of freshly cleaved HOPG (ZYB Quality, mosaic spread 0.8–1.2 degrees, NT-MDT, Russia) at ∼20 °C. The resulting periodic lamellar structure (nanotemplate) was stable in aqueous solutions used in this study (ESI, file 1, Fig. S1†).Prior to AFM imaging, a 20 μl portion of phage T7 DNA (GeneON, Germany) diluted to a concentration of 1 μg ml−1 in either Milli-Q water, NaCl (p.a.) or MgCl2 (p.a.) solution, was deposited onto the modified HOPG surface, which was mounted on the sample stage of MultiMode 8 atomic force microscope with a Nanoscope V controller (Bruker, USA). AFM imaging was performed at ∼20 °C in tapping mode and PeakForce mode using tapping mode fluid cell equipped with a fluorosilicone o-ring to minimize evaporation. The time interval between DNA deposition and start of AFM scanning was 5–15 minutes. We have used cantilevers “E” (spring constant 0.1 N m−1) and “F” (spring constant 0.6 and 0.5 N m−1) of MSCT and MLCT series (Bruker, USA), which were operated in liquid at a tapping frequency of ∼30 (for “E” cantilever) or ∼10 (for “F” cantilever) kHz. In tapping mode AFM height, phase and amplitude images were obtained simultaneously. Phase images usually were less noisy and of higher contrast than height images. The range of the color scale of the presented AFM phase images was chosen individually for each image to obtain good visual contrast. In order to minimize the effective force applied by a cantilever the setpoint amplitude was maximized. The line scan rate was typically 2.1 Hz for tapping mode and 0.5 Hz for PeakForce mode, with 512 × 512 pixels per image. AFM images were processed in NanoScope Analysis (Bruker, USA) and Femtoscan software (Advanced Technologies Center, Russia).
The contours of DNA molecules were traced using a tool “selection of a lengthy object” implemented in the FemtoScan software (the sum of the lengths of all analyzed DNA molecules was at least 50 μm). Calculation of the mean square distance 〈R2〉 as a function of length l along the DNA contour was performed in Scilab 5.3.3.30 For this purpose, the distances between all pairs of points on the DNA strand separated by a length along the contour l (with a step length equal to l) were averaged for each l ranging from 10 nm to the maximal traced DNA length with a 10 nm step (this is a slightly modified algorithm suggested by Frontali et al.31). The scaling exponent ν was determined from the linear regression of the experimental dependence ln(〈R2〉) = f(ln(l)) for l > 100 nm (which does not correspond to the rodlike regime). The corresponding errors were estimated by the standard deviation of the regression coefficients.
Results and discussion
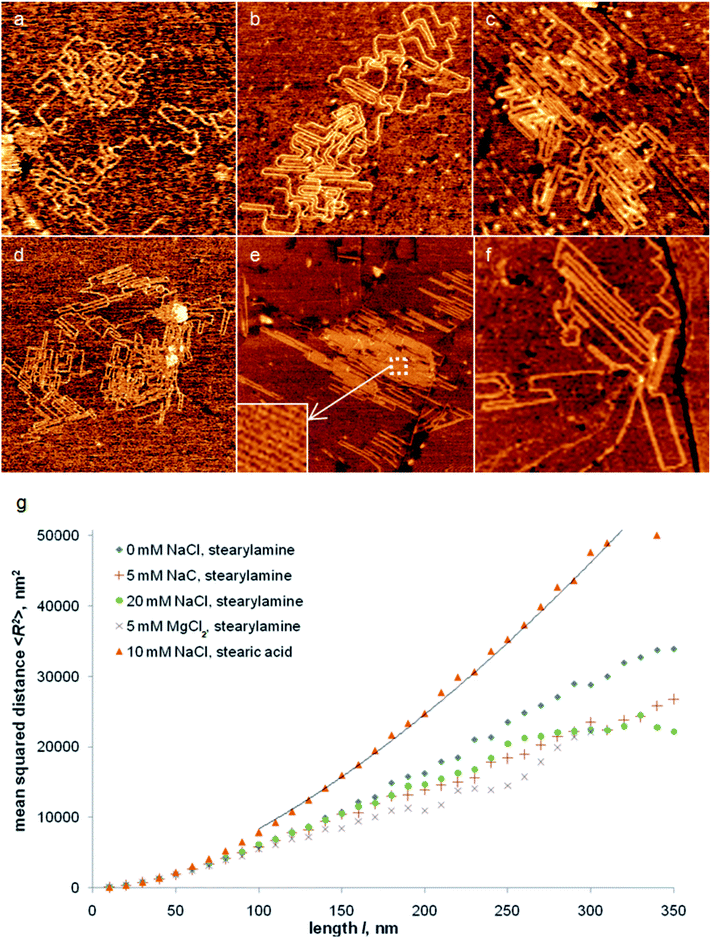
DNA alignment along preferred directions, which are defined by lamellas of a stearylamine nanotemplate, was observed in water and different aqueous solutions (Fig. 1a–e). This is in agreement with previous AFM studies in air that have shown an ordering effect of DNA on stearylamine nanotemplates on graphite.20–22 This agreement indicates that drying does not significantly change the shape of DNA adsorbed on a stearylamine nanotemplate. The mean height of DNA molecules measured from AFM images is around 1 nm (ESI, file 1, Fig. S2†), again in agreement with previous AFM studies.20–22 Aligned DNA can be packed on the surface very tightly, especially when adsorbed from 20 mM NaCl (Fig. 1c) or 5–10 mM MgCl2 solution (Fig. 1d and e). In the latter case DNA may form sheet-like structures of several hundred nanometers in size, composed of densely packed DNA molecules (Fig. 1e). The periodicity of such DNA structures (6.3 ± 0.2 nm) corresponds to one of the most often observed periods of a stearylamine nanotemplate (ESI,† file 1) and, therefore, indicates that DNA molecules are packed there as close as adjacent stearylamine lamellas.Denser packing of DNA molecules adsorbed on a stearylamine nanotemplate from salt containing solutions may be also seen from the analysis of the mean square distance 〈R2〉 between points of the DNA molecule separated by the length l (Fig. 1g). For small separations (l < 70 nm) the difference between the different traces is negligible. This can be explained by the presence of straight DNA segments in all studied samples, which leads to the same parabolic dependence of 〈R2〉 on l. A similar dependence for small separations was observed in ref. 22 for DNA ordered on molecular nanotemplates on graphite. At l > 70 nm, 〈R2〉 values for DNA adsorbed from water and salt solutions deviate from each other. For a stearylamine nanotemplate, DNA adsorbed from water has larger 〈R2〉 values than DNA from salt solutions, indicating that DNA adsorption from salt solutions leads to a more compact conformation on the surface. This is probably a consequence of the Debye screening effect in electrolyte solutions, which decreases the mutual repulsion of negatively charged DNA segments. The difference of 〈R2〉 obtained for various NaCl and MgCl2 concentrations above 5 mM is not well pronounced. The estimated Debye lengths of the solutions used in this study are 1.5 nm (10 mM MgCl2)–4 nm (5 mM NaCl),32 whereas the nearest possible distance between two DNA strands adsorbed on a stearylamine nanotemplate is about 4 nm (taking into account a period of a dense DNA structure of ∼6.3 nm and a DNA diameter of ∼2 nm). Moreover, an effective Debye length may be even smaller in the proximity of a stearylamine nanotemplate due to the high density of positively charged amine groups on the surface. Therefore, there is no significant influence of the NaCl or MgCl2 concentration (above ≈5 mM) on the conformation of the adsorbed DNA segments, whose adsorption is directed by the lamellas of the nanotemplate. This result may be important for utilization of nanotemplates on graphite for investigation of biomolecular processes at physiological conditions, e.g., DNA–protein interaction.
The dependences in Fig. 1g allow defining a scaling exponent ν, which relates 〈R2〉 to l by the formula: 〈R2〉 = const × l2ν.33 The scaling exponent ν obtained for an ensemble of polymeric molecules adsorbed on a substrate together with an estimation of a degree of suppression of their self-crossings may serve as criteria for an assignment of a polymer conformation to a certain type.34 For example, ν = 1 corresponds to a rigid rod conformation, ν = 0.5 (without self-crossings) to the two-dimensional compact globule conformation and ν = 0.75 (without self-crossings) to the two-dimensional coil with excluded volume conformation (the model of a self-avoiding walk).35–37 An example fitting curve for estimation of ν is shown as a solid line in Fig. 1g. The ν values (obtained for l > 100 nm) for DNA, adsorbed from solution on a stearylamine nanotemplate are close to 0.5 (Table 1). Though self-crossings of DNA contours are occasionally observed in AFM images, they are rare even in dense DNA structures. Also, DNA ordering on the nanotemplates supports the notion that DNA reorients on the surface after initial adsorption and, therefore, final DNA conformation on the nanotemplate is not a projection of its three-dimensional conformation in solution (which is characterized by ν = 0.59 and allowed self-crossings34). Thus, we conclude that the conformation of DNA adsorbed on a stearylamine nanotemplate is close to the compact globule conformation. This is consistent with our previous results using AFM imaging in air.22
| Nanotemplate | Solution | ν |
|---|---|---|
| Stearylamine | 5 mM NaCl | 0.54 ± 0.01 |
| Stearylamine | 20 mM NaCl | 0.51 ± 0.01 |
| Stearylamine | 5 mM MgCl2 | 0.54 ± 0.02 |
| Stearic acid | 10 mM NaCl | 0.77 ± 0.01 |
The presence of preferred directions of alignment of adsorbed biomolecules was also observed on a stearic acid nanotemplate (Fig. 1f). Previous AFM studies in air, however, demonstrated another orientation configuration of DNA on this nanotemplate with one preferred direction of biopolymer alignment.22 Such DNA alignment was explained by a possible effect of molecular combing connected with the drying procedure.38,39 DNA molecules adsorbed on a stearic acid nanotemplate reveal significantly larger surface coverage than for a stearylamine nanotemplate (Fig. 1g). The obtained ν value (0.77 ± 0.01, Table 1) and the observation of a low number of self-crossings allow describing the DNA conformation by the model of a self-avoiding walk. The conformation of a polymer molecule on a surface is defined by the interplay of the internal rigidity of the molecule and its interaction with the surface.40 Therefore, the less compact DNA conformation on a stearic acid nanotemplate may be connected with a weaker attractive interaction of the negatively charged phosphate groups of the DNA double helix with a stearic acid nanotemplate, which does not possess positively charged groups (as amine groups in a stearylamine nanotemplate), and with a larger repulsive monomer–monomer interaction. A similar conformation was observed on the optical scale for DNA molecules electrostatically bound to fluid cationic lipid bilayers.36
To get deeper insight on the adsorption and ordering effect of DNA molecules on the nanotemplates, we have investigated the dynamical behavior of single adsorbed biomolecules in liquid. Fig. 2 shows successive AFM images of the thermal movement of a segment of a T7 DNA molecule adsorbed on a stearylamine nanotemplate in 5 mM NaCl solution (see also the movies in ESI,† file 2.1 and 2.2). We observe the following characteristics of this movement. First, any segment of DNA has moved from its initial position after ∼30 minutes (compare the first and last images of the series). This suggests moderate adsorption forces and an absence of trapping of the DNA molecule by the surface. Second, thermal movement of the DNA molecule takes place mostly along the preferred directions of DNA alignment (along lamellas of the nanotemplate). To illustrate this in detail, we demonstrate the movements of short (within few hundred nanometers) DNA segments (Fig. 3). This observation is in discrepancy with DNA diffusion reported for DNA molecules (or their segments) adsorbed on mica,27,41,42 where no preferred directions of the movement were revealed. We may conclude that a molecular nanotemplate assigns the directions of DNA 2D diffusion.
The “nanorails” of lamellas complicate the movement of DNA molecules across them. The movement we mostly observed was along the “nanorails” (Fig. 3 and the movies in ESI†), which suggests a motional confinement owing to the “nanorails”. Since DNA contours are mostly aligned along the “nanorails” (Fig. 1–4), this movement may be considered as movement of DNA segments along themselves, which is often referred to as reptational movement. Formally, reptational movement is defined as free movement of a polymer between fixed obstacles.43 A molecular nanotemplate on graphite represents a two-dimensional example of a system with reptational movement, where lamellas take the role of these obstacles. In previously described systems with reptational movements the obstacles were distributed randomly.27–29 In contrast, molecular lamellas on graphite have a linear geometry and well defined symmetry, which causes the observed peculiarities of DNA movement on molecular nanotemplates on graphite.
Besides reptational movement, we also observed segmental hopping of DNA to an adjacent lamella of the nanotemplate (black arrows in Fig. 3). Also, out-of-plane movement of small DNA segments (temporary desorption) occurred, which can be confirmed by the following observations: temporary loss of contrast of some DNA segments in an AFM image (solid white arrows in Fig. 2 and 3) and formation (or disappearance) of small loops in the middle of DNA molecules (dashed white arrows in Fig. 2). In summary, DNA thermal motion on a stearylamine nanotemplate has a complex character, including reptation, hopping between lamellas and out-of-plane movement (desorption of segments).
In contrast to DNA molecules adsorbed from 5–20 mM NaCl solutions, sheet-like DNA structures, which were formed in MgCl2 solution, are much more stabilized on the surface (Fig. 4 and the movies in ESI,† file 3.1 and 3.2). Movement of DNA segments within a dense, sheet-like structure was not observed, owing to the adjacent lamellas being occupied. However, DNA segmental motion along the lamellas of the nanotemplate (reptational movement) and out-of-plane movement was detected (Fig. 4, insets).
The motion of DNA molecules on a stearic acid nanotemplate comprised of the same three components as for stearylamine; however, DNA segment hopping to adjacent lamellas seems more pronounced (Fig. 5 and the movie in ESI,† file 4). DNA molecules are more mobile on a stearic acid nanotemplate: almost each DNA segment changed its position between consecutive AFM frames (Fig. 5, insets). As noted above, the absence of positively charged groups on a stearic acid nanotemplate weakens its interaction with DNA. This favors DNA segmental motion and leads to a more extended DNA conformation on this nanotemplate, which is close to the self-avoiding walk model. Moreover, relatively weak polymer–substrate interaction is probably a factor, which allows previously observed molecular combing of DNA on a stearic acid nanotemplate upon drying (such an effect was absent for a stearylamine nanotemplate).22
We note that the observed motion of DNA segments did not depend on the scan direction of the cantilever. Moreover, similar nanotemplate directed DNA segmental motion was observed in PeakForce mode, in which the tip is repeatedly withdrawn from the surface during lateral scanning of the sample (ESI, file 1, Fig. S3†).
We can conclude that DNA adsorption on molecular nanotemplates formed on HOPG from stearylamine or stearic acid has characteristic features such as local DNA straightening along the lamellas of the nanotemplates. DNA surface coverage is larger and adsorbed DNA molecules are more mobile on a stearic acid nanotemplate, owing to a weaker DNA–template interaction. The thermal motion of DNA molecules on the nanotemplates is strongly influenced by their lamellas, which defines the preferred directions of DNA segmental movement.
Though all our experiments were performed at a room temperature (∼20 °C), we expect that the nanotemplate directed character of DNA segmental thermal motion will also remain upon increase of temperature until thermal disintegration of the nanotemplate.20,44
Conclusions
The investigated nanotemplates represent good model systems for studying two-dimensional conformations of DNA at the nanoscale, such as compact globule and self-avoiding walk. These systems will allow directly exploring conformational relaxation times of DNA on a single molecule level, especially with the use of high-speed AFM, which is capable of obtaining about 10 AFM images per second in liquid media.45 Moreover, both investigated surfaces are ideal candidates for utilization as model substrates for AFM studies of the dynamics of DNA–protein interactions, owing to the following reasons: (1) extended and straightened shape of DNA molecules, which is convenient for analysis of long DNA molecules and protein position on DNA; (2) moderate strength of adsorption of DNA to the substrate, which allows AFM imaging, but at the same time DNA motion across the surface; (3) the presence of “out-of-plane” segmental motion, which will allow protein interaction with DNA above the surface, in “bulk” solution. The effective speed of DNA–protein interaction (e.g., movement of an enzyme along DNA), which is significantly slower on the surface than in bulk solution, may be tuned by adjusting the ionic composition of the solution and by the choice of nanotemplate.Acknowledgements
The work was supported by the joint Lomonosov Moscow State University – DAAD Program “Vladimir Vernadskij-2015”.Notes and references
- D. Porath, A. Bezryadin, S. de Vries and C. Dekker, Nature, 2000, 403, 635–638 CrossRef CAS PubMed.
- T. R. Strick, J. F. Allemand, D. Bensimon, A. Bensimon and V. Croquette, Science, 1996, 271, 1835–1837 CAS.
- C. Bustamante, S. B. Smith, J. Liphardt and D. Smith, Curr. Opin. Struct. Biol., 2000, 10, 279–285 CrossRef CAS PubMed.
- A. D. Chepelianskii, D. Klinov, A. Kasumov, S. Gueron, O. Pietrement, S. Lyonnais and H. Bouchiat, New J. Phys., 2011, 13, 063046 CrossRef.
- M. I. Pividori, A. Merkoci and S. Alegret, Biosens. Bioelectron., 2000, 15, 291–303 CrossRef CAS PubMed.
- A. A. Ensafi, E. Heydari-Bafrooei, M. Dinari and S. Mallakpour, J. Mater. Chem. B, 2014, 2, 3022–3028 RSC.
- E. Winfree, F. R. Liu, L. A. Wenzler and N. C. Seeman, Nature, 1998, 394, 539–544 CrossRef CAS PubMed.
- A. Kuzuya and M. Komiyama, Nanoscale, 2010, 2, 309–321 RSC.
- J. Rabe and S. Buchholz, Science, 1991, 253, 424–427 CAS.
- T. Yang, S. Berber, J.-F. Liu, G. P. Miller and D. Tomanek, J. Chem. Phys., 2008, 128, 124709 CrossRef PubMed.
- D. Bléger, A. Bocheux, D. Kreher, F. Mathevet, A.-J. Attias, G. Metgé, L. Douillard, C. Fiorini-Debuisschert and F. Charra, Nanoscale, 2013, 5, 1452–1455 RSC.
- M. Hibino, A. Sumi, H. Tsuchiya and I. Hatta, J. Phys. Chem. B, 1998, 102, 4544–4547 CrossRef CAS.
- I. De Cat, C. Gobbo, B. Van Averbeke, R. Lazzaroni, S. De Feyter and J. van Esch, J. Am. Chem. Soc., 2011, 133, 20942–20950 CrossRef CAS PubMed.
- F. Tao, Pure Appl. Chem., 2008, 80, 45–57 CrossRef CAS.
- A. J. Groszek, Proc. R. Soc. London, Ser. A, 1970, 314, 473–498 CrossRef.
- N. Severin, J. Barner, A. A. Kalachev and J. P. Rabe, Nano Lett., 2004, 4, 577–579 CrossRef CAS.
- N. Severin, I. M. Okhapkin, A. R. Khokhlov and J. P. Rabe, Nano Lett., 2006, 6, 1018–1022 CrossRef CAS PubMed.
- N. Severin, W. Zhuang, C. Ecker, A. A. Kalachev, I. M. Sokolov and J. P. Rabe, Nano Lett., 2006, 6, 2561–2566 CrossRef CAS PubMed.
- G. Binnig, C. Quate and C. Gerber, Phys. Rev. Lett., 1986, 56, 930–933 CrossRef PubMed.
- J. Adamcik, S. Tobenas, G. Di Santo, D. Klinov and G. Dietler, Langmuir, 2009, 25, 3159–3162 CrossRef CAS PubMed.
- E. V. Dubrovin, J. W. Gerritsen, J. Zivkovic, I. V. Yaminsky and S. Speller, Colloids Surf., B, 2010, 76, 63–69 CrossRef CAS PubMed.
- E. V. Dubrovin, S. Speller and I. V. Yaminsky, Langmuir, 2014, 30, 15423–15432 CrossRef CAS PubMed.
- S. Granick, S. K. Kumar, E. J. Amis, M. Antonietti, A. C. Balazs, A. K. Chakraborty, G. S. Grest, C. J. Hawker, P. Janmey, E. J. Kramer, R. Nuzzo, T. P. Russell and C. R. Safinya, J. Polym. Sci., Part B: Polym. Phys., 2003, 41, 2755–2793 CrossRef CAS.
- H. Hansma, M. Bezanilla, F. Zenhausern, M. Adrian and R. Sinsheimer, Nucleic Acids Res., 1993, 21, 505–512 CrossRef CAS PubMed.
- S. Mantelli, P. Muller, S. Harlepp and M. Maaloum, Soft Matter, 2011, 7, 3412–3416 RSC.
- L. S. Shlyakhtenko, V. N. Potaman, R. R. Sinden and Y. L. Lyubchenko, J. Mol. Biol., 1998, 280, 61–72 CrossRef CAS PubMed.
- W. Vanderlinden and S. De Feyter, Nanoscale, 2013, 5, 2264–2268 RSC.
- J. Kumaki, T. Kawauchi and E. Yashima, Macromolecules, 2006, 39, 1209–1215 CrossRef CAS.
- J. Kumaki, T. Kawauchi and E. Yashima, Macromol. Rapid Commun., 2008, 29, 406–411 CrossRef CAS.
- Scilab Enterprises https://www.scilab.org, accessed May 2016.
- C. Frontali, E. Dore, A. Ferrauto, E. Gratton, A. Bettini, M. Pozzan and E. Valdevit, Biopolymers, 1979, 18, 1353–1373 CrossRef CAS PubMed.
- J. N. Israelachvili, Intermolecular and Surface Forces. With Applications To Colloidal And Biological Systems, Academic Press, London, 1985 Search PubMed.
- B. Stokke and D. Brant, Biopolymers, 1990, 30, 1161–1181 CrossRef CAS PubMed.
- M. O. Gallyamov, Macromol. Rapid Commun., 2011, 32, 1210–1246 CrossRef CAS PubMed.
- M. Joanicot and B. Revet, Biopolymers, 1987, 26, 315–326 CrossRef CAS PubMed.
- B. Maier and J. O. Radler, Phys. Rev. Lett., 1999, 82, 1911–1914 CrossRef CAS.
- P.-K. Lin, C.-C. Fu, Y.-L. Chen, Y.-R. Chen, P.-K. Wei, C. H. Kuan and W. S. Fann, Phys. Rev. E: Stat., Nonlinear, Soft Matter Phys., 2007, 76, 011806 CrossRef PubMed.
- J. W. Li, C. L. Bai, C. Wang, C. F. Zhu, Z. Lin, Q. Li and E. H. Cao, Nucleic Acids Res., 1998, 26, 4785–4786 CrossRef CAS PubMed.
- C. Y. Khripin, M. Zheng and A. Jagota, J. Colloid Interface Sci., 2009, 330, 255–265 CrossRef CAS PubMed.
- C. Rivetti, M. Guthold and C. Bustamante, J. Mol. Biol., 1996, 264, 919–932 CrossRef CAS PubMed.
- C. Bustamante, M. Guthold, X. S. Zhu and G. L. Yang, J. Biol. Chem., 1999, 274, 16665–16668 CrossRef CAS PubMed.
- Y. Suzuki, Y. Higuchi, K. Hizume, M. Yokokawa, S. H. Yoshimura, K. Yoshikawa and K. Takeyasu, Ultramicroscopy, 2010, 110, 682–688 CrossRef CAS PubMed.
- P. G. De Gennes, J. Chem. Phys., 1971, 55, 572–579 CrossRef.
- L. Askadskaya and J. P. Rabe, Phys. Rev. Lett., 1992, 69, 1395–1398 CrossRef CAS PubMed.
- C. Braunsmann and T. E. Schaeffer, Nanotechnology, 2010, 21, 225705 CrossRef PubMed.
Footnote |
| † Electronic supplementary information (ESI) available: AFM height images of nanotemplates and DNA on stearylamine nanotemplate (ESI file 1). The movies of DNA segmental dynamics on a stearylamine nanotemplate in 5 mM NaCl solution, composed of AFM phase images acquired with a frequency of 1 image per min (ESI file 2.1, the size of the frame is 950 × 890 nm and ESI file 2.2, the size of the frame is 870 × 930 nm). The movie of DNA segmental dynamics on a stearylamine nanotemplate in 10 mM MgCl2 solution, composed of AFM height images acquired with a frequency of 1 image/4 min (the size of the frame is 520 × 440 nm) (ESI file 3.1). The movie of DNA segmental dynamics on a stearylamine nanotemplate in 10 mM MgCl2 solution, composed of AFM phase images acquired with a frequency of 1 image/4 min (the size of the frame is 870 × 510 nm) (ESI file 3.2). The movie of DNA segmental dynamics on a stearylamine nanotemplate in 10 mM MgCl2 solution, composed of AFM height images acquired with a frequency of 1 image/2 min (the size of the frame is 630 × 650 nm) (ESI file 3.3). The movie of DNA segmental dynamics on a stearic acid nanotemplate in 5 mM NaCl solution, composed of AFM height images acquired with a frequency of 1 image/5 min (the size of the frame is 1.5 × 1.3 μm) (ESI file 4). See DOI: 10.1039/c6ra14383k |
| This journal is © The Royal Society of Chemistry 2016 |