Identification of novel cathepsin K inhibitors using ligand-based virtual screening and structure-based docking†
Yali Wanga,
Ruolan Lib,
Zhihui Zhengb,
Hong Yia and
Zhuorong Li*a
aInstitute of Medicinal Biotechnology, Chinese Academy of Medical Science and Peking Union Medical College, Beijing 100050, China. E-mail: lizhuorong@imb.pumc.edu.cn; Tel: +86-10-83152017
bNew Drug Research & Development Center, North China Pharmaceutical Group Corporation, Shijiazhuang 050015, China
First published on 5th August 2016
Abstract
A combination of virtual screening and biological testing was used to identify novel cathepsin K (Cat K) inhibitors. A ligand-based pharmacophore model with five features was constructed and used for the virtual screening, which was followed by structure-based docking. Compounds were selected for biological testing based on fit values in the pharmacophore matching, structural diversity, binding pocket location and docking score in the screening. Four (compounds 3, 4, 5 and 21) out of twenty-two initial in silico hit compounds showed promising Cat K inhibition with IC50 values of 19.21, 24.83, 21.43 and 17.73 μM, respectively. Three compounds also showed anti-proliferative activity against one or more cancer cell lines with micromolar IC50 values. Compound 21 was the most promising hit with IC50 values of 28.25, 16.35 and 5.05 μM against HCT116, MCF7 and 143B cancer cell lines, respectively. This pioneering work has identified novel Cat K inhibitors using virtual screening in combination with pharmacophore modeling and docking approaches. Compound 21 could be used as a scaffold for the development of novel Cat K inhibitors with anti-proliferative activities.
Introduction
Cathepsin K (Cat K), a cysteine protease that is selectively and abundantly expressed within osteoclasts,1–5 is believed to be crucial for the resorption of bone matrix. The ability to degrade type I collagen allows Cat K to make a unique contribution to the balance between bone resorption (performed by osteoclasts) and bone formation (performed by osteoblasts).6,7 Inhibitors of Cat K could prevent bone resorption and may provide a promising approach for the treatment of osteoporosis,2–4 which is characterized by low bone mass and micro-architectural deterioration. Apart from collagen metabolism, Cat K has been shown to be important in other physiological and pathological processes. Increased Cat K activity contributes to cardiovascular diseases, arthritic diseases and cancer progression. Cat K is expressed in melanoma, prostate cancer, giant cell tumors and basal cell carcinoma, and has also been implicated in breast cancer and osteosarcoma.8–13 Inhibition of Cat K has, therefore, been proposed as a promising strategy for the treatment of osteoporosis, cancer and other diseases but there are, as yet, no inhibitors on the market.Several Cat K inhibitors, including odanacatib, MIV-711, relacatib and balicatib, have progressed into clinical trials. Odanacatib is a selective inhibitor with good pharmacokinetic properties and is currently finishing phase III clinical trials for osteoporosis.14–16 It has also been shown to have good therapeutic effects in patients with breast cancer.17 MIV-711 reduces biomarkers of bone resorption and has being studied in clinical trials.18–20 Unfortunately, some inhibitors have been abandoned because of side effects. Phase II clinical studies of balicatib were discontinued because of adverse dermatological effects and relacatib was discontinued following a phase I study that showed possible drug–drug interactions with paracetamol, ibuprofen and atorvastatin.16,21–23
In the present study, novel Cat K inhibitors were identified among commercially available compounds using a combination of virtual screening and in vitro tests. Firstly, a ligand-based pharmacophore model was generated based on common features of known Cat K inhibitors; secondly, the pharmacophore model was used to virtually screen the Specs compound library of >500![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 compounds (available online http://www.specs.net) and, thirdly, molecular docking screening was carried out. From a list of retrieved compounds, twenty-two molecules were selected for enzymatic assays and four were tested for anti-proliferative activity in vitro.
000 compounds (available online http://www.specs.net) and, thirdly, molecular docking screening was carried out. From a list of retrieved compounds, twenty-two molecules were selected for enzymatic assays and four were tested for anti-proliferative activity in vitro.
Results and discussion
Common features of pharmacophore modeling and validation
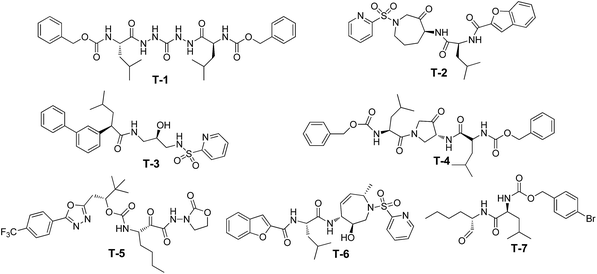
Common features of pharmacophore models were generated using Discovery Studio 4.5 (DS 4.5, Neo trident Technology LTD) software. For the training set, we chose seven structurally diverse Cat K inhibitors for which co-crystal structures with the enzyme were available (Fig. 1). The potencies of the inhibitors against Cat K are listed in Table 1.| Comp. | Entry | Potency measure | Potency human Cat Ka | Fit values | |
|---|---|---|---|---|---|
| Hypo-02 | Hypo-09 | ||||
| a Expressed in nM. | |||||
| T-1 | 1AYU | Ki (ref. 24) | 0.7 | 3.525 | 4.297 |
| T-2 | 1NLJ | Ki (ref. 25) | 0.16 | 4.028 | 3.912 |
| T-3 | 1BGO | Ki (ref. 26) | 3.5 | 5.000 | 5.000 |
| T-4 | 1AU3 | Ki (ref. 27) | 2.3 | 3.194 | 3.451 |
| T-5 | 1YT7 | IC50 (ref. 28) | 0.13 | 2.933 | 4.049 |
| T-6 | 2FTD | Ki (ref. 29) | 0.041 | 3.669 | 4.188 |
| T-7 | 1SNK | IC50 (ref. 30) | 0.25 | 3.246 | 4.463 |
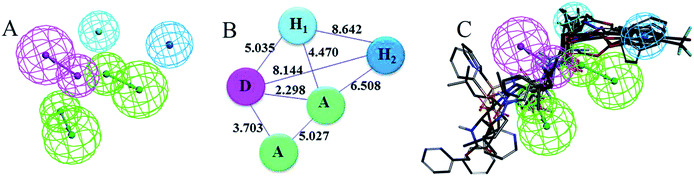
Ten pharmacophore models were generated based on common features (H-bond acceptors (A), H-bond donors (D), alkyl hydrophobic groups (H1) and aromatic hydrophobic groups (H2)) of the compounds in the training set. The ten hypotheses could be classified into two groups according to the pharmacophore features: HHDAA (hypotheses 01, 02, 04, 06, 09 and 10) and HHAAA (hypotheses 03, 05, 07 and 08). The fit values were obtained by mapping the pharmacophore models onto the training set compounds (Tables 1 and S1†). High fit values indicate a good match between the pharmacophore features and the ligands. Hypothesis 02 (hypo-02) and hypothesis 09 (hypo-09) had higher fit values than the other hypotheses and were further validated using the Güner–Henry (GH) scoring method, which is used to evaluate the performance of models in virtual screening (Table 2). The results were analyzed using the following parameters: yield of activities, enrichment factor (E) and GH score. For hypo-02, the number of predicted positives was 70 and the number of hits was 28, giving a yield of 80.0% (28 out of 35 inhibitors). The E and GH scores were 11.8 and 0.48, respectively. For hypo-09, the number of predicted positives was 50 and the yield was 88.6% (31 out of 35 inhibitors). An E of 16.3 and a GH of 0.72 indicated the high quality of hypo-09, which consists of two hydrophobic groups (H1 and H2), one hydrogen bond donor (D) and two hydrogen bond acceptors (A).
| Parameters | Values | |
|---|---|---|
| Hypo-02 | Hypo-09 | |
| a [Ha/(4 × Ht × A)] × (3 × A + Ht) × [1 − (Ht − Ha)/(D − A)]. | ||
| Total molecules in database (D) | 1035 | 1035 |
| Total number of actives in database (A) | 35 | 35 |
| Total hits (Ht) | 70 | 50 |
| Active hits (Ha) | 28 | 31 |
| % Yield of actives [(Ha/Ht) × 100] | 40.0% | 62.0% |
| % Ratio of actives [(Ha/A) × 100]] | 80.0% | 88.6% |
| Enrichment factor (E) [(Ha × D)/(Ht × A)] | 11.8 | 18.3 |
| False negatives [A − Ha] | 7 | 4 |
| False positives [Ht − Ha] | 42 | 19 |
| Güner–Henry (GH) scorea | 0.48 | 0.72 |
The pharmacophore features and geometric parameters of hypo-09 are shown in Fig. 2A and B. All features of hypo-09 that were mapped onto the seven inhibitors in the training set are depicted in Fig. 2C. The hydrophobic groups (H1 and H2) are aliphatic groups (isopropyl) and aromatic groups (phenyl); the hydrogen bond donors (D) are hydrogen atoms and oxygen atoms in carbonyl or hydroxyl groups. The selected model was then prepared for virtual screening. As shown in Fig. 2C, the pharmacophore cannot be extended to the left hand side (as drawn) of the inhibitors in the training set because of the random orientation of the hydrophobic groups. The combined docking method was used to complement the pharmacophore model during virtual screening.
Virtual screening of specs library using the pharmacophore model and molecular docking
Virtual screening has been used extensively in the development of pharmacological agents.31–33 We performed a combination of ligand-based and structure-based virtual screening to identify novel lead compounds. Pharmacophore screening was carried out using the Ligand Profiler protocol in DS 4.5 and molecular docking was implemented using Sybyl-X2.0 (Tri-I Biotech, Shanghai Inc).Firstly, the ligand-based pharmacophore model, hypo-09, was used as a 3D query to search the Specs library, which contains 536![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 683 compounds. The Ligand Profiler protocol with flexible fitting was used to screen this library. Fit values were obtained for the compounds and a set of 2017 hits with fit values ≥3 were selected. Next, the crystal structure of Cat K was processed using the Protein Preparation protocol in Sybyl-X2.0 and the location of the ligand in the co-crystal structure was defined as the active site. The hits obtained from pharmacophore screening were docked into the active site of Cat K. For each ligand, 10 docking poses were generated and only the best scoring pose was retained for the overall ranking. Following this procedure, 103 potential test compounds with total scores ≤−7 kcal mol−1 were identified. The final selection of test compounds was based on fit values of pharmacophore matching, structural diversity, binding pocket location, docking score and commercial availability. Twenty-two compounds (Fig. 3) were bought and tested for biological activity in vitro.
683 compounds. The Ligand Profiler protocol with flexible fitting was used to screen this library. Fit values were obtained for the compounds and a set of 2017 hits with fit values ≥3 were selected. Next, the crystal structure of Cat K was processed using the Protein Preparation protocol in Sybyl-X2.0 and the location of the ligand in the co-crystal structure was defined as the active site. The hits obtained from pharmacophore screening were docked into the active site of Cat K. For each ligand, 10 docking poses were generated and only the best scoring pose was retained for the overall ranking. Following this procedure, 103 potential test compounds with total scores ≤−7 kcal mol−1 were identified. The final selection of test compounds was based on fit values of pharmacophore matching, structural diversity, binding pocket location, docking score and commercial availability. Twenty-two compounds (Fig. 3) were bought and tested for biological activity in vitro.
Biological evaluation
In vitro enzymatic assays
The ability of the compounds identified by virtual screening to inhibit Cat K enzymatic activity was evaluated in vitro, using odanacatib as the positive control. Initial screening was performed at a concentration of 200 μM and IC50 values were then determined for the compounds that showed the greatest inhibition at 200 μM. The results are expressed as inhibition rates (%) and IC50 values (Table 3).| Compound | Specs ID | Inhibition (200 μM, %) | IC50 (μM) | Docking scorec | Fit value |
|---|---|---|---|---|---|
| a Used as positive control.b ND = not determined.c Expressed in kcal mol−1. | |||||
| 1 | AN-329/15538231 | 65.09 | 138.63 | −7.15 | 3.943 |
| 2 | AG-205/41006057 | 77.18 | 62.47 | −7.21 | 3.795 |
| 3 | AO-022/43452566 | 80.33 | 19.21 | −7.46 | 4.299 |
| 4 | AJ-292/40711443 | 77.14 | 24.83 | −7.27 | 4.571 |
| 5 | AL-281/15328108 | 80.18 | 21.43 | −7.79 | 4.209 |
| 6 | AG-690/15432266 | 54.95 | 184.15 | −7.06 | 3.340 |
| 7 | AG-690/15432263 | 40.01 | NDb | −7.21 | 3.376 |
| 8 | AG-670/40735611 | 24.99 | ND | −8.36 | 3.190 |
| 9 | AG-690/11571116 | 46.65 | ND | −7.05 | 4.168 |
| 10 | AG-690/11571105 | 50.09 | ND | −7.23 | 3.879 |
| 11 | AK-968/12971204 | 45.83 | ND | −7.14 | 4.094 |
| 12 | AM-879/40777400 | 45.62 | ND | −7.20 | 3.164 |
| 13 | AM-879/40777423 | 50.70 | ND | −7.65 | 3.108 |
| 14 | AJ-030/12105219 | 49.94 | ND | −7.18 | 3.520 |
| 15 | AK-968/41016994 | 53.71 | 124.39 | −7.08 | 3.196 |
| 16 | AK-968/41017104 | 48.61 | ND | −7.71 | 3.125 |
| 17 | AP-970/41682693 | 54.96 | 149.86 | −7.37 | 3.060 |
| 18 | AN-329/15538232 | 44.33 | ND | −7.36 | 3.921 |
| 19 | AN-979/41713878 | 30.53 | ND | −8.08 | 4.095 |
| 20 | AN-329/43385980 | 37.89 | ND | −7.34 | 3.941 |
| 21 | AP-124/41029494 | 87.80 | 17.73 | −7.48 | 4.451 |
| 22 | AN-329/41563502 | 39.35 | ND | −7.27 | 3.724 |
| Odanacatiba | — | 100 | 0.001 | −7.59 | 4.326 |
At the initial concentration of 200 μM, five compounds (2, 3, 4, 5 and 21) showed >70% inhibition, six compounds (1, 6, 10, 13, 15 and 17) showed 50–70% inhibition, and the remaining eleven compounds showed <50% inhibition (Table 4). Odanacatib showed 100% inhibition at this concentration. Following IC50 determinations, compounds 1, 2, 6, 15 and 17 were found to be relatively weak inhibitors, with IC50 values in the range 62.47–184.15 μM. Compounds 3, 4, 5 and 21 emerged as the most potent inhibitors, with IC50 values of 19.21, 24.83, 21.43 and 17.73 μM, respectively. All of the compounds possessed novel scaffolds compared with known Cat K inhibitors.
| Compound | IC50b (μM) ± SD | ||
|---|---|---|---|
| HCT116 | MCF7 | 143B | |
| a Used as positive control.b Data presented is the mean ± SD value of three dependent determinations. | |||
| 3 | >100 | 26.31 ± 3.46 | 15.54 ± 0.65 |
| 4 | >100 | >100 | >100 |
| 5 | >100 | >100 | 11.26 ± 1.38 |
| 21 | 28.25 ± 2.34 | 16.35 ± 0.887 | 5.05 ± 0.77 |
| Odanacatiba | >100 | 45.38 ± 3.09 | 12.71 ± 1.41 |
| 5-FUa | 8.64 ± 1.64 | 32.63 ± 2.78 | 25.22 ± 1.06 |
In order to observe the possible binding modes of compounds 3, 4, 5 and 21 in the active site of Cat K, the docking results were analyzed (Fig. 4). Classical H-bonds, non-classical carbon H-bonds, and σ–π and π–π hydrophobic interactions between the compounds and Cat K revealed that compounds 3, 4, 5 and 21 could interact with amino acid residues surrounding the active site of Cat K including Gln19, Gly23, Ser24, Cys25, Trp26, Asp61, Gly65, Gly66, Trp67, Gln76, Arg108, Ala134, Ala137, Ser138, Gln143, Leu160, Asn161, His162, Lys173 and Trp184.
A suggested binding mode for compound 3 inside the Cat K active site is shown in Fig. 4(A1) and (A2). The hydroxyl group serves as a donor to form a classical H-bond interaction with Gly66 and also forms a non-classical H-bond interaction with Trp26. The amide and methyl groups form non-classical H-bond interactions with Asn161 and Ser138, respectively. The phenyl group at the right terminal in Fig. 4(A1) forms hydrophobic interactions with Asn161, Tyr67 and Ala134. A suggested binding mode for compound 4 is shown in Fig. 4(B1) and (B2). Two carbonyl groups make classical H-bond interactions with Gln19 and Gly66. The amide group acts as a classical H-bond donor to Asn161. The oxygen atoms of two amide groups were also observed to form non-classical H-bond interactions with Gly23 and Trp26. The methyl and phenyl groups form hydrophobic interactions with Cys25, Try67 and Ala137. The proposed binding mode of compound 5 is shown in Fig. 4(C1) and (C2). The carbonyl group serves as both H-bond donor and acceptor, forming interactions with Gln19 and His162. It also forms a non-classical H-bond interaction with Gly23. Two amide groups donate H-bonds to Asn161 and the carbonyl group connected to the 3-bromophenyl moiety forms a classical H-bond interaction with Gly66. Two phenyl groups also form hydrophobic interactions with Tyr67 and Ala134. The proposed binding mode of compound 21 is shown in Fig. 4(D1) and (D2). Three hydroxyl groups serve as classical H-bond donors to form interactions with Ala137, Gln143 and Gln19. Compound 21 makes a classical H-bond interaction and a non-classical H-bond interaction with Cys25 and Asn161, respectively.
Hydrophobic interactions were also observed with Gly65 and Tyr67.
The binding mode of odanacatib is shown in Fig. 4(E1) and (E2). Odanacatib accepts a classical H-bond from Trp184 and forms hydrophobic interactions with Cys25 and Tyr67. It is well-established that H-bond and hydrophobic interactions are essential for biological activity28,30 and compounds 3, 4, 5 and 21, which have been identified to be Cat K inhibitors, make these types of interaction with the enzyme. Compounds 4, 5 and 21 fit into the pocket formed by Asp61 and Tyr 67 and, since this pocket shows different characteristics from pockets in cathepsins B, L and S,34 compounds 4, 5 and 21 might be selective for Cat K. Understanding the binding modes of these compounds in the active site of Cat K will be useful for the development of more potent Cat K inhibitors. The anti-proliferative activities of compounds 3, 4, 5 and 21 were evaluated in the following experiments.
In vitro activities against tumor cells
The anti-proliferative activities of compounds 3, 4, 5 and 21 were evaluated against HCT116 (human colon cancer), MCF7 (human breast cancer) and 143B (human osteosarcoma) cell lines using the MTT assay. Odanacatib and 5-FU were used as the positive controls. Results are the mean of three experiments, expressed as IC50 values (Table 4).Compound 21 showed clear inhibitory activity against all three cancer cell lines. The IC50 values of compound 21 were 28.25, 16.35 and 5.05 μM against HCT116, MCF7 and 143B cell lines, respectively. Compound 21 was more potent than odanacatib against all three cell lines and more potent than 5-FU against MCF7 and 143B cells. Interestingly, compound 21 showed superior activity against 143B cells than against the other two cell lines. Compounds 3 and 5 also showed greater activity against the 143B cell line. Compound 4 had no activity against any of the three cell lines. Three Cat K inhibitors (3, 5 and 21), identified using virtual screening, thus had selective anti-proliferative activity against 143B cells. These inhibitors provide a sound basis for the development of novel agents to treat osteosarcoma.
No biological activities for compounds 3, 4 and 5 have, so far, been reported. Compound 21 was identified by Rajesh35 during a study to discover anti-inflammatory CCR2 antagonists using virtual screening, but no biological test results were reported. As the most potent Cat K inhibitor identified in the present study, compound 21 is characterized, for the first time, as a lead compound for the discovery of novel anti-tumor agents. Surprisingly, compound 21 showed higher anti-proliferative activity than odanacatib, despite having much lower Cat K inhibitory activity. This suggests that compound 21 may have additional biological activities that should be investigated further.
Conclusion
In summary, the integrative application of virtual screening and experimental tests has successfully identified new potent Cat K inhibitors. In this study, a ligand-based pharmacophore model with five features was generated for the first time and successfully used for virtual screening. Biological testing of twenty-two selected hit compounds in enzymatic assays identified four inhibitors (compounds 3, 4, 5 and 21), with IC50 values of 19.21, 24.83, 21.43 and 17.73 μM, respectively. The binding modes of these four compounds in the enzyme active site were also analyzed. Compounds 3, 4, 5 and 21 were then progressed to cell-based bioassays using the MTT method. Compound 21 showed the most potent anti-proliferative activity with IC50 values of 28.25, 16.35 and 5.05 μM against HCT116, MCF7 and 143B cell lines, respectively. Surprisingly, compounds 3, 5 and 21 showed selective activity against the human osteosarcoma cell line, 143B. These compounds may thus provide good starting points for the development of novel Cat K inhibitors for the treatment of osteosarcoma. The combined use of computational and experimental approaches in the present study has generated new leads and is thus an effective tool for the rational identification of lead compounds.Experimental section
Establishment and validation of the common feature pharmacophore model
Common feature pharmacophore models were established using the Common Feature Pharmacophore Generation protocol of the DS 4.5 software. Seven inhibitors, for which co-crystal structures were available, were selected as the training set to perform the experiments. All of the ligands used in the training set were extracted from their crystal structures and checked for bond orders. Since the protein-bound conformations of the ligands in the training set were taken into consideration, an initial diverse conformation generation step was not performed. H-Bond acceptor (A), H-bond donor (D), hydrophobic aliphatic (H1) and hydrophobic aromatic (H2) features were specified as the pharmacophore features. The maximum number of pharmacophore hypotheses was set to ten. The minimum feature option was 4 and the maximum feature option was 6. The minimum inter-feature distance was set to 2.5 Å. The values of MaxOmitFeat and Principal were set to 0 and 2, respectively. All other parameters were set at their default values.The validity of the models was evaluated using fit values and GH scores. The Ligand Profiler module in DS 4.5 was used to map the training set ligands onto the pharmacophore models. Fit values were obtained by mapping the structural features of the molecules to the pharmacophore features. High fit values indicated an excellent model. The GH score method36 quantified the validity of the pharmacophore model by retrieving active compounds from a test set of compounds containing 35 active Cat K inhibitors and 1300 inactive decoy molecules that were randomly selected from the ZINC database (available online at http://zinc.docking.org). The active Cat K inhibitors in the test set were different from the ligands in the training set. The GH scores were in the range of 0.6 to 1, indicating that the pharmacophore model was valid.
Virtual library screening using the pharmacophore model
Compounds from the Specs library (536![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 683 compounds) were virtually screened using the Ligand Profiler protocol in DS 4.5. To narrow down the list of possible hits for in vitro biological testing, the following criteria were applied: log
683 compounds) were virtually screened using the Ligand Profiler protocol in DS 4.5. To narrow down the list of possible hits for in vitro biological testing, the following criteria were applied: log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) P < 5, molecular weight < 600, number of hydrogen bond acceptors ≤ 10, number of hydrogen bond donors ≤ 5, number of rotatable bonds ≤ 10 and polar surface area < 140 Å2. Compounds that met these criteria were converted from 2D formatting to 3D structures using the Ligand Preparation program in DS 4.5, which includes protonation and tautomer generation. Final coordinates were energy minimized using CHARMM force fields. The selected pharmacophore model was used as the screening template to identify novel Cat K inhibitors. The Fitting Method was set to flexible and Maximum Omitted Features was set to −1. To avoid removal of potential inhibitors, two pharmacophore features were allowed to be missing when mapping the ligands to the model. Compounds with high fit values and drug-like properties were selected and saved in an SD file for subsequent molecular docking studies.
P < 5, molecular weight < 600, number of hydrogen bond acceptors ≤ 10, number of hydrogen bond donors ≤ 5, number of rotatable bonds ≤ 10 and polar surface area < 140 Å2. Compounds that met these criteria were converted from 2D formatting to 3D structures using the Ligand Preparation program in DS 4.5, which includes protonation and tautomer generation. Final coordinates were energy minimized using CHARMM force fields. The selected pharmacophore model was used as the screening template to identify novel Cat K inhibitors. The Fitting Method was set to flexible and Maximum Omitted Features was set to −1. To avoid removal of potential inhibitors, two pharmacophore features were allowed to be missing when mapping the ligands to the model. Compounds with high fit values and drug-like properties were selected and saved in an SD file for subsequent molecular docking studies.
Molecular docking
The X-ray crystal structure of Cat K (PDB code 1NLJ) was used for the docking studies. The reason for selecting 1NLJ is illustrated in the ESI.† 1NLJ was processed using the Protein Preparation protocol in Sybyl-X2.0. Firstly, the ligand was extracted from the complex to determine the location of the active site. Secondly, backbones and sidechains of the protein were repaired. Thirdly, all hydrogen atoms were added and the protein was protonated at pH 7.4. Fourthly, the Amber7 FF99 force field was applied to the biopolymer. A Surflex-Dock Control File containing information about the protein and the active site was then defined with a threshold value of 0.5. The maximum number of poses per ligand was set to 10. The ligand in the crystal structure was defined as the reference molecule. Fifthly, ligands obtained from the pharmacophore virtual screening were minimized using the AMBER7 FF99 force field and charged with Gasteiger–Huckel method. All other parameters were set at their default values. Finally, structure-based docking was carried out using the Surflex-Dock Geom mode of Sybyl-X2.0.Biological evaluation
Enzyme inhibition studies
The Cat K inhibitory activity of the compounds was determined using a previously described method37 with minor modifications. Compounds were firstly screened at 200 μM and those which showed >50% inhibition at 200 μM were then screened at eight concentrations (0.091–200 μM) to obtain IC50 values.Recombinant human Cat K (5.5 ng mL−1) in reaction buffer (50 μL) containing sodium acetate (50 mM), EDTA (2.5 mM) and DTT (1 mM) was added to a 96-well plate containing the test compounds at pH 6.0. The plate was incubated at 37 °C for 15 min and the reaction was then initiated by adding a solution of the substrate, Z-Phe-Arg-AMC (20 μL, 10 μM). After 30 min, the fluorescence intensity was measured (excitation at 355 nm; emission at 460 nm) using a microplate reader (Wallac 1420 Victor2, Perkin-Elmer, USA). The inhibition rate was determined using the fluorescence intensity and IC50 values were calculated from the inhibition curves.
Cell culture experiments
The anti-proliferative activities of compounds 3, 4, 5 and 21 were evaluated against HCT116, MCF7 and 143B cell lines (ATCC, USA) in vitro using the standard MTT assay, with odanacatib and 5-FU as the positive controls. Compounds were tested at six concentrations (0.001–100 μM).The cancer cell lines were cultured in RPMI 1640 medium (Sigma-Aldrich, USA), supplemented with 10% fetal bovine serum (FBS, Sigma-Aldrich, USA). Approximately 4 × 103 cells, suspended in RPMI 1640 medium, were plated into each well of a 96-well plate and incubated at 37 °C for 24 h in 5% CO2. The test compounds were added to the culture medium and the cells were incubated for 48 h. Fresh MTT was added to each well at the terminal concentration (0.5 mg mL−1) and the cells were incubated at 37 °C for 4 h in 5% CO2. DMSO (150 μL) was then added to each well to dissolve the formazan crystals and the absorbance at 540 nm was measured using a microplate reader (HEALES MB-580, China). The results, expressed as IC50 values, were the average of three determinations and were calculated using SPSS software.
Acknowledgements
The work was supported by the National Natural Science Foundation of China (81473097), Student Fund of Innovation Project of Peking Union Medical College (2015-1007-22), and the National Mega-Project for Innovative Drugs (2012ZX09301002-003).Notes and references
- T. Inaoka, G. Bilbe, O. Ishibashi, K. Tezuka, M. Kumegawa and T. Kokubo, Biochem. Biophys. Res. Commun., 1995, 206, 89–96 CrossRef CAS PubMed.
- A. G. Dossetter, H. Beeley, J. Bowyer, C. R. Cook, J. J. Crawford, J. E. Finlayson, N. M. Heron, C. Heyes, A. J. Highton, J. A. Hudson, A. Jestel, P. W. Kenny, S. Krapp, S. Martin, P. A. MacFaul, T. M. McGuire, P. M. Gutierrez, A. D. Morley, J. J. Morris, K. M. Page, L. R. Ribeiro, H. Sawney, S. Steinbacher, C. Smith and M. Vickers, J. Med. Chem., 2012, 55, 6363–6374 CrossRef CAS PubMed.
- J. J. Crawford, P. W. Kenny, J. Bowyer, C. R. Cook, J. E. Finlayson, C. Heyes, A. J. Highton, J. A. Hudson, A. Jestel, S. Krapp, S. Martin, P. A. Macfaul, B. P. McDermott, T. M. McGuire, A. D. Morley, J. J. Morris, K. M. Page, L. R. Ribeiro, H. Sawney, S. Steinbacher, C. Smith and A. G. Dossetter, J. Med. Chem., 2012, 55, 8827–8837 CrossRef CAS PubMed.
- J. Borisek, M. Vizovisek, P. Sosnowski, B. Turk, D. Turk, B. Mohar and M. Novic, J. Med. Chem., 2015, 58, 6928–6937 CrossRef CAS PubMed.
- F. Lecaille, D. Bromme and G. Lalmanach, Biochimie, 2008, 90, 208–226 CrossRef CAS PubMed.
- P. Garnero, O. Borel, I. Byrjalsen, M. Ferreras, F. H. Drake, M. S. McQueney, N. T. Foged, P. D. Delmas and J. M. Delaisse, J. Biol. Chem., 1998, 273, 32347–32352 CrossRef CAS PubMed.
- W. Kafienah, D. Bromme, D. J. Buttle, L. J. Croucher and A. P. Hollander, Biochem. J., 1998, 331, 727–732 CrossRef CAS PubMed.
- M. J. Quintanilla-Dieck, K. Codriansky, M. Keady, J. Bhawan and T. M. Runger, J. Invest. Dermatol., 2008, 128, 2281–2288 CrossRef CAS PubMed.
- M. K. Herroon, E. Rajagurubandara, D. L. Rudy, A. Chalasani, A. L. Hardaway and I. Podgorski, Oncogene, 2013, 32, 1580–1593 CrossRef CAS PubMed.
- J. H. N. Lindeman, R. Hanemaaijer, A. Mulder, P. D. S. Dijkstra, K. Szuhai, D. Bromme, J. H. Verheijen and P. C. W. Hogendoorn, Am. J. Pathol., 2004, 165, 593–600 CrossRef CAS PubMed.
- M. Ishida, F. Kojima and H. Okabe, J. Eur. Acad. Dermatol. Venereol., 2013, 27, 128–130 CrossRef PubMed.
- A. J. Littlewood-Evans, G. Bilbe, W. B. Bowler, D. Farley, B. Wlodarski, T. Kokubo, T. Inaoka, J. Sloane, D. B. Evans and J. A. Gallagher, Cancer Res., 1997, 57, 5386–5390 CAS.
- K. Husmann, R. Muff, M. E. Bolander, G. Sarkar, W. Born and B. Fuchs, Mol. Carcinog., 2008, 47, 66–73 CrossRef CAS PubMed.
- J. Y. Gauthier, N. Chauret, W. Cromlish, S. Desmarais, L. T. Duong, J. P. Falgueyret, D. B. Kimmel, S. Lamontagne, S. Leger, T. LeRiche, C. S. Li, F. Masse, D. J. McKay, D. A. Nicoll-Griffith, R. M. Oballa, J. T. Palmer, M. D. Percival, D. Riendeau, J. Robichaud, G. A. Rodan, S. B. Rodan, C. Seto, M. Therien, V. L. Truong, M. C. Venuti, G. Wesolowski, R. N. Young, R. Zamboni and W. C. Black, Bioorg. Med. Chem. Lett., 2008, 18, 923–928 CrossRef CAS PubMed.
- S. A. Stoch, S. Zajic, J. A. Stone, D. L. Miller, L. van Bortel, K. C. Lasseter, B. Pramanik, C. Cilissen, Q. Liu, L. Liu, B. B. Scott, D. Panebianco, Y. Ding, K. Gottesdiener and J. A. Wagner, Br. J. Clin. Pharmacol., 2013, 75, 1240–1254 CrossRef CAS PubMed.
- J. Schroder, A. Klinger, F. Oellien, R. J. Marhofer, M. Duszenko and P. M. Selzer, J. Med. Chem., 2013, 56, 1478–1490 CrossRef PubMed.
- A. B. Jensen, C. Wynne, G. Ramirez, W. He, Y. Song, Y. Berd, H. Wang, A. Mehta and A. Lombardi, Clin. Breast Cancer, 2010, 10, 452–458 CrossRef CAS PubMed.
- U. Grabowska, I. Henderson, M. Edlund, L. Vrang, S. Sedig, Y. Terelius, K. Wikstrom, B. L. Sahlberg, T. Chambers and E. Lindstrom, Bone, 2012, 50, 164–165 CrossRef.
- U. Grabowska, M. Jerling, D. Bottiger, T. Larsson, K. Danielson, E. Lindstrom and C. Edenius, J. Bone Miner. Res., 2013, 28, presentation number SU0381, see http://www.asbmr.org/education/2013-abstracts Search PubMed.
- E. Lindstrom, U. Grabowska, M. Jerling and C. Edenius, Osteoarthr. Cartil., 2014, 22, 197 CrossRef.
- S. Kumar, L. Dare, J. A. Vasko-Moser, I. E. James, S. M. Blake, D. J. Rickard, S. M. Hwang, T. Tomaszek, D. S. Yamashita, R. W. Marquis, H. Oh, J. U. Jeong, D. F. Veber, M. Gowen, M. W. Lark and G. Stroup, Bone, 2007, 40, 122–131 CrossRef CAS PubMed.
- J. P. Falgueyret, S. Desmarais, R. Oballa, W. C. Black, W. Cromlish, K. Khougaz, S. Lamontagne, F. Masse, D. Riendeau, S. Toulmond and M. D. Percival, J. Med. Chem., 2005, 48, 7535–7543 CrossRef CAS PubMed.
- T. M. Runger, S. Adami, C. L. Benhamou, E. Czerwinski, J. Farrerons, D. L. Kendler, L. Mindeholm, G. Realdi, C. Roux and V. Smith, J. Am. Acad. Dermatol., 2012, 66, 89–96 CrossRef PubMed.
- S. K. Thompson, S. M. Halbert, M. J. Bossard, T. A. Tomaszek, M. A. Levy, B. Zhao, W. W. Smith, S. S. Abdel-Meguid, C. A. Janson, K. J. D'Alessio, M. S. McQueney, B. Y. Amegadzie, C. R. Hanning, R. L. DesJarlais, J. Briand, S. K. Sarkar, M. J. Huddleston, C. F. Ijames, S. A. Carr, K. T. Garnes, A. Shu, J. R. Heys, J. Bradbeer, D. Zembryki, L. Lee-Rykaczewski, I. E. James, M. W. Lark, F. H. Drake, M. Gowen, J. G. Gleason and D. F. Veber, Proc. Natl. Acad. Sci., 1997, 94, 14249–14254 CrossRef CAS.
- R. W. Marquis, Y. Ru, S. M. LoCastro, J. Zeng, D. S. Yamashita, H. J. Oh, K. F. Erhard, L. D. Davis, T. A. Tomaszek, D. Tew, K. Salyers, J. Proksch, K. Ward, B. Smith, M. Levy, M. D. Cummings, R. C. Haltiwanger, G. Trescher, B. Wang, M. E. Hemling, C. J. Quinn, H. Y. Cheng, F. Lin, W. W. Smith, C. A. Janson, B. Zhao, M. S. McQueney, K. D'Alessio, C. P. Lee, A. Marzulli, R. A. Dodds, S. Blake, S. M. Hwang, I. E. James, C. J. Gress, B. R. Bradley, M. W. Lark, M. Gowen and D. F. Veber, J. Med. Chem., 2001, 44, 1380–1395 CrossRef CAS PubMed.
- R. L. DesJarlais, D. S. Yamashita, H.-J. Oh, I. N. Uzinskas, K. F. Erhard, A. C. Allen, R. C. Haltiwanger, B. G. Zhao, W. W. Smith, S. S. Abdel-Meguid, K. D. Alessio, C. A. Janson, M. S. McQueney, T. A. Tomaszek, M. A. Levy and D. F. Veber, J. Am. Chem. Soc., 1998, 120, 9114–9115 CrossRef CAS.
- R. W. Marquis, D. S. Yamashita, Y. Ru, S. M. LoCastro, H.-J. Oh, K. F. Erhard, R. L. DesJarlais, M. S. Head, W. W. Smith, B. G. Zhao, C. A. Janson, S. S. Abdel-Meguid, T. A. Tomaszek, M. A. Levy and D. F. Veber, J. Med. Chem., 1998, 41, 3563–3567 CrossRef CAS PubMed.
- D. G. Barrett, V. M. Boncek, J. G. Catalano, D. N. Deaton, A. M. Hassell, C. H. Jurgensen, S. T. Long, R. B. McFadyen, A. B. Miller, L. R. Miller, J. A. Payne, J. A. Ray, V. Samano, L. M. Shewchuk, F. X. Tavares, K. J. Wells-Knecht, D. H. Willard Jr, L. L. Wright and H. Q. Zhou, Bioorg. Med. Chem. Lett., 2005, 15, 3540–3546 CrossRef CAS PubMed.
- D. S. Yamashita, R. W. Marquis, R. Xie, S. D. Nidamarthy, H.-J. Oh, J. U. Jeong, K. F. Erhard, K. W. Ward, T. J. Roethke, B. R. Smith, H.-Y. Cheng, X. Geng, F. Lin, P. H. Offen, B. Wang, N. Nevins, M. S. Head, R. C. Haltiwanger, A. A. N. Sarjeant, L. M. Liable-Sands, B. Zhao, W. W. Smith, C. A. Janson, E. Gao, T. Tomaszek, M. McQueney, I. E. James, C. J. Gress, D. L. Zembryki, M. W. Lark and D. F. Veber, J. Med. Chem., 2006, 49, 1597–1612 CrossRef CAS PubMed.
- E. E. Boros, D. N. Deaton, A. M. Hassell, R. B. McFadyen, A. B. Miller, L. R. Miller, M. G. Paulick, L. M. Shewchuk, J. B. Thompson, D. H. Willard Jr and L. L. Wright, Bioorg. Med. Chem. Lett., 2004, 14, 3425–3429 CrossRef CAS PubMed.
- A. N. Jain, Curr. Opin. Drug Discovery Dev., 2004, 7, 396–403 CAS.
- U. Rester, Curr. Opin. Drug Discovery Dev., 2008, 11, 559–568 CAS.
- K. H. Kim, N. D. Kim and B. L. Seong, Expert Opin. Drug Discovery, 2010, 5, 205–222 CrossRef CAS PubMed.
- E. Altmann, S. W. Cowan-Jacob and M. Missbach, J. Med. Chem., 2004, 47, 5833–5836 CrossRef CAS PubMed.
- R. Singh, A. Balupuri and M. E. Sobhia, Mol. Simul., 2013, 39, 49–58 CrossRef CAS.
- L. L. Yang, G. B. Li, H. X. Yan, Q. Z. Sun, S. Ma, P. Ji, Z. R. Wang, S. Feng, J. Zou and S. Y. Yang, Eur. J. Med. Chem., 2012, 56, 30–38 CrossRef CAS PubMed.
- D. Bromme, A. Steinert, S. Friebe, S. Fittkau, B. Wiederanders and H. Kirschke, Biochem. J., 1989, 264, 475–481 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available: Hit values of compounds in the training set mapped to hypotheses 01–10; the reason for selecting 1NLJ as receptor for virtual screening. See DOI: 10.1039/c6ra14251f |
| This journal is © The Royal Society of Chemistry 2016 |