In vitro toxicity, apoptosis and antimicrobial effects of phyto-mediated copper oxide nanoparticles†
Abstract
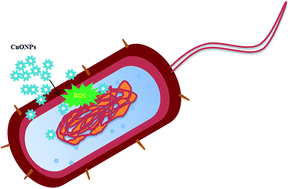
Herein, copper oxide nanoparticles (CuONPs) are proposed for widespread use in emerging biomedical applications. The aim of this study is to synthesize and characterize CuONPs using the aqueous dried fruit extract of Tribulus terrestris and assess their potential in vitro cytotoxicity and antibacterial activity. The CuONPs bio-physical and morphological properties are characterized via different analytical techniques. The synthesized particles are highly stable and spherical with particle sizes in the range of 5–22 nm. Confocal microscopy reveals that ROS generation is the potent mechanism behind the role of CuONPs mediated antimicrobial toxicity. In addition, MTT and apoptotic analyses illustrate that the CuONPs are relatively cytofriendly toward human mesenchymal stem cells and also exhibit anti-cancer activity on human adenocarcinoma AGS cell lines. These results clearly indicate that the prepared copper oxide nanoparticles are safe and their application can be expanded as effectual antimicrobial and anticancer agents for pharmaceutical applications.

 Please wait while we load your content...
Please wait while we load your content...