Open Access Article
Open Access ArticleCrystalline pyrazine-2-amidoxime isolated by diffusion method and its structural and behavioral analysis in the context of crystal engineering and microbiological activity†
Agnieszka
Chylewska
*a,
Małgorzata
Ogryzek
a,
Angelika
Głębocka
a,
Artur
Sikorski
a,
Katarzyna
Turecka
b,
Ewa. D.
Raczyńska
c and
Mariusz
Makowski
*a
aFaculty of Chemistry, University of Gdansk, W. Stwosza 63, 80-308 Gdansk, Poland. E-mail: agnieszka.chylewska@ug.edu.pl; mariusz.makowski@ug.edu.pl; Tel: +48 58 523 50 52 Tel: +48 58 523 50 55
bFaculty of Pharmacy with Subfaculty of Laboratory Medicine, Al. Hallera 107, 80-416 Gdansk, Poland
cDepartment of Chemistry, Warsaw University of Life Sciences, Nowoursynowska 159c, 02-776 Warszawa, Poland
First published on 28th June 2016
Abstract
Pyrazine-2-amidoxime (PAOX) is a structural analogue of a popular drug, i.e. pyrazine-2-carboxamide (PZA). The crystalline PAOX was obtained by diffusion as a method of crystallization. Various types of intermolecular interactions between the H-bond donors (CH, NH, and OH) and H-bond acceptors (hydroxyl O, imino N, aza N) were found between PAOX molecules in X-ray diffraction studies. It was observed in the crystal structure that PAOX forms not only dimers but also stable helical-like polymers, stabilized by intermolecular interactions between two neighbouring molecules. Their geometric, energetic and spectroscopic properties were also characterised by DFT methods. Thermal decomposition of PAOX was examined with the use of a TG-IR analysis (20–1000 °C), and the results were further resolved. Electrochemical behaviour of the compound studied in acetonitrile in the absence or presence of methanol was described in detail, and mechanisms of the anodic oxidation and cathodic reduction were proposed. The complexometric properties of PAOX were examined against selected d-block metal ions in acetonitrile, as well as in aqueous solution. Biological assay of PAOX was performed to determine the antimicrobial activity and potential pharmaceutical applications. The minimum inhibitory (MIC) and minimal bactericidal (or fungicidal) concentrations (MBC/MFC) for PAOX were determined against six microorganisms.
Introduction
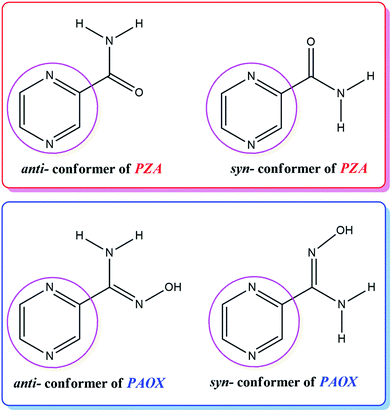
The discovery of the biological activity of pyrazine derivatives1–3 led to the systematic search for potential drugs within the discussed class of compounds. The pyrazine ring system has proven to be an interesting class in heterocyclic chemistry but it has received little attention in the literature so far. Some of its derivatives are important as anticancer agents4 with low toxicity and blood platelet aggregation inhibitors.5 They also show anti-inflammatory, antifungal, antiparasitic and antioxidant activities.6–11 In addition, pyrazine derivatives are used as bone metabolism improvers, as adenosine antagonists, and as controlling herbicides.12Amidoximes retain a complementary position in drug design and discovery, being individually considered as pharmacophores and playing a role as parent compounds in various transformations.13 Moreover, amidoximes can serve as starting materials for the synthesis of multiple heterocyclic compounds and could be useful as building blocks for a number of heteroarenes.14,15 It has also been reported that many drugs containing the amidoxime group are biologically active and display antihypertensive, antibacterial, antitrypanocidic and cytostatic properties.16,17 They were found as NO generators in vivo, which had neuromodulatory and neurotransmitory effects.18 The cyclic amidoximes were also reported to possess anamnestic, antihypoxive and hypotensive activities.19,20 Homo- and heterometallic oximate complexes have played a considerable role in coordination chemistry and crystal engineering, because of their fascinating topological geometries and interesting properties.21 One of the most appropriate examples may be pyrazine-2-amidoxime (PAOX), which structure corresponds to the structure of pyrazine-2-carboxamide (PZA) (Fig. 1).
 | ||
| Fig. 1 Molecule structure of pyrazine-2-amidoxime (PAOX) as analogue of known pyrazine-2-carboxamide (PZA). | ||
The choice of PAOX for our studies was based on the following reasons. The ability of this molecule to coordinate metal ions through one or both N and O donor atoms of the amidoxime group as well as one or both N donor atoms of the pyrazine ring.22 In this case, PAOX can act as a chelating terminal or bridging ligand (Fig. 2), and shows the interesting mode of self-activation by metal ions, which could be used in synthetic inorganic chemistry.23 It is worth pointing out here that the structure of pyrazine-2-amidoxime as a ligand should directly influence the structure of complexes in the presence of such an organic ligand in the coordination sphere; the formation of clusters or two-dimensional molecular brick–wall complexes with Ni(II) or Fe(II) ions has already been examined.24,25 The same relation can be possible in the case of its magnetic or other properties.24,25 Because solubility, polymorphism, crystallinity and crystal habits are solvent-dependent, thus solvent screening is fundamental and essential, especially in the pharmaceutical industry.24,25 That is why the study in solution is a very important assay and allows the determination of optimal conditions of crystallization and complexometric titrations.
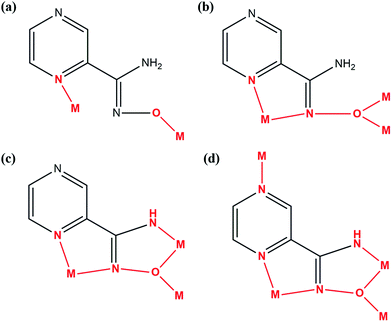 | ||
| Fig. 2 Coordination modes of metal – PAOX in different types of complexes: (a) and (b) dinuclear; (c) trinuclear; (d) tetranuclear.25 | ||
The designing of chelators with high selectivity for metal ions are very important because of their potential analytical applications; they are significant for the removal, separation and increasing concentration of metallic species. They can be potentially applied in the recycling of resources in metallurgy or in the waste water treatment. Transition metal ions such as Cd(II), Pb(II) or Hg(II) are recognized as highly toxic and their presence in the environmental waters or soils is undesirable.24,25 Cu(II), Ni(II) and Co(II) are bio-essential and there are numerous metalloenzymes containing these elements. However, even essential metal ions can be toxic when present in excess.26,27 Therefore, separation of these trace metals is necessary due to their potential health and ecological hazard. The complexation and absorption process is one of the ways which can be used for the removal of metals from waste water. For this purpose, PAOX has been examined and its complexometric properties investigated in aqueous and acetonitrile solutions.
Some oxime derivatives were indicated as antioxidants with scavenging activity towards different forms of reactive species28–30 (e.g. hydrogen peroxide, hydroxyl or nitric oxide radicals), which was also related to their metal chelating properties.31 The investigation of the redox behaviour of PAOX, using electrochemical technique, leads to better understanding of its biological redox reactions or generally – for this class of compounds. The mechanism of pyridinium oxime reduction was investigated and published about twenty years ago.32–34 Although this area for pyridinium oximes is well understood in organic electrochemistry, no experimental data are available on the redox properties of PAOX.
The structural properties of pyrazine-2-amidoxime were investigated here in the solid state and in solution as well as in the gas phase by performing density functional theory (DFT) calculations. Our study of inter- and intramolecular interactions between side chain of PAOX individual units resulted in the new knowledge about the three-dimensional helix-like formation. Due to the above, PAOX can play the role of biomimetic foldamer, which is generally an artificial model for molecular folding and could express other functions (catalyst, receptor). The crystal forms of each foldamer need to be resolved at first, in order to provide the information for the following step of studies about its interaction properties or functional abilities. Here, we investigate co-crystallization structural studies of other pyrazine derivatives.35,36 Although, we focused here on untypical properties, such as aromatic amidoxime as foldamer, its conformation, thermal, redox and complexometric properties.
Results and discussion
Crystallographic studies
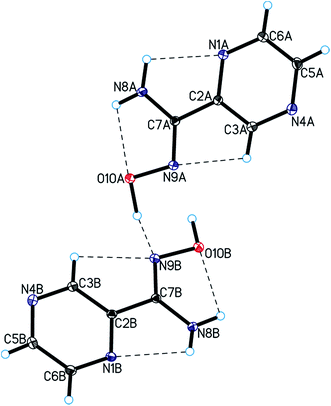
Single-crystal X-ray diffraction measurements showed that pyrazine-2-amidoxime crystallizes in the monoclinic P21 space group with two PAOX molecules (denote as A and B) in the asymmetric unit (Fig. 3, Table S1†).The bond lengths, valence and torsion angles (Tables S2–S4†) characterizing the geometry of the pyrazine ring and amidoxime moieties are consistent with the geometric parameters observed for these groups of compounds.37,38 In the PAOX crystals, pyrazine rings are nearly planar with an average deviation from planarity of 0.001(2) and 0.012(2)° for molecules A and B, respectively, and form a dihedral angle of 10.2(2) and 3.6(2)° with the amidoxime group in molecules A and B, respectively. In the crystal structure of pyrazine-2-amidoxime, intramolecular N–H⋯O, N–H⋯N, and C–H⋯N and intermolecular O–H⋯N, N–H⋯N, and C–H⋯O hydrogen bonds stabilize the structure (Fig. 3, Table 1). An analysis of the crystal packing of the title compound showed that adjacent molecules of PAOX are linked by O10–H10⋯N9 hydrogen bonds and interact by π(pyrazine)–π(pyrazine) interactions (Fig. 4, Table 1).
| D–H⋯A | d(D–H) (Å) | d(H⋯A) (Å) | d(D⋯A) (Å) | <D–H⋯A (°) |
|---|---|---|---|---|
| a Symmetry code: (i) −1 + x, y, z; (ii) −x, −1/2 + y, −z; (iii) 1 − x, 1/2 + y, 1 − z; (iv) x, −1 + y, z; (v) x, 1 + y, z; (vi) 1 − x, −1/2 + y, 1 − z; (vii) 1 − x, 1/2 + y, −z. | ||||
| Intramolecular | ||||
| N8A–H8A⋯O10A | 0.92(3) | 2.18(3) | 2.535(2) | 102(2) |
| N8B–H8B⋯O10B | 0.89(3) | 2.31(3) | 2.599(2) | 99(2) |
| N8A–H8C⋯N1A | 0.96(3) | 2.33(3) | 2.717(2) | 103(2) |
| N8B–H8D⋯N1B | 0.87(3) | 2.35(2) | 2.730(3) | 106(2) |
| C3A–H3A⋯N9A | 0.93 | 2.52 | 2.817(3) | 99 |
| C3B–H3B⋯N9B | 0.93 | 2.47 | 2.781(3) | 100 |
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) |
||||
| Intermolecular | ||||
| O10A–H10A⋯N9B | 0.92(3) | 1.82(3) | 2.734(2) | 170(3) |
| O10B–H10B⋯N9Ai | 0.99(3) | 1.86(3) | 2.849(2) | 174(2) |
| N8A–H8A⋯N4Bii | 0.92(3) | 2.42(3) | 3.316(3) | 165(3) |
| N8B–H8B⋯N4Aiii | 0.89(3) | 2.45(3) | 3.266(3) | 153(2) |
| N8A–H8C⋯N1Biv | 0.96(3) | 2.22(3) | 3.054(3) | 144(2) |
| N8B–H8D⋯N1Av | 0.87(3) | 2.48(3) | 3.275(3) | 151(2) |
| C5A–H5A⋯O10Bvi | 0.93 | 2.51 | 3.273(3) | 139 |
| C5B–H5B⋯O10Avii | 0.93 | 2.50 | 3.374(3) | 156 |
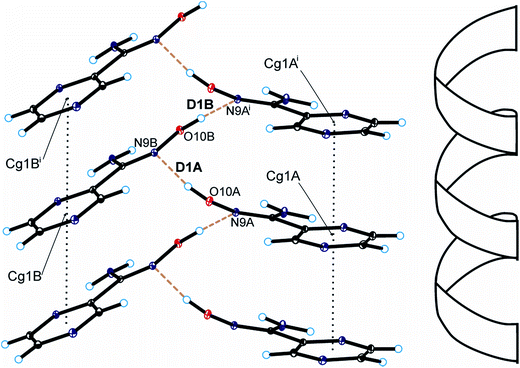 | ||
| Fig. 4 Helical arrangement of molecules of pyrazine-2-amidoxime in the crystal packing of the title compound (hydrogen bonds are represented by dashed lines and π–π interactions by dotted lines; symmetry codes are the same as in Table S2†). | ||
As a result, we can observe a helical-like arrangement of the molecules of PAOX in the crystals. The neighbouring helical-like structures are linked by N8–H8⋯N1 hydrogen bonds (dimer D2) to produce blocks along the a-axis (Fig. 5, Table 1). The adjacent blocks are linked by N8–H8⋯N4 and C5–H5⋯O10 hydrogen bonds to form a three-dimensional framework structure (Fig. 6, Table 1).
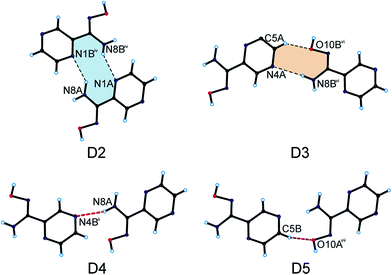 | ||
| Fig. 5 Dimers formed by PAOX molecules in the crystal packing of the title compound (hydrogen bonds are represented by dashed lines; symmetry codes are the same as in Table S2†). | ||
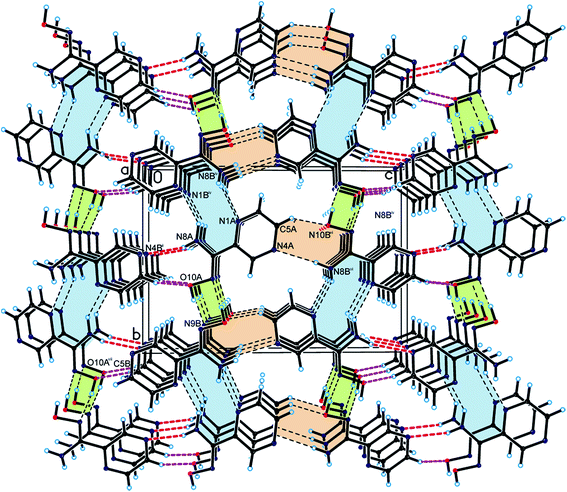 | ||
| Fig. 6 Crystal packing of the title compound viewed along the a-axis (helical arrangement is highlighted in green; hydrogen bonds are represented by dashed lines). | ||
Because of the existence of several possible interaction sites in the structure of PAOX36 by itself and crystallographic results included in this work formation energies of five possible dimers (D1 in Fig. 3 and D2-D5 in Fig. 5, respectively) and one tetramer (T1) to model helical-like turn (Fig. 4) were considered. The T1 form was chosen as a smallest fragment of the helical-like structure to in order check its stability in comparison to dimers. The frequencies were calculated, first to prove that the structures correspond to the energy minima, and then to estimate the corresponding zero-point energies and to assess their theoretical IR, Raman, and UV spectra for all the structures discussed in this paper. These calculations allowed to assess the most stable and probable organizations of PAOX molecules in its crystal structure. According to the results of calculations performed in this work dimers D2, D4, and D5 are alike. For this reason, only one structure D2 was considered in the following discussion. Free energies and atom coordinates of optimized structures of polymers (D1, D2, D3, and T1), obtained by using the DFT method (B3LYP/6-311+G**) in vacuo were collected in Table S5 in the ESI.† The counterpoise procedure was considered in the energy calculations. The formation energies of polymers were collected in Table 2.
The crystallographic data (coordinates) obtained in this work were used in the calculations as starting points and allowed to assess the formation energies of the systems studied. Based on the results in Table 2 it was found that the most stable, and probable interaction system in the crystal structures of PAOX is D1 with the complexation energy −11.31 kcal mol−1. This result is in agreement with the crystal analysis. As shown in Table 1, the shortest hydrogen bonds were found in the case when two PAOX molecules were planar, interacted through amidoxime group (Fig. 3), and were stabilized by two hydrogen bonds N9A⋯O10B, and O10A⋯N9B, with the distances experimentally found at about 2.85 and 2.73 Å, respectively. The second energetically preferred system found in the crystal structure of PAOX was its tetramer, modelling the helical-like fragment, with the formation energy −7.63 kcal mol−1.
The experimental results showed that four PAOX molecules in the tetramer (T1) could form a helical-like turn. All possible hydrogen bond distances were longer for T1 than those in the case of D1 but shorter than those for D2, respectively. In terms of complexation energy D3 was slightly more stable than D2. For D2 and D3, the formation energies were about −4.00 kcal mol−1. The knowledge of the PAOX's crystal structure may have a cognitive importance for supramolecular chemistry and can be implemented from the crystal engineering's point of view in the designing of complexes containing this compound.
Spectroscopic characterization
The infrared and Raman spectroscopies together with quantum-chemical calculations performed in parallel are important techniques of analysis used for confirming the structure of compounds. The vibrational band assignments of PAOX have been made in accordance with their position, magnitude, nature and relative intensities. Also the assignments have been made in analogy with the structurally related molecules. On this basis, characteristic bands of aromatic and aliphatic groups of the C–H–N–O scaffold were identified. The results of the analyses of the theoretical and experimental FT-IR and Raman spectra for monomer, the most energetically stable dimer D1, and solid pyrazine-2-amidoxime are collected in Table S6 and shown in Fig. S1 and S2, respectively, in the ESI.† The vibrational frequencies obtained by DFT calculation are also shown in Table S6† for comparison.The comparison of infrared and Raman spectra of PAOX and structural data provide specific vibrations. In the experimental FT-IR range 3437–3331 cm−1, the peaks generated via symmetric and asymmetric stretching modes of NH2 group are observed as strong and sharp for solid PAOX. They correspond to multiple interactions possible in the solid state. The calculated FT-IR frequencies for monomeric PAOX and the dimer D1 are equal to 3587, 3414 (monomer), and 3531, 3410 cm−1 (dimer) respectively. Similarly, the same signals can be found in the Raman spectra with their values being of 3537 and 3425 cm−1 for monomeric PAOX, and 3532 and 3410 cm−1 for D1. The associated O–H stretching vibration generated in all cases studied, i.e. experimental broad band at 3144, and calculated for monomer and D1 band at 3657 and 3294 cm−1, respectively, in their infrared spectra, and 3060, 3657, 3294 cm−1 in their Raman spectra. The C–H stretching vibrations of hetero aromatic structure occur in the region 3100–3000 cm−1 for asymmetric and 2990–2900 cm−1 for symmetric stretching modes of vibration.39,40 In many cases, they are too weak for detection. Hence in the present study, the experimentally observed C–H symmetric stretching peaks were found at 2816 cm−1, and those calculated at 3016, and 3017 cm−1 for isolated D1.
The band observed at 3059 cm−1 (calculated 3037–2999, and 3017 cm−1, respectively) is associated with the asymmetric stretching vibration C–H in the FT-Raman spectrum. The C–H-out-of-plane bending modes occur in the spectral range41 900–667 cm−1. The bands observed in the region 1153–1018 cm−1 are assigned to ™(CH) in-plane vibrations in experimental FT-IR spectrum. The ™(CH) out-of-plane modes generated sharp bands 862 and 844 cm−1. The corresponding peaks appeared at 1188–1056 cm−1 and 845 cm−1 in the FT-Raman spectrum. Based on the DFT calculated vibrational spectra, the C–H stretching vibrations are assigned to the calculated values 1128–1043 (1130 for D1), 825–805 (823 for D1), and 1134–1020 (1130 for D1) cm−1 in the case of FT-IR and FT-Raman spectra, respectively.
In the present investigations, a very strong, sharp band was found at 1659 cm−1 in the experimental FT-IR along with 1662 cm−1 in the experimental FT-Raman. These two signals are associated with C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N stretching vibration of oxime group. In our case the experimentally observed values of peak due to pyrazine ring C
N stretching vibration of oxime group. In our case the experimentally observed values of peak due to pyrazine ring C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N stretching vibrations were found at 1590 cm−1 (FT-IR) and 1576 cm−1 (Raman). The theoretically calculated values of C
N stretching vibrations were found at 1590 cm−1 (FT-IR) and 1576 cm−1 (Raman). The theoretically calculated values of C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N stretching vibration of oxime group were found at 1619 (monomer), 1627 (D1) in both types of spectra.
N stretching vibration of oxime group were found at 1619 (monomer), 1627 (D1) in both types of spectra.
The absorption peaks in the range 1470–1525 cm−1 in the heterocyclic compounds are ascribed to carbon vibrations (C–C).42 In the present investigation, the FT-IR bands appeared at 1520, 1483 cm−1 (calculated for monomer at 1510 and 1472, and D1 at 1426 cm−1) and FT-Raman bands were observed at 1521 and 1491 cm−1 [calculated 1493 (isolated), 1495 (D1), and 1432 (isolated) cm−1] were caused by asymmetric and symmetric carbon–carbon ring stretching mode, respectively.
The experimental IR spectrum of PAOX presents characteristic band which confirmes the presence of oxime group in the structure of examined pyrazine derivative.43 The sharp, strong peak found at 953 cm−1 in the experimental FT-IR spectrum can be attributed to the stretching mode44 of ν(N–O)oxime. Based on the DFT calculated vibrational spectra, this band appeared in the region of 914 (monomer) and 940 cm−1 (D1). This indicates the presence of oxime group, in accordance with the crystallographic data.
The electronic spectrum of the PAOX shows in water two absorption bands at 252 nm and 290 nm which are ascribed to π → π* and n → π* transitions, respectively (Fig. 7). Dissolution of PAOX in a different solvent leads to a change in the position of the maxima of absorption. Changing the solvent from water to acetonitrile (at constant PAOX concentration, 6 × 10−5 M) causes blue shift of π → π* absorption band to shorter wavelengths – hypsochromic effect, with significant change in its intensity (ΔA about −0.24 – hypochromic effect). Moreover, the value of molar absorption coefficient (ε) is nearly double reduced when the solvent was changed from water (εPAOX = 9500 dm3 mol−1 cm−1) to acetonitrile (εPAOX = 5500 dm3 mol−1 cm−1). The n → π* absorption band of PAOX aqueous solution (290 nm) moves in MeCN to longer wavelengths – red shift and changes intensity from 0.68 in water to 0.48 in acetonitrile. The value of ε decreased from 11![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 333 dm3 mol−1 cm−1 (H2O) to 8000 dm3 mol−1 cm−1 (MeCN).
333 dm3 mol−1 cm−1 (H2O) to 8000 dm3 mol−1 cm−1 (MeCN).
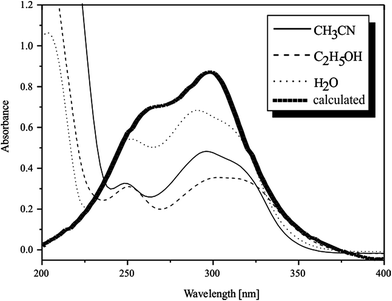 | ||
| Fig. 7 UV spectra of pyrazine-2-amidoxime obtained in three solvents: acetonitrile (MeCN), ethanol (EtOH), water (H2O), and from calculation (B3LYP, 6-311+G**). | ||
Maintaining the constant concentration of PAOX (6 × 10−5 M) and changing the solvent from water to ethanol changes the values of absorption and wavelength of the examined compound. In the case of maximum assigned to π → π* transition, the slightly hypsochromic effect of absorption band with a significant decrease in intensity (ΔA about −0.26) was observed. The value of PAOX molar absorption coefficient in ethanolic solution is about 60% lower than the value of εPAOX in aqueous solvent (εPAOX = 9500 dm3 mol−1 cm−1 in water; εPAOX = 5167 dm3 mol−1 cm−1 in ethanol). The second maximum ascribed to n → π* transition in the spectrum of ethanolic solution of PAOX appeared at 308 nm. The bathochromic shift to longer wavelength with a significant decrease in intensity (ΔA about −0.33) could be observed when the solvent was changed from water to ethanol (hypochromic effect). Furthermore, ε is nearly twice lower in ethanol (εPAOX = 5833 dm3 mol−1 cm−1) as compared to water (εPAOX = 11![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 333 dm3 mol−1 cm−1).
333 dm3 mol−1 cm−1).
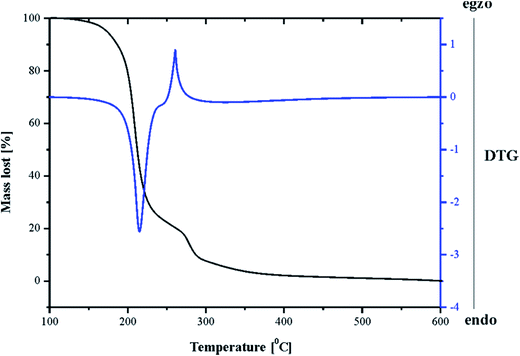
Thermogravimetry investigations
Thermal analyses of solid pyrazine-2-amidoxime were carried out by the TG and DTG techniques. The experimental results revealed that the degradation occurred in two stages (see Fig. 8). The study of thermal decomposition of PAOX showed an increased heat endurance of this compound, which did not contain crystal water and generally decomposed at temperatures exceeding 170–400 °C.The first step between 153 and 270 °C (the black line in Fig. 8) was accompanied by an 82.04% mass loss, attributed to the loss of a pyrazine molecule, which was identified as IR spectra of products (Fig. 9). The second step (271–316 °C) was accompanied by a 13.22% mass loss, which might be due to the decomposition of amidoxime group. The residual mass of 4.74% above 320 °C was probably caused by simultaneous formation of nitrogen species: nitrogen (N2), nitrous oxide (N2O) and ammonia (NH3), continued to the moment of physical capacity of the TG curve registration (range of heating). The DTA curve (the blue line in Fig. 9) for PAOX shows two peaks. The first one is endo- and the second is exothermic at 153–270 °C and 271–316 °C, respectively.
Electrochemical studies
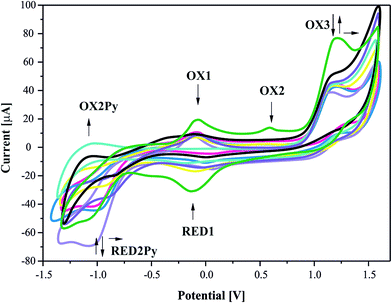
The redox behaviour of the pyrazine-2-amidoxime was studied in acetonitrile solution at a platinum working electrode. The relevant electrochemical data are summarized in Table 3 and cyclic voltammogram (CV) is shown in Fig. 10.| Symbol of the reaction | Electrode nature | Potential value Epa or Epc [V] | CV reversible nature | CV irreversible nature | ||
|---|---|---|---|---|---|---|
| ΔEp [V] | E 1/2 | I a/Ic | I a [μA] | |||
| a The presented representative potential was observed after an addition the first portion of methanol to PAOX acetonitrile solution (1 mM); reference electrode – SCE; E1/2 = 0.5(Epa + Epc) where Epa and Epc are anodic and cathodic peak potential, respectively; ΔEp as mathematical subtract both electrode potential values. | ||||||
| OX1 | Anodic | −0.058 | 0.067 | −0.092 | 0.603 | — |
| RED1 | cathodic | −0.125 | — | |||
| OX2 | Anodic | 0.619 | — | — | — | 8.35 |
| OX3 | Anodic | 1.231 | — | — | — | 76.59 |
| RED2Py | cathodic | −1.060 | 0.072 | −1.096 | 0.688 | −55.28 |
| OX2Py a | Anodic | −1.132 | — | |||
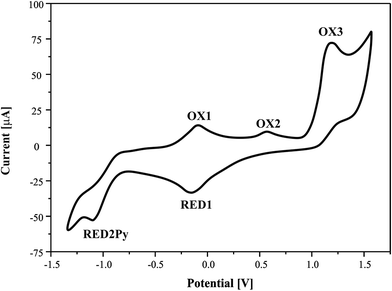 | ||
| Fig. 10 Cyclic voltammograms of pyrazine-2-amidoxime (1 mM) in acetonitrile solution, 0.1 M TBP. Scan rate = 100 mV s−1. | ||
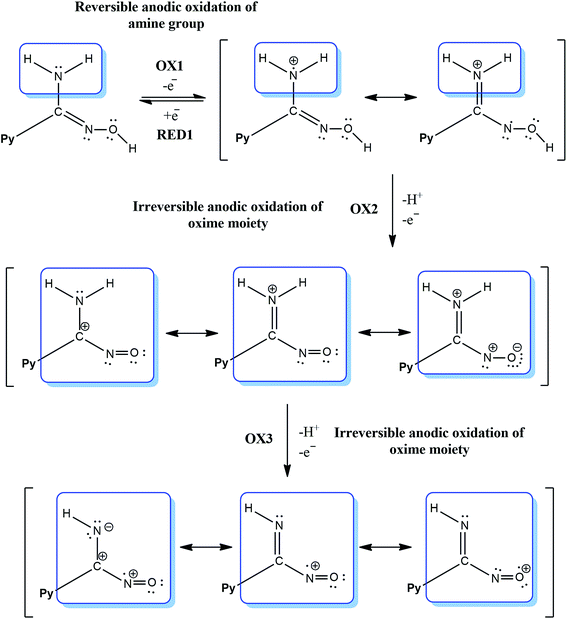
It can be seen that CV in acetonitrile solvent showed the occurrence of three consecutive anodic reactions: the first, reversible reaction of amine group oxidation OX1 and RED1, the second, main and well-defined peak at OX3, and a broad and ill-defined peak OX2. Due to electrochemical data, we suggest that oxidation of reaction (positive potentials), involving a proton and a single-electron abstractions, forms the anion oxime group, leading to the formation of iminoxy radical (see Schemes 1 and 2). It is known that a large number of biologically active compounds exhibit reduction potentials greater than −0.5 V, like RED2Py for irreversible reaction of PAOX, which can permit electron acceptance from in vivo donors.45
The electrochemical process in negative potential region is due to the reduction of pyrazine ring – Py (potential marked as RED1Py in Fig. 10) in PAOX. Pyrazine decomposition occurs simultaneously with hydrogen evolution, and a reduction peak RED1Py can be observed at −1.060 V (vs. SCE). After electrochemical reduction of Py in PAOX, the potential sweep was switched to the positive direction. No oxidation peak was observed for this reaction, due to the above it can be concluded that the reduction of pyrazine ring as part of PAOX is an irreversible process.
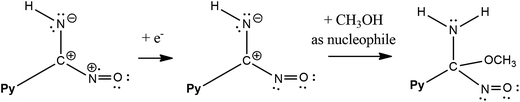
The follow-up chemical reaction is directly related to the nucleophilic attack of the molecule presented in the solution under study, e.g. methanol (Scheme 3). The addition of methanol to the acetonitrile solutions of the investigated PAOX resulted in substantial change in CVs (Fig. 11). With an increase in methanol concentration (from 0.001 M to 0.5 M), the potentials of reduction (RED2Py) and oxidation (OX3) shifted towards more positive values. RED2Py potential values increased initially and then decreased, but in the case of OX3 potential values – at the beginning they decreased and subsequently increased (all obtained changes were related with successive, additional portions of methanol).
The methanol is a nucleophile, therefore the shift of the potential OX3 and disappearance of the potential OX2 with the increase in the methanol concentration indicate that the chemical reaction preceding the second electrode stage is accompanied by the proton abstraction as an irreversible reaction (see Scheme 2). Moreover, a new very weak oxidation peak OX2Py was observed (Fig. 11), which described the re-oxidation reaction of DHPAOX+ during the addition of methanol (see Scheme 4). All the proposed mechanisms are modifications taking into account the data obtained from our own research, they are based, however, on our best scientific knowledge.46–49
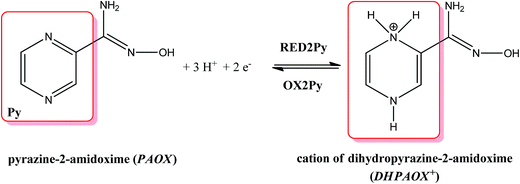 | ||
| Scheme 4 The proposed reversible electrochemical equation for pyrazine ring of PAOX (observed in negative potentials). | ||
Complexometric properties
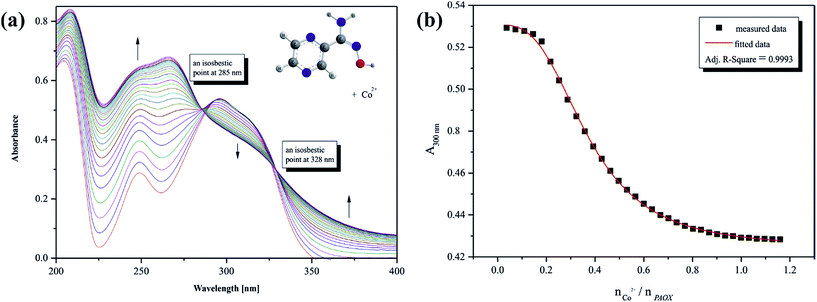
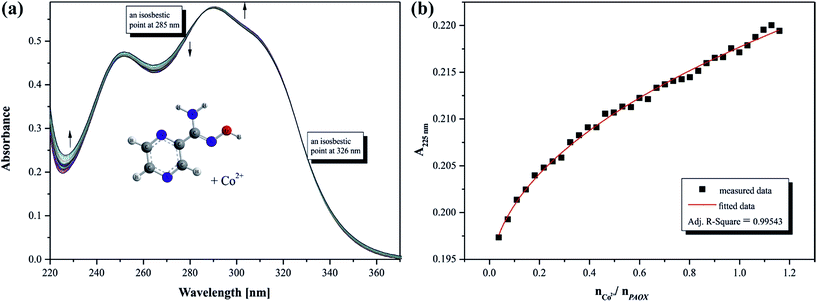
The presence of chromophore groups in a molecule of a ligand, whose absorption spectra change upon complexation, allows for the use of spectroscopy methods to study the chelation process. On the basis of the results from spectroscopy measurements it is possible to show a quantity of equilibria existing in a particular solution. Furthermore, this investigation can provide complete information about species formed during the titration process.Changes in the position and intensity of the absorption peak after addition of metal ion to pyrazine-2-amidoxime solution show which ions form the most stable complex in the selected solution. Consequently, the influence of the metal ions Co(II), Mn(II), Fe(II) and Cr(III) on the electronic spectrum of PAOX in acetonitrile and in water was examined. Significant changes in the position and intensity of bands were observed for all analyzed ions mainly in acetonitrile (see ESI, Fig. S3–S6 present the result for MeCN, and Fig. S7–S9† for H2O, respectively).
Based on the analysis of the spectrophotometric titration curves, the number of equilibria and stoichiometry of complexes formed with cobalt(II) ion were determined for both solvent systems (Fig. 12a and 13a). The intensities of absorption bands gradually increase and slight bathochromic shift can be observed at MeCN. At the same time two isosbestic points appeared at 285 nm and 328 nm, respectively. These effects suggest the existence of metal ion interactions with donor atom of PAOX in the system studied. The stoichiometric diagram (the absorbance at 300 nm as a function of Co2+–ligand molar ratio) was plotted to illustrate specific quantity stoichiometry in the system studied (Fig. 12b – MeCN and Fig. 13b – water). The proposed complexing equilibria models (eqn (1) and (2)) correspond to the formation of two metal ion–ligand complexes in aqueous solution.
| [Co(OH2)6]2+ + PAOX ⇆ [Co(PAOX)(OH2)4]2+ | (1) |
| [Co(PAOX)(OH2)4]2+ + PAOX ⇆ [Co(PAOX)2(OH2)2]2+ | (2) |
On the basis of the results from spectrophotometric measurements, the values of gradual and cumulative formation constants for selected metal ion complexes were determined in acetonitrile and water systems (Table 4). It must be pointed out that acetonitrile is a better solvent for the observation of the complexometric process between d-block metal ions and PAOX. It corresponds to PAOX better solubility in non-aqueous solvents due to the absence of weak intermolecular interactions with the solvent.
| Metal ion | Molar ratio | Spectrophotometric technique | |||||
|---|---|---|---|---|---|---|---|
| MeCN | H2O | ||||||
log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) K1 K1 |
log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) K2 K2 |
log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) β β |
log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) K1 K1 |
log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) K2 K2 |
log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) β β |
||
| Cr3+ | 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2 2 |
3.51 (±0.02) | 4.91 (±0.01) | 8.42 (±0.01) | 5.75 (±0.18) | 7.67 (±0.35) | 13.42 (±0.26) |
| Co2+ | 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2 2 |
3.65 (±0.03) | 6.74 (±0.03) | 10.39 (±0.03) | 3.42 (±0.09) | 4.74 (±0.09) | 8.16 (±0.13) |
| Fe2+ | 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2 (MeCN)/1 2 (MeCN)/1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 (H2O) 1 (H2O) |
3.98 (±0.04) | 4.99 (±0.04) | 8.97 (±0.04) | 4.79 (±0.12) | — | 4.79 (±0.12) |
| Mn2+ | 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2 2 |
4.10 (±0.03) | 3.97 (±0.05) | 8.07 (±0.04) | 3.79 (±0.14) | 4.35 (±0.07) | 8.14 (±0.11) |
Complexometric properties of pyrazine-2-amidoxime were also reported in order to obtain the full information about the behaviour of the compound studied as a bio-ligand towards d-block metal ions. The complexes formed in acetonitrile solution can be put to order according to their increasing stability, which is consistent with the Irving–Williams theoretical predictions:
| [Mn(PAOX)2aq]2+ < [Cr(PAOX)2aq]3+ < [Fe(PAOX)2aq]2+ < [Co(PAOX)2aq]2+ |
The order of the value stability constants of complexes under study can be established also for aqueous solution:
| [Fe(PAOX)2aq]2+ < [Mn(PAOX)2aq]2+ ≈ [Co(PAOX)2aq]2+ < [Cr(PAOX)2aq]3+ |
Microbiological assay
The activity of pyrazine-2-amidoxime was examined for the reference strains, provided by the Institute of Immunology and Experimental Therapy of the Polish Academy of Sciences, Wroclaw, Poland. The selected reference strains include: Gram-positive bacteria: Enterococcus hirae ATCC 8043 and Staphylococcus aureus ATCC 6538; Gram-negative bacteria such as Escherichia coli ATCC 8739, Proteus vulgaris ATCC 6896, Pseudomonas aeruginosa ATCC 9027 and yeast Candida albicans ATCC 4635. The values of the Minimal Bactericidal (fungicidal) Concentration (MBC) and Minimal Inhibitory Concentration (MIC) were determined. In the microbiological studies quantitative methods, such as microdilution in liquid (MIC) and solid (MBC) culture media, were applied respectively.The results of biological assays show that the investigated compound exhibited bactericidal activity against both Gram negative and Gram positive bacteria strains. Pyrazine-2-amidoxime showed killing and inhibiting properties at a concentration of 5.79 mM. Moreover, studied pyrazine derivative had about 10-fold higher inhibitory activity against C. albicans than other investigated strains. The PAOX inhibited growth of selected yeast at the concentration of 0.58 mM. The present studies will indicate the possibility of using PAOX as a new antimicrobial agent. Since C. albicans is the major fungal pathogen in humans (carried by over 50% of the population), in this context further investigations in this field are reasonable.
Conclusions
On the basis of our research, PAOX was found to be stable in the experiment conditions at a room temperature. Its decomposition at higher temperatures (>150 °C) was observed. The structural studies in the solid state, solution,36 and gas phase36 showed the same tautomeric preferences, anti-conformation on the C2–C7 single bond, and Z-configuration on the amidoxine C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N double bond.36
N double bond.36
There was no possibility of observing the self-association of PAOX in solution with the use of spectroscopic methods in our earlier studies36 because of the low solubility of PAOX in nonpolar solvents. Finally, the crystallographic methods showed what interaction types occurred and what groups were involved in it. These interactions were also found in the experimental IR and Raman spectra, recorded for solid PAOX, and confirmed computationally at the DFT level for selected polymers of PAOX. Two anti-conformers were observed in the crystal structure of PAOX (hereinafter referred to as A and B molecules). They differ between each other only by an angle in the pyrazine ring and –(NH2)C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) (NH–OH) group (molecule A showed larger value of this angle than B); both with nearly planar pyrazine ring. The controlled use of the H-bonds formed between PAOX-A molecules chain and PAOX-B molecules chain, and π–π stacking between the same types of PAOX molecules in two chains (πA–πA or πB–πB) as a component in foldamers is challenging. The PAOX ability to create channel structures can be used better as a co-crystal host or antimicrobial agent.
(NH–OH) group (molecule A showed larger value of this angle than B); both with nearly planar pyrazine ring. The controlled use of the H-bonds formed between PAOX-A molecules chain and PAOX-B molecules chain, and π–π stacking between the same types of PAOX molecules in two chains (πA–πA or πB–πB) as a component in foldamers is challenging. The PAOX ability to create channel structures can be used better as a co-crystal host or antimicrobial agent.
The electrochemical properties of pyrazine-2-amidoxime were discussed, as well as the mechanisms of cathodic reduction of pyrazine part and anodic oxidations of amidoxime moieties (in the absence or presence of methanol) were proposed in detail, based on our own research and knowledge acquired from scientific literature. It should be pointed out that the acquired knowledge about electrochemical behaviour of PAOX can implicate its subsequent application (as a biologically active compound). Our biological assay indicated the antifungal activity of pyrazine-2-amidoxime against C. albicans. PAOX is active against both Gram-positive and Gram-negative bacteria. The PAOX's binding ability to d-block metal ions was examined in aqueous and acetonitrile solutions. The obtained results showed that pyrazine-2-amidoxime behaves like a bidentate ligand and it coordinates selected metal ions by azomethine and amine nitrogen atoms, respectively.
Experimental protocols
Materials and methods
All starting materials were commercially available and used as such unless otherwise noted. Pyrazine-2-amidoxime of analytical purity grade (≥97%) was purchased from a commercial source (Sigma Aldrich). Twice distilled water (Hydrolab-Reference purified) with conductivity not exceeding 0.09 μS cm−1 was used in the preparation of all the solutions under study.X-ray crystallography
X-ray quality crystals of pyrazine-2-amidoxime (Fig. 14) were obtained from diffusion crystallization. 2 mg of pyrazine-2-amidoxime was dissolved in 1 mL of acetonitrile (MeCN), and 2.5 mL of tetrahydropyran (THP) was used as an antisolvent in a closed container. The sample was left to crystallize at room temperature for a period of over two weeks. All solvents used during crystallization were purified before by standard procedures. The elemental analysis confirmed the composition of the obtained PAOX (as white crystals) with ±0.3% of the theoretical values {calcd for C5H6N4O (%) C, 43.48; H, 4.38; N, 40.56 and found C, 43.50; H, 4.41; N, 40.59}. | ||
| Fig. 14 Crystals of pyrazine-2-amidoxime obtained by diffusion crystallization (microscopic image, 40× zoom). | ||
The X-ray diffraction (XRD) measurement was carried out on an Oxford Diffraction Gemini R ULTRA Ruby CCD diffractometer with CuKα (λ = 1.54184) radiation source. Data were reduced using CrysAlis RED software.50 The structural resolution procedure was carried out using the SHELX package (for details see: ESI†).51 All interactions were determined using the PLATON program.52 The ORTEPII53 and Mercury54 programs were used to prepare the molecular graphics.
Computational details
Geometries of five dimers and one tetramer were fully optimized with the use of the DFT method along with B3LYP functional,55–57 and the 6-311+G(d,p) basis set.58 Basis set superposition error (BSSE) was also accounted for in the calculations of energy formations for dimers and a tetramer.59 All calculations were performed in vacuo without symmetry constraints, according to the procedures included in the Gaussian-03 series of programs.60Analytical instrumentations
The percentage composition of the elements of the examined compounds was determined with the use of an element analyzer Carlo Erba EA 1108 CHNS. The infrared spectra of pyrazine-2-amidoxime was recorded as potassium bromide (KBr) pellets using Brüker Infrared Spectrometer in the range of 4500–500 cm−1. The electronic spectra were recorded on a Perkin Elmer Lambda 650 spectrophotometer in the range of 220–400 nm with a spectral band width 2 nm. The concentrations of compounds in acetonitrile solution considered in spectrophotometric measurements were around 10−6 M. Raman FRA 106 – laser NdYAG (1060 nm) with MCT detector was used to register Raman's spectra of solid state PAOX under study.The thermogravimetric and IR spectra were measured by means of a thermal equaliser TG209 Nietzsche equipment with FT-IR. All experiments were carried out in an argon atmosphere. The analyzer was equipped with a programmed temperature controller, which automatically maintains constant temperature during thermal events. The TG weight-loss measurements were made at 20–1000 °C at a heating rate of 15 °C min−1. The infrared spectra were registered in Nujol mulls using a Brüker IFS 66 spectrophotometer. All measurements were verified at least twice.
CV experiments were done on an Gamry Potentiostat/Galvanostat (model 600). Data were collected and were analysed by the model Framework Analysis System Software on a PC computer. Platinum wire electrodes were used as both counter and working electrodes, and combined calomel electrode was used as a reference electrode (acetonitrile with an electrolyte inside). Solution cyclic voltammetry was carried out in a acetonitrile as a solvent containing tetrabutylammonium perchlorate TBP (0.1 M) as an electrolyte for PAOX (10−3 M). All solutions in the cell were purged with ultra-purity argon (Ar) for 10–15 minutes before each experiment, and a blanket of Ar was used during the experiment.
Minimal bactericidal or fungicidal concentrations
The procedure of microbiological tests was described in our previous paper.35 For details see ESI.†Conflict of interest
The authors declare no competing financial interest.Acknowledgements
DFT calculations were carried out with the use of the computational resources at the Faculty of Chemistry University of Gdansk – cluster Piasek, the Information Center of the Metropolitan Academic Network (CI TASK) in Gdansk, and the Interdisciplinary Mathematical Center (ICM) in Warsaw. This work was financially supported by grants from the Polish Ministry of Science and Higher Education (DS/530-8236-D601-16, BMN 538-8236-B022-15, BMN 538-8236-B022-16, BMN 538-8236-B065-15, and BMN 538-8236-B065-16).References
- M. L. Dubuissona, J. F. Reesa and J. Marchand-Brynaert, Mini-Rev. Med. Chem., 2004, 4, 421 CAS.
- K. D. Tripathi, Essentials of medical pharmacology, Jaypee brother’s medical publishers, New Delhi, 6th edn, 2008, pp. 39–50 Search PubMed.
- S. S. Kadam, K. R. Mahadik and K. G. Bothara, Principles of medicinal chemistry, Nirali Prakashan, 6th edn., 1999, pp. 169–175 Search PubMed.
- S. Suzuki and A. Inoue, Indian Chem. Abstr., 1990, 113, 218276t Search PubMed.
- T. L. El-Emary, J. Chin. Chem. Soc., 2006, 53, 391 CrossRef CAS.
- H. S. El-Kashef, T. I. El-Emary, M. Gasquet, P. Timon-David, J. Maldonado and P. Vanelle, Pharmazie, 2000, 55, 572 CAS.
- L. Tumova, J. Tuma and M. Dolezal, Molecules, 2011, 16, 9142 CrossRef CAS PubMed.
- L. Tumova, J. Tuma, K. Megusar and M. Dolezal, Molecules, 2010, 15, 331 CrossRef CAS PubMed.
- B. A. Ellsworth, Y. Wang, Y. Zhu, A. Pendri, S. W. Gerritz, C. Sun, K. E. Carlson, L. Kang, R. A. Baska, Y. Yang, Q. Huang, N. T. Burford, M. J. Cullen, S. Johnghar, K. Behnia, M. A. Pelleymounter, W. N. Washburn and W. R. Ewing, Bioorg. Med. Chem. Lett., 2007, 17, 3978 CrossRef CAS PubMed.
- M. Dolezal, L. Palek, J. Vinsova, V. Buchta, J. Jampilek and K. Kralova, Molecules, 2006, 11, 242 CrossRef CAS PubMed.
- M. Dolezal, J. Zitko, D. Kesetovicová, J. Kunes and M. Svobodova, Molecules, 2009, 14, 4180 CrossRef CAS PubMed.
- A. W. Cuthbert and J. M. Edwardson, J. Pharm. Pharmacol., 1979, 31, 382 CrossRef CAS PubMed.
- K. Gobis, H. Foks, A. Kedzia, M. Wierzchowska, E. Kwapisz, Z. Zwolska and E. A. Kopec, Acta Poloniae Pharmaceutica. Drug Research, 2006, 63, 39 CAS.
- A. Y. Il’chenko, V. I. Krokhtyak and L. M. Yagupolskii, Chem. Heterocycl. Compd., 1981, 17, 1236 CrossRef.
- W. Szczepankiewicz, J. Suwiński and R. Bujok, Tetrahedron, 2000, 56, 9343 CrossRef CAS.
- N. Buu-Hoi, M. Welsch, N. Xuong and K. Thang, Esperientia, 1954, 10, 169 CrossRef CAS.
- D. N. Nicolaides and E. A. Varella, in The Chemistry of Amidoximes. Supplement B. The chemistry of acid derivatives. Interscience, ed. S. Patai, New York, 1992, vol. 2, p. 876 Search PubMed.
- O. M. Glozman, T. A. Voronina, E. K. Orlova, I. K. Meshcheryakova, A. A. Kazanskaya, L. D. Smirnov, A. Rostock and H. Siegemund, Zh. Nauchn. Khim.-Farm. Inst., 1989, 23, 1147 CAS.
- S. M. Ray and S. C. Lahiri, J. Indian Chem. Soc., 1991, 68, 549 CAS.
- A. Havemeyer, F. Bittner, S. Wollers, R. Mendel, T. Kunze and B. Clement, J. Biol. Chem., 2006, 281, 34796 CrossRef CAS PubMed.
- C. J. Marmion, D. Griffith and K. B. Nolan, Eur. J. Inorg. Chem., 2004, 15, 3003 CrossRef.
- G. Y. An, C. M. Ji, A. L. Cui and H. Z. Kou, Inorg. Chem., 2011, 50, 1079 CrossRef CAS PubMed.
- L. An, L. J. Zhong, J. Feng, S. Z. Du and X. M. Lu, Inorg. Chem. Commun., 2012, 22, 108 CrossRef CAS.
- C. R. Cagin, M. Diloy-Puray and M. P. Westerman, Ann. Intern. Med., 1978, 89, 509 CrossRef CAS PubMed.
- J. C. Clifton, Pediatr. Clin. North Am., 2007, 54, 237 CrossRef PubMed.
- N. N. Greenwood and A. Earnshaw, Chemistry of the elements, Pergamon Press, Oxford, 1984 Search PubMed.
- C. Pfeiffer and R. Mailloux, Journal of Orthomolecular Medicine, 1987, 2, 171 Search PubMed.
- J. P. Ley and H. J. Bertram, Eur. J. Lipid Sci. Technol., 2002, 104, 319 CrossRef CAS.
- G. O. Puntel, P. Gubert, G. L. Peres, L. Bresolin, J. B. T. Rocha, M. E. Pereira, V. S. Carratu and F. Antunes Soares, Mol. Toxicol., 2008, 82, 755 CAS.
- T. Özen and M. Tas, J. Enzyme Inhib. Med. Chem., 2009, 24, 1141 CrossRef PubMed.
- G. O. Puntel, N. R. de Carvalho, P. Gubert, A. S. Palma, C. Lenz Dalla Corte, D. Silva Avila, M. E. Pereira, V. S. Carratu, L. Bresolin, J. B. Teixeira da Rocha and F. A. Antunes Soares, Chem.-Biol. Interact., 2009, 177, 153 CrossRef CAS PubMed.
- A. P. da Silva, M. Farina, J. L. Franco, A. L. Dafre, J. Kassa and K. Kuca, Neurotoxicology, 2008, 29, 184 CrossRef CAS PubMed.
- M. Pohanka, J. Z. Karasova, K. Musilek, K. Kuca, Y. S. Jung and J. Kassa, J. Enzyme Inhib. Med. Chem., 2011, 26, 93 CrossRef CAS PubMed.
- A. Antunes dos Santos, D. Bonfanti dos Santos, R. Pietsch Ribeiro, D. Colle, K. C. Peres, J. Hermes, A. Machado Barbosa, A. L. Dafre, A. F. de Bem, K. Kuca and M. Farina, Neurotoxicology, 2011, 32, 888 CrossRef PubMed.
- A. Chylewska, A. Sikorski, M. Ogryzek and M. Makowski, J. Mol. Struct., 2016, 1105, 96 CrossRef CAS.
- A. Głębocka, E. D. Raczyńska, A. Chylewska and M. Makowski, J. Phys. Org. Chem., 2016, 29, 326 CrossRef.
- K. N. Jarzembska, A. A. Hoser, R. Kamiński, A. O. Madsen, K. Durka and K. Woźniak, Cryst. Growth Des., 2014, 14, 3453 CAS.
- K. S. Eccles, R. E. Morrison, A. R. Maguire and S. E. Lawrence, Cryst. Growth Des., 2014, 14, 2753 CAS.
- S. Gunasekaran, R. K. Natarajan, R. Rathikha and D. Syamala, Indian J. Pure Appl. Phys., 2005, 43, 509 Search PubMed.
- S. Breda, I. D. Reva, L. Lapinski, M. J. Nowak and R. Fausto, J. Mol. Struct., 2006, 786, 193 CrossRef CAS.
- H. Endredi, F. Billes and S. Holly, J. Mol. Struct., 2003, 633, 73 CrossRef CAS.
- R. B. Venkatram and R. Ramana, Indian J. Pure Appl. Phys., 2002, 76, 473 Search PubMed.
- F. Huang, Y. Xu, S. Liao, D. I. Yang, Y. Hsieh and Q. Wei, Materials, 2013, 6, 969 CrossRef CAS.
- N. Bilba, D. Bilba and G. Moroi, J. Appl. Polym. Sci., 2004, 92, 3730 CrossRef CAS.
- D. M. Frank, P. K. Arora, J. L. Blumer and L. M. Sayre, Biochem. Biophys. Res. Commun., 1987, 147, 1095 CrossRef CAS PubMed.
- S. I. Kulakovskaya, A. V. Kulikov and A. F. Shestakov, Russ. J. Electrochem., 2012, 48, 1023 CrossRef CAS.
- C. J. Tonzola, M. M. Alam, W. Kaminski and S. A. Jenekhe, J. Am. Chem. Soc., 2003, 125, 13548 CrossRef CAS PubMed.
- I. Novak, S. Komorsky-Lovric, S. Zunec and A. L. Vrdoljak, Int. J. Electrochem. Sci., 2013, 8, 9818 CAS.
- A. G. Brolo and D. E. Irish, J. Chem. Soc., Faraday Trans., 1997, 93, 419 RSC.
- CrysAlis CCD and CrysAlis RED (Version 1.171.36.24), Oxford Diffraction Ltd, Yarnton, England, 2012 Search PubMed.
- G. M. Sheldrick, Acta Crystallogr., Sect. A: Found. Crystallogr., 2008, 64, 112 CrossRef CAS PubMed.
- A. L. Spek, Acta Crystallogr., Sect. D: Biol. Crystallogr., 2009, 65, 14 CrossRef PubMed.
- C. K. Johnson, ORTEP II, Report ORNL-5138, Oak Ridge National Laboratory, Oak Ridge, TN, USA, 1976 Search PubMed.
- C. F. Macrae, I. J. Bruno, J. A. Chisholm, P. R. Edgington, P. McCabe, E. Pidcock, L. Rodriguez-Monge, R. Taylor, J. van de Streek and P. A. Wood, J. Appl. Crystallogr., 2008, 41, 466 CrossRef CAS.
- R. G. Parr and W. Yang, Density Functional Theory of Atoms and Molecular Orbital Theory, Oxford University Press, New York, 1989 Search PubMed.
- A. D. J. Becke, Chem. Phys., 1993, 98, 5648 CAS.
- C. Lee, W. Yang and R. G. Parr, Phys. Rev. B: Condens. Matter Mater. Phys., 1988, 37, 785 CrossRef CAS.
- W. J. Hehre, L. Radom, P. V. R. Schleyer and J. A. Pople, Ab initio Molecular Theory, Wiley, New York, 1986 Search PubMed.
- S. F. Boys and F. Bernardi, Mol. Phys., 1970, 19, 553 CrossRef CAS.
- M. J. Frisch, G. W. Trucks, H. B. Schlegel, G. E. Scuseria, M. A. Robb, J. R. Cheeseman, J. A. Montgomery, T. Vreven, K. N. Kudin, J. C. Burant, J. M. Millam, S. S. Yengar, J. Tomasi, V. Barone, B. Mennucci, M. Cossi, G. Scalmani, N. Rega, R. Petersson, H. Nakatsuji, M. Hada, M. Ehara, K. Toyota, R. Fukuda, J. Hasegawa, M. Ishida, T. Nakajima, Y. Honda, O. Kitao, H. Nakai, M. Klene, X. Li, J. E. Knox, H. P. Hratchian, J. B. Cross, V. Bakken, C. Adamo, R. Jaramillo, R. Gomperts, R. E. Stratmann, O. Yazyev, A. J. Austin, R. Cammi, C. Pomelli, J. W. Ochterski, P. Y. Ayala, K. Morokuma, G. A. Voth, P. Salvador, J. J. Dannenberg, V. G. Zakrzewski, S. Dapprich, A. D. Daniels, M. C. Strain, O. Farkas, D. K. Malick, A. D. Rabuck, K. Raghavachari, J. B. Foresman, J. V. Oritz, Q. Cui, A. G. Baboul, S. Clifford, J. Cioslowski, B. B. Stefanov, G. Liu, A. Liashenko, P. Piskorz, I. Komaromi, R. L. Martin, D. J. Fox, T. Keith, M. A. Al-Laham, C. Y. Peng, A. Nanayakkara, M. Challacombe, P. M. W. Gill, B. Johnson, W. Chen, M. W. Wong, C. Gonzalez and J. A. Pople, GAUSSIAN-03 (Revision E.01), Gaussian, Inc., Wallingford, CT, 2004 Search PubMed.
Footnote |
| † Electronic supplementary information (ESI) available: File includes both experimental, and theoretical (calculated) IR, Raman 1H NMR spectra, experimental protocols details (X-ray, microbiological tests) and titration spectra, as well as atom Cartesian coordinates, and formation free energies of three dimers D1–D3, and a tetramer T1. CCDC 1474463 full crystallographic details, excluding structure features, have been deposited for pyrazine-2-amidoxime. For ESI and crystallographic data in CIF or other electronic format see DOI: 10.1039/c6ra10537h |
| This journal is © The Royal Society of Chemistry 2016 |