Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Synthetic m3G-CAP attachment necessitates a minimum trinucleotide constituent to be recognised as a nuclear import signal†
M. Honcharenko‡
*a,
B. Bestas‡b,
M. Jezowska‡a,
B. A. Wojtczakc,
P. M. D. Morenob,
J. Romanowskaaf,
S. M. Bächled,
E. Darzynkiewiczce,
J. Jemielity c,
C. I. E. Smithb and
R. Strömberg*a
c,
C. I. E. Smithb and
R. Strömberg*a
aDepartment of Biosciences and Nutrition, Karolinska Institutet, Novum, SE-14183 Huddinge, Sweden. E-mail: malgorzata.honcharenko@ki.se; roger.stromberg@ki.se
bClinical Research Center, Department of Laboratory Medicine, Karolinska Institutet, Karolinska University Hospital Huddinge, SE-141 86 Huddinge, Sweden
cCentre of New Technologies, University of Warsaw, Banacha 2c, 02-097 Warsaw, Poland
dCenter for Infectious Medicine, Department of Medicine, Karolinska Institutet, Karolinska University Hospital Huddinge, 14186 Stockholm, Sweden
eDivision of Biophysics, Institute of Experimental Physics, Faculty of Physics, University of Warsaw, Zwirki i Wigury 93, 02-089 Warsaw, Poland
fInstitute of Bioorganic Chemistry, Polish Academy of Sciences, Noskowskiego 12/14, 61-704 Poznań, Poland
First published on 18th May 2016
Abstract
Achieving higher nuclear concentrations by active transport may give potent therapeutic effects at lower doses for many drugs. A method of increasing nuclear uptake is the use of naturally existing Nuclear Localization Signals (NLS) by conjugating NLS structures to the cargo. We have synthesized a set of 2,2,7-trimethylguanosine cap (m3G-CAP)-containing structures (and their biotin conjugates) as artificially attached analogs of a naturally found NLS. The origin of a naturally found NLS is a uridine rich, small nuclear ribonucleoprotein (U snRNP) that employs Snurportin1 as a nuclear transport protein. In this report the NLS activity of various m3G-CAP biotin constructs was studied. We have shown that a minimal requirement for nuclear uptake is the inclusion of a trinucleotide sequence between the m3G-CAP and the artificial linker.
Introduction
Accessing the nucleus through the surrounding membrane (nuclear envelope) is one of the major barriers for therapeutic molecules. For medicinal applications, such as plasmid DNA, the large size of the nucleic acid hampers access to the nuclear compartment.1 On the other hand, also for small oligonucleotides (ONs), achieving higher nuclear concentrations by active transport might give the advantage of having a therapeutic effect at lower doses.The nuclear envelope consists of control gates known as nuclear pore complexes (NPCs), which selectively regulate nuclear transport.2,3 Small molecules such as metabolites and ions can diffuse through the NPC but molecules bigger than 40 kDa such as most proteins and RNA, ribosomal subunits and viral particles need to be actively transported.4,5 The majority of the large molecules have to be either interacting directly with NPCs or be actively transported with soluble carrier molecules collectively named as the karyopherin-β family (importins/exportins and transportins).6 The Ran-GTP/GDP gradient across the NPC regulates the transport of the cargo molecules between the nucleoplasm and the cytoplasm.7 Ran is a GTPase that can hydrolyse GTP into GDP. RanGTP binds to a carrier importin and helps to release the cargo in the nucleus. Conversely, when a cargo is to be transported to the cytoplasm, exportin–RanGTP–cargo complex is hydrolysed in the cytoplasm releasing the cargo.8,9
Large proteins need to have a nuclear localization signal (NLS) in order to be transported through the NPC. The one most widely studied, is the classical NLS (cNLS) sequence, which contains a short stretch of basic residues.10,11 cNLS containing cargo is recognized by the adapter protein importin-α and the complex is further bound by importin-β that interacts with NPC and translocate the cargo into the nucleus by the help of RanGTP.12,13 On the other hand, the non-classical NLSs do not need an adaptor protein and bind directly to a transporter protein.6 Earlier studies showed that non-covalent or covalent complexation of the NLS peptide with a plasmid DNA or an oligonucleotide promoted nuclear delivery including in vivo.14–20 Moreover, a more recent study shows the increased transfection efficiency of plasmid DNA by using PEIs (polyethylenimine) conjugated to an NLS peptide in adipose derived stem cell.21
Another NLS type is the 2,2,7-trimethylguanosine cap (m3G-CAP) which is naturally found in the uridine rich, small nuclear ribonucleoproteins (U snRNPs) that are involved in pre-mRNA splicing.22 The RNA (snRNA) part of the snRNPs mainly consist of U1, U2, U5 and U4/U6. Among the snRNAs, U1, U2, U4 and U5 are transcribed by RNA polymerase II and co-transcriptionally capped with an m7G-CAP. The m7G-CAP is recognized by the heterodimeric cap binding complex (CBC)23 and recruits the phosphorylated adaptor of export protein (PHAX).24 This complex is further recognized by an export receptor CRM1 for cytoplasmic localization and maturation.24 Following the transport, a complex of proteins assemble around the Sm core domain of the snRNAs,25 which creates a binding platform for trimethylguanosine synthase 1 Tgs1 (PIMT). Tgs1 catalyzes the methylation of the m7G-CAP to an m3G-CAP.26,27 Subsequently, the m3G-CAP is recognized by an adapter protein Snurportin1, which then binds to importin-β for nuclear relocalization, where the final maturation of the snRNPs occurs.28,29
Investigation of the crystal structure of Snurportin1 binding domain (residues 97–300) with bound m3GpppG, revealed that Snurportin1 binds both the hyper-methylated cap and the first nucleotide of the RNA in a stacked conformation.30 The investigation of the Kd-value for m3GpppG dinucleotide (1.0 μM) was determined to be lower than for the m3GpppA dinucleotide (12.1 μM), when using the full length Snurportin1. Moreover, binding of full-length Snurportin1 was found to be even stronger (Kd = 0.23 mM) for the synthetic capped trinucleotide m3GpppA(OMe)U(OMe)A, which more closely resembles the natural UsnRNAs. Since the crystallized domain of Snurportin1 lacked 61 residues from the C-terminus, the authors speculate that the real RNA binding surface of Snurportin1 may be larger and the 2′-O-methylated riboses of the RNA stem could have additional interaction.30
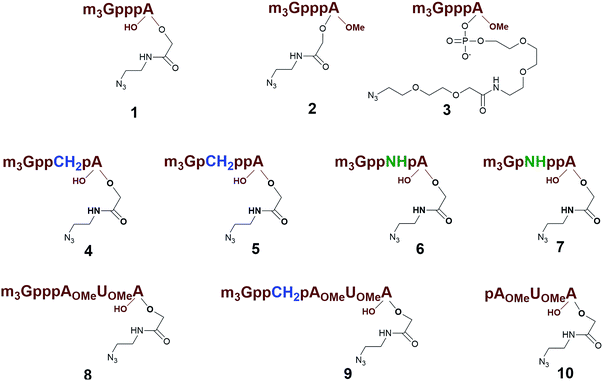
Previously, we have reported that use of the m3G-CAP as a synthetic RNA 5′-end NLS signal increases the nuclear transport of oligonucleotides as well as a cargo protein.31 The use of the m3G-CAP as an NLS signal for increased nuclear delivery of a therapeutic compound is likely to require additional stability towards enzymatic degradation. For that reason introduction of stabilizing modifications into the triphosphate bridge should be beneficial. In addition, the m3G-CAP can serve as an adaptor for delivery in vivo, which requires not only convenient synthesis but also a method that is readily scaled up. Thus, we decided to design artificially attachable synthetic m3G-CAPs, in order to investigate the minimum structural requirements for retained nuclear transport ability. A method for preparation of different constructs was necessary for scanning reaction requirements and allowing attachment of different cargos. We have recently reported the procedure for the synthesis of the m3G-CAP analogues equipped with an azide handle that can be conjugated to oligonucleotides via “click reaction”.32 Importantly, these m3G-CAP-oligonucleotide constructs contain native and modified triphosphate bridges that would stabilize these and reduce degradation.33 Here we report on methodology and synthesis of different m3G-CAP-biotin constructs (1–10, Fig. 1), which have been synthesized to evaluate the minimum structural requirement for retained nuclear transport ability in vitro.
Results and discussion
Synthesis of m3G-CAP-biotin constructs
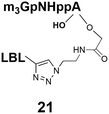
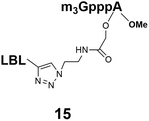
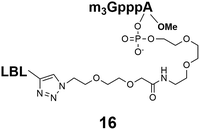
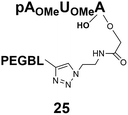
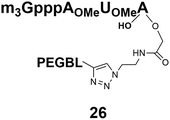
Different “clickable” m3G-CAP constructs (1–8, Fig. 1) with or without triphosphate modifications and equipped with an azide handle for attachment to a cargo molecule, were designed and synthesized according to procedures reported earlier.32,34 In addition, a capped trinucleotide construct with a methylene-modified triphosphate bridge and an uncapped trinucleotide control was also synthesized (9 and 10, Fig. 1).The most simplified structure contains a 2,2,7-trimethyl-guanosine connected via a triphosphate bridge to the adenosine residue (1). The functional azide linker protrudes from the 2′-position of the adenosine nucleoside (structure 1, Fig. 1). Since the 2′-O-methyl position may have additional effects on the nuclear transport, m3G-CAP construct 2 is equipped with a functional linker at the 3′-position. The 2′-position is O-methylated, and thus a step closer to the native m3G-CAP (structure 2, Fig. 1). The m3G-CAP construct 3 is equipped with both a 2′-O-methylated adenosine residue and a 3′-phosphate as a part of a longer linker attachment. Cap analogues 4–7 contain modification between α and β phosphates of the triphosphate bridge since this can stabilize the m3G-CAPs towards degradation.33,34 Finally, the m3G-CAP constructs 8 and 9 contain a 2,2,7-trimethylguanosine connected via triphosphate bridge to the AUA sequence, where A and U are 2′-O-methylated (structure 8 and 9, Fig. 1). In 9 the same methylene modification of the triphosphate as in 4 is included. The 5′-phosphorylated trimer (structure 10, Fig. 1) can also be attached to cargos and used as a non-capped control of conjugates in the studies of nuclear transport.
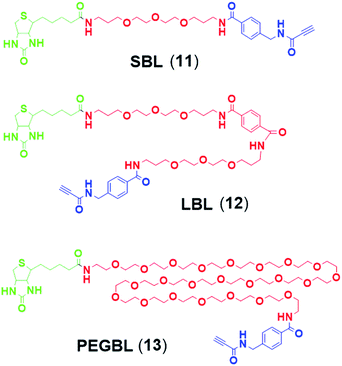
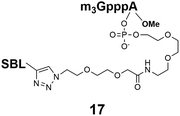
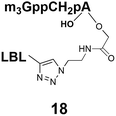
Synthesis of 9 and 10 was performed as reported in the synthesis of 8 with the exception that for 10 the capping step was performed with the methylene(bisphosphonate) analog of N2,N2,N7-trimethylguanosinepyrophosphoryl imidazolide. In order to assess the nuclear transport with a sizeable cargo, the different cap constructs were attached via linkers to biotin that in turn can be bound to a fluorescent streptavidin cargo (STV-Alexa488). From our experience in conjugating ONs to peptides it seems that low concentration of the reacting biomolecules requires more active functional groups to achieve sufficient conversion.35 Thus, biotin constructs with different linkers attached to an activated triple bond donor, p-(N-propynoylamino)toluic acid PATS,35 were synthesized (11, 1236 and 13, Fig. 2).
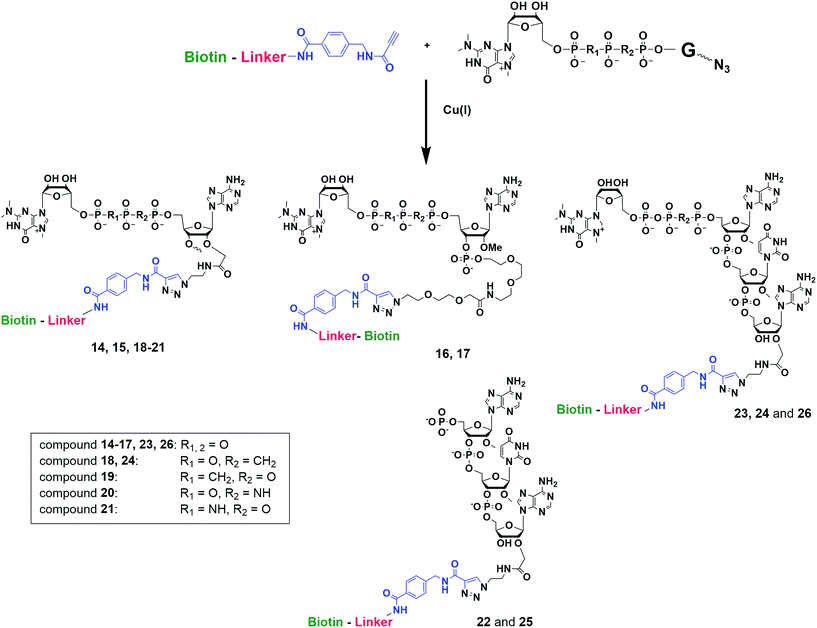
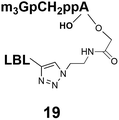
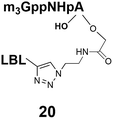
Synthesis of m3G-CAP-biotin and uncapped trinucleotide constructs (Scheme 1) were performed in solution using a tert-butanol/water mixture as solvent using Cu(I) catalyzed Hüisgen dipolar [3 + 2]-cycloaddition (“click chemistry”) (Scheme 1) which resulted in the products 14–26 (Table 1).
Biological evaluation of synthetic m3G-CAP-biotin constructs
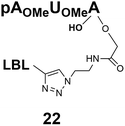
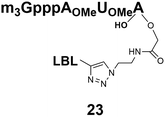
The synthesized m3G-CAP-biotin analogues were then evaluated with respect to localization within cells. Streptavidin-Alexa488 (STV-488) was incubated with 3.5 to 4 molar excess of the constructs 14–26. STV has a molecular weight of ca. 56 kDa. Together with the biotinylated constructs the total molecular weight adds up to 60–70 kDa, which is well above the threshold for passive diffusion through the nuclear pore channels (NPCs). For evaluation of possible nuclear localization, STV-complexes with 14–26 were transfected into U2OS cells by the PULSin protein transfection reagent. After 6 hours, cells were analyzed by fluorescent microscopy in order to detect the localization of the STV-Alexa488 complexes.All constructs with the m3G-CAP connected to a mononucleotide, whether the triphosphate was modified or not, failed to show up in the nucleus (see ESI†). However, cells treated with the construct having a capped trinucleotide (23) and to a lesser extent the same cap construct conjugated to biotin with a different linker (26) displayed fluorescence signals from the nucleus in the form of speckles (Fig. 3), in line with our previous publication where longer oligonucleotides were equipped with biotin linkers at the 3′-end.31
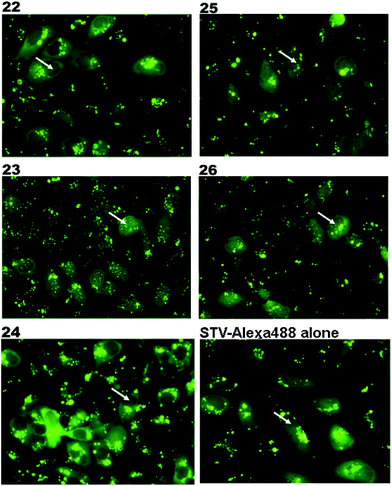 | ||
| Fig. 3 Fluorescence microscopy images of the cells treated with STV-Alexa488 complexes with constructs 22–26 (Table 1). Nuclear region of the cells are marked with a white arrow. Nuclear transport is evaluated by the Alexa-488 signal from the nucleus. | ||
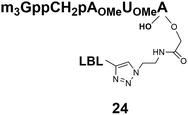
Somewhat surprising is that no nuclear localization was observed (Fig. 3) with the m3G-CAP carrying a methylene(bisphosphonate)-modified triphosphate bridge even when this was connected to a trinucleotide (24). As expected control cap analogues 22, 25 and STV-Alexa488 alone showed no nuclear accumulation (Fig. 3).
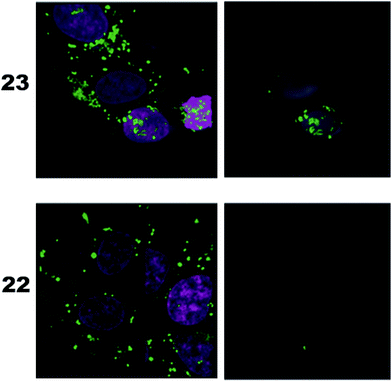
As a complementary study we also ran confocal microscopy in order to confirm that the origin of the signal comes from the nucleus. For this study we decided to compare the compound 23 to control 22. Using 3D image processing software, we defined the volume of the nucleus based on the nuclear staining. Fig. 4 shows the confocal microscope image to the left, and the processed image with the defined nuclear volume and the signal coming from STV-Alexa488 within that volume to the right. Cells treated with STV-Alexa488 complex with 23 showed a clear signal within the defined nuclear volume, whereas control 22 did not.
Conclusions
That, none of the “shorter” mononucleotide m3G-CAP-biotin constructs seem to end up in the nucleus (caps 1–7, constructs 14–21) despite the fact that these “short” CAPs bind well to Snurportin137 suggests that more is required for nuclear transport than just Snurportin binding. It is not clear why the methylene(bisphosphonate) modified m3G-capped trinucleotide is not transported, but suggests that it there is a sensitivity even to minor alterations. These studies demonstrate that efficient transport of a cargo is highly sensitive to the exact design of the Snurportin1-interacting m3G-CAP. Owing to that the only structure, which shows the interaction30 is based on a truncated form of Snurportin1, it is not straightforward to predict the conformations, which are tolerated, let alone to use this structure as a basis for further improvements. Thus, while the existing structure can be used as a starting point, currently the only way to study this interaction is by testing new constructs in biological assays. It should also be mentioned that simply enhancing the binding interaction may not directly predict biological activity, since apart from binding to Snurportin1, interaction with importin β and the release of the cargo may be equally important. One could speculate that this could be related to the interaction of the snurportin1-complex with importin β when this is recruited to obtain nuclear transport.38 In any case, our studies clearly demonstrate that not only the m3G-CAP, but also the adjacent nucleotides are crucial for efficient nuclear transport within cells. The findings suggest that a minimum requirement is three nucleotides connected to the CAP and nuclear transport could well be more efficient with even longer oligonucleotide attached to the m3G-CAP. Further studies on such constructs would be valuable for further exploration of m3G-CAPs as adaptors for transport of therapeutic cargos into the nucleus.Experimental
Materials and methods
Acetonitrile (MeCN, HPLC grade, VWR), methanol (MeOH, Fisher Scientific), dichloromethane (DCM), dimethylformamide (DMF, Merck) and pyridine (Py, Merck) were of commercial grade and dried additionally over 4 Å molecular sieves. Reverse phase HPLC was carried out on a Jasco HPLC system using the Discovery BIO Wide Pore c18-5, 5 μm (250 × 4.6 mm) with 1 ml min−1 flow rate. The buffers used for reversed phase chromatography were as follows: (A): 50 mM TEAA pH 6.5; (B): 50 mM TEAA pH 6.5 in 50% MeCN. Mass spectra (TOF-MS, ES) were obtained using a Micromass LCT electrospray time-of-flight (ES-TOF) instrument. Molecular weights of conjugates were reconstructed from m/z values of the multiply charged ions using the mass deconvolution program (MAXENT) of the instrument, which gave values with no decimal place. N-methyl morpholine (NMM), copper iodide (CuI) and N,N-diisopropylethylamine (DIPEA) were purchased from Aldrich and used without further purification. Following compounds were also obtained commercially and used without further purification EDTA (Fluka), tert-butanol (tBuOH, Merck), dichloroacetic acid (DCA, Merck), iodine (I2, ACROS), and N,N,N′,N′-tetramethyl-O-(1H-benzotriazol-1-yl)uronium hexafluorophosphate (HBTU) (Senn Chemicals). m3G CAPs, 2-(2-azidoethoxy)ethoxyacetic acid and PATA were prepared using known procedures.35General procedure for conjugating m3G CAPs with biotin linkers (“click” reaction in solution)
All reactions were carried out in the Eppendorf tube with a screw cap. Each reagent, unless stated differently, was added to the tube with a syringe or pipette. The lyophilized Biotin linker equipped with triple bond donor PATA was dissolved in 70 μL of tBuOH![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) water (1
water (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) and the solution was added to the Eppendorf tube the lyophilized Cap dissolved in water (1.1 eq., 20 μL). Next, 2 eq. of DIPEA in 5 μL of water (stock solution: 3.5 μL in 996.5 μL water) were added, followed by 2 eq. of copper iodide in 5 μL of DMSO (stock solution: 3.9 mg ml−1). The reaction was placed on vortex and left to stir overnight. The conjugate was purified by RP-HPLC using linear gradient of buffer B in A respectively:
1) and the solution was added to the Eppendorf tube the lyophilized Cap dissolved in water (1.1 eq., 20 μL). Next, 2 eq. of DIPEA in 5 μL of water (stock solution: 3.5 μL in 996.5 μL water) were added, followed by 2 eq. of copper iodide in 5 μL of DMSO (stock solution: 3.9 mg ml−1). The reaction was placed on vortex and left to stir overnight. The conjugate was purified by RP-HPLC using linear gradient of buffer B in A respectively:
14: 0% to 80%, 25 min, tR = 22.3 min, RT, detection at 254 nm;
15: 0% to 80%, 25 min, tR = 21.0 min, RT, detection at 254 nm;
16: 0% to 80%, 25 min, tR = 21.1 min, RT, detection at 254 nm;
17: 0% to 35%, 30 min, tR = 18.6 min, RT, detection at 254 nm;
18: 0% to 80%, 25 min, tR = 21.9 min, RT, detection at 254 nm;
19: 0% to 80%, 25 min, tR = 21.8 min, RT, detection at 254 nm;
20: 0% to 80%, 25 min, tR = 21.9 min, RT, detection at 254 nm;
21: 0% to 80%, 25 min, tR = 22.0 min, RT, detection at 254 nm;
22: 0% to 80%, 25 min, tR = 21.1 min, RT, detection at 254 nm;
23: 0% to 80%, 25 min, tR = 20.1 min, RT, detection at 254 nm;
24: 0% to 80%, 25 min, tR = 22.0 min, RT, detection at 254 nm;
25: 0% to 100%, 15 min, tR = 11.1 min, RT, detection at 254 nm;
26: 0% to 80%, 25 min, tR = 21.1 min, RT, detection at 254 nm.
STV-m3G-CAP-biotin complexation and cell transfections
U2OS cells were plated in a 24-well plate with DMEM + 10% FCS the day before transfection. The day of the transfection, Streptavidin-Alexa488 (Molecular Probes) and m3G-CAP-biotin analogues were incubated for 2 hours at RT. Streptavidin was reconstituted in 1× PBS (pH 7.4). For the complexation, 2 mg of STV (Streptavidin) together with 3.5–4 times molar excess of the m3G-CAP-biotin analogues were used. After the incubation, 20 mM HEPES buffer (pH 7.4) was used to bring the total volume up to 100 μl and then 3.5 μl of PULSin transfection reagent (PolyPlus-Transfection, NY, USA) was added. The mixture was incubated for 15 min at RT. During the incubation time, cells were washed twice with PBS 1× and 900 ml of pre-warmed Opti-Mem I (Invitrogen) was added to the wells. The PULSin complexes were then added to the cells and left to incubate for 4 hours. After 4 hours, the medium was discarded and cells were washed twice with pre-warmed Opti-MEM I. Finally, 400 ml of Opti-MEM I was added to the wells after which the cells were further incubated for 2–3 h. Cells were then analyzed by fluorescence microscopy for nuclear transport.Confocal microscopy
For the confocal analysis, glass microscope cover slips were coated with poly-lysine solution (Sigma Aldrich, 0.1% (w/v)) for 1 hour at RT. Subsequently, 50![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 cells were plated onto coated cover slips in a 24-well plate. 6 hours post-transfection with STV-CAP complexes, the cells were fixed using 4% PFA for 10 min at 37 °C. Then cells were washed with PBS 1× twice. After this, DRAQ5 nuclear stain (Abcam) was added onto the slides and incubated for 5 min at 37 °C. After washing the cells with PBS 1× twice, the cells were mounted using ProLong Antifade mounting medium (ThermoFisher Scientific). Images were obtained on a Nikon A1R confocal system with a 60×/1.49 oil objective, acquired using NIS-Elements A1R software (Version 3.2; Nikon Instruments) and analyzed with Imaris 3D image processing software (Bitplane).
000 cells were plated onto coated cover slips in a 24-well plate. 6 hours post-transfection with STV-CAP complexes, the cells were fixed using 4% PFA for 10 min at 37 °C. Then cells were washed with PBS 1× twice. After this, DRAQ5 nuclear stain (Abcam) was added onto the slides and incubated for 5 min at 37 °C. After washing the cells with PBS 1× twice, the cells were mounted using ProLong Antifade mounting medium (ThermoFisher Scientific). Images were obtained on a Nikon A1R confocal system with a 60×/1.49 oil objective, acquired using NIS-Elements A1R software (Version 3.2; Nikon Instruments) and analyzed with Imaris 3D image processing software (Bitplane).
Acknowledgements
We gratefully acknowledge funding from Karolinska Institute, the Swedish Research Council and the Polish National Center of Research and Development (02/EuroNanoMed/2011) and The National Science Centre (Poland, UMO-2012/05/E/ST5/03893).Notes and references
- K. M. Wagstaff and D. A. Jans, Biochem. J., 2007, 406, 185–202 CrossRef CAS PubMed.
- E. Grossman, O. Medalia and M. Zwerger, Annu. Rev. Biophys., 2012, 41, 557–584 CrossRef CAS PubMed.
- G. Kabachinski and T. U. Schwartz, J. Cell Sci., 2015, 128, 423–429 CrossRef CAS PubMed.
- C. M. Feldherr and D. Akin, J. Cell Sci., 1997, 110(24), 3065–3070 CAS.
- O. Keminer and R. Peters, Biophys. J., 1999, 77, 217–228 CrossRef CAS PubMed.
- B. Cautain, R. Hill, P. N. de and W. Link, FEBS J., 2015, 282, 445–462 CrossRef CAS PubMed.
- P. Kalab, K. Weis and R. Heald, Science, 2002, 295, 2452–2456 CrossRef CAS PubMed.
- A. H. Kettler and L. H. Goldberg, Am. Fam. Physician, 1986, 34, 147–152 CAS.
- R. Budai, O. Galaktion-Nitselv and E. Churia, Zh. Nevropatol. Psikhiatr. im. S. S. Korsakova, 1965, 65, 1139–1142 CAS.
- D. Kalderon, B. L. Roberts, W. D. Richardson and A. E. Smith, Cell, 1984, 39, 499–509 CrossRef CAS PubMed.
- D. S. Goldfarb, J. Gariepy, G. Schoolnik and R. D. Kornberg, Nature, 1986, 322, 641–644 CrossRef CAS PubMed.
- A. Lange, R. E. Mills, C. J. Lange, M. Stewart, S. E. Devine and A. H. Corbett, J. Biol. Chem., 2007, 282, 5101–5105 CrossRef CAS PubMed.
- M. Soniat and Y. M. Chook, Biochem. J., 2015, 468, 353–362 CrossRef CAS PubMed.
- L. J. Branden, A. J. Mohamed and C. I. E. Smith, Nat. Biotechnol., 1999, 17, 784–787 CrossRef CAS PubMed.
- P. Collas, H. Husebye and P. Alestrom, Transgenic Res., 1996, 5, 451–458 CrossRef CAS PubMed.
- W. Ritter, C. Plank, J. Lausier, C. Rudolph, D. Zink, D. Reinhardt and J. Rosenecker, J. Mol. Med., 2003, 81, 708–717 CrossRef CAS PubMed.
- J. J. Ludtke, G. Zhang, M. G. Sebestyen and J. A. Wolff, J. Cell Sci., 1999, 112(12), 2033–2041 CAS.
- T. Shiraishi, R. Hamzavi and P. E. Nielsen, Bioconjugate Chem., 2005, 16, 1112–1116 CrossRef CAS PubMed.
- H. Zhang, A. Mitin and S. V. Vinogradov, Bioconjugate Chem., 2009, 20, 120–128 CrossRef CAS PubMed.
- L. J. Branden, B. Christensson and C. I. Smith, Gene Ther., 2001, 8, 84–87 CrossRef CAS PubMed.
- E. Park, H.-B. Cho and K. Takimoto, Cytotherapy, 2015, 17, 536–542 CrossRef CAS PubMed.
- C. L. Will and R. Luhrmann, Cold Spring Harbor Perspect. Biol., 2011, 3(7), a003707 CAS.
- E. Izaurralde, J. Lewis, C. Gamberi, A. Jarmolowski, C. McGuigan and I. W. Mattaj, Nature, 1995, 376, 709–712 CrossRef CAS PubMed.
- S. Kitao, A. Segref, J. Kast, M. Wilm, I. W. Mattaj and M. Ohno, Mol. Cell. Biol., 2008, 28, 487–497 CrossRef CAS PubMed.
- U. Fischer, C. Englbrecht and A. Chari, Wiley Interdiscip. Rev.: RNA, 2011, 2, 718–731 CrossRef CAS PubMed.
- G. Plessel, U. Fischer and R. Luhrmann, Mol. Cell. Biol., 1994, 14, 4160–4172 CrossRef CAS PubMed.
- J. Mouaikel, U. Narayanan, C. Verheggen, A. G. Matera, E. Bertrand, J. Tazi and R. Bordonne, EMBO Rep., 2003, 4, 616–622 CrossRef CAS PubMed.
- J. Huber, A. Dickmanns and R. Luhrmann, J. Cell Biol., 2002, 156, 467–479 CrossRef CAS PubMed.
- J. Huber, U. Cronshagen, M. Kadokura, C. Marshallsay, T. Wada, M. Sekine and R. Luhrmann, EMBO J., 1998, 17, 4114–4126 CrossRef CAS PubMed.
- A. Strasser, A. Dickmanns, R. Luehrmann and R. Ficner, EMBO J., 2005, 24, 2235–2243 CrossRef CAS PubMed.
- P. M. D. Moreno, M. Wenska, K. E. Lundin, O. Wrange, R. Stroemberg and C. I. E. Smith, Nucleic Acids Res., 2009, 37, 1925–1935 CrossRef CAS PubMed.
- M. Honcharenko, J. Romanowska, M. Alvira, M. Jezowska, M. Kjellgren, C. I. Edvard Smith and R. Stroemberg, RSC Adv., 2012, 2, 12949–12962 RSC.
- M. Honcharenko, M. Zytek, B. Bestas, P. Moreno, J. Jemielity, E. Darzynkiewicz, C. I. E. Smith and R. Stroemberg, Bioorg. Med. Chem., 2013, 21, 7921–7928 CrossRef CAS PubMed.
- B. A. Wojtczak, M. Warminski, J. Kowalska, M. Lukaszewicz, M. Honcharenko, C. I. E. Smith, R. Stromberg, E. Darzynkiewicz and J. Jemielity, RSC Adv., 2016, 6, 8317–8328 RSC.
- M. Wenska, M. Alvira, P. Steunenberg, A. Stenberg, M. Murtola and R. Stroemberg, Nucleic Acids Res., 2011, 39, 9047–9059 CrossRef CAS PubMed.
- M. Jezowska, J. Romanowska, B. Bestas, U. Tedebark and M. Honcharenko, Molecules, 2012, 17, 14174–14185 CrossRef CAS PubMed.
- M. Zytek, J. Kowalska, M. Lukaszewicz, B. A. Wojtczak, J. Zuberek, A. Ferenc-Mrozek, E. Darzynkiewicz, A. Niedzwiecka and J. Jemielity, Org. Biomol. Chem., 2014, 12, 9184–9199 CAS.
- G. Mitrousis, A. S. Olia, N. Walker-Kopp and G. Cingolani, J. Biol. Chem., 2008, 283, 7877–7884 CrossRef CAS PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c6ra09568b |
| ‡ Authors equally contributed to the work. |
| This journal is © The Royal Society of Chemistry 2016 |