Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Impact of different alginate lyases on combined cellulase–lyase saccharification of brown seaweed†
D.
Manns
a,
C.
Nyffenegger
a,
B.
Saake
b and
A. S.
Meyer
*a
aCenter for BioProcess Engineering, Department of Chemical and Biochemical Engineering, Technical University of Denmark, DK-2800 Kgs. Lyngby, Denmark. E-mail: am@kt.dtu.dk
bChemical Wood Technology, Department of Wood Science, University of Hamburg, Hamburg, Germany
First published on 4th May 2016
Abstract
Two bacterial polysaccharide lyase (PL) family 7 alginate lyases (EC 4.2.2.-) from Sphingomonas sp. (SALy) and Flavobacterium sp. (FALy), respectively, were selected for heterologous, monocomponent expression in Escherichia coli. The thermal stability, pH, and temperature reaction optima and substrate preferences of the enzymes on different alginate polymers were assessed and compared to those of a commercially available microbial alginate lyase (SigmALy). The optimal pH range for SALy was pH 5.5–7.0; for FALy and SigmALy it was pH 7.5. Reaction temperatures of 30–50 °C had no influence on the activity of any of the enzymes, but the thermal stability was reduced above 50 °C. The FALy enzyme preferred poly-mannuronic acid as substrate, but exhibited activity also on poly-guluronic acid, whereas the SALy had highest activity on poly-guluronic acid, and the SigmALy was active only on poly-guluronic acid. When applied together with a fungal cellulase preparation (Cellic®CTec2) at pH 6 and 40 °C on a glucan rich brown seaweed Laminaria digitata the viscosity decreased in the initial minutes while measurable alginate degradation occurred primarily within the first 1–2 hours of reaction. Whereas FALy and SALy addition catalyzed degradation of more alginate in L. digitata than SigmALy addition, only the SigmAly enabled release of 90% of the available glucose within 8 hours of combined enzyme treatment. The level of mannuronic acid moieties released was inversely proportional to the glucose release, indicating that the degradation of mannuronic acid blocks inhibited cellulase catalyzed glucose release from L. digitata. Nevertheless, combined alginate lyase and cellulase treatment for 24 hours released all potential glucose regardless of the applied lyase. The enzymatic treatment moreover induced solubilization of sulfated fucoidan, whereas most of the nitrogen was recovered in the residual seaweed solids.
1 Introduction
There is a growing interest in using macroalgae, i.e. seaweeds, as a potential new biomass resource for bioenergy and biomass derived chemicals production.1,2 The brown seaweed Laminaria digitata harvested in late summer in the Danish North Sea (off Hanstholm) has been found to have a low ash content and a very high glucan content with glucose moieties constituting 51% by weight of the dry matter.3L. digitata is therefore considered a particularly suitable brown seaweed glucose feedstock when harvested at the right time and place.3Brown seaweeds are also rich in alginate, and application of alginate lyases (EC 4.2.2.-) for brown seaweed saccharification4 and for improving enzymatic glucose release has been evaluated recently.5,6 When combined with fungal cellulases, alginate lyase addition appears to induce viscosity decrease and alginate removal from the cell wall matrix of the brown seaweed to help cellulase catalyzed saccharification of glucan (laminarin and cellulose) to enhance glucose release.5
Alginate polysaccharides are unique to brown seaweeds and consist of 1,4-glycosidically linked α-L-guluronic acid (G) and β-D-mannuronic acid (M) in varying proportions forming linear chains with M/G ratios ranging from 1.2 to 3. The linear chains are made up of alternating long blocks of guluronic (GG) and mannuronic (MM) acids with DP 90–100, but less crystalline and shorter MG/GM blocks may also occur.7,8 Alginate lyases catalyze depolymerization of alginates via a β-elimination reaction. Alginate lyases are classified as EC 4.2.2.-. Preference towards G-blocks (poly-guluronate lyase) is classified as EC 4.2.2.11 and specificity towards M-blocks (poly-mannuronate lyase) as EC 4.2.2.3.4,9 Alginate lyases are mainly divided into two polysaccharide lyase (PL) families, PL 5 and 7, exhibiting preference for poly-(M) and poly-(G) blocks, respectively, but enzymes categorized in PL family 6, 14, 15, 17, and 18 have also been categorized to have alginate lyase activity (with different substrate specificities including “MG-specific”). Additionally, alginate lyases with high activity on both homopolymers have been isolated from varies sources.10 Even if an alginate lyase is classified as M or G specific, the catalytic degradation of alternating blocks and activity towards “the other” homopolymer may take place. Endolytic alginate lyases have been reported to have higher activity than exolytic lyases making them promising catalysts for alginate degradation.11,12
The objective of this study was to examine substrate specificity and substrate viscosity impact of different microbial alginate lyases to improve the knowledge base for their use in seaweed biorefining to support enzymatic glucose release.
2 Experimental
2.1 Alginate lyase cloning, expression and purification
Genes encoding two different alginate lyases (EC 4.2.2.3), namely A1-II′ from Sphingomonas sp. (SALy) and Alg2A from Flavobacterium sp. (FALy), were selected from a literature search.12–14 The Protein Data Bank accession number for SALy, in fact a recombinantly expressed truncated A1-II′ alginate lyase lacking 80 terminal amino acid residues12 is 2CWS and the GenBank accession number for FALy is JF412659.For both alginate lyases, DNA constructs were engineered to also encode an N-terminal His6-tag that was linked to the ORF via a thrombin recognition site. Constructs were codon optimized for expression in Escherichia coli (E. coli), synthesized and inserted into the vectors pET21b (T7 promotor) and pET21a, respectively, by DNA2.0 (Menlo Park, CA, USA). E. coli BL21 (DE3), E. coli C41 and C43 (DE3) and E. coli Tuner (DE3) were transformed with the resulting plasmids and selected for ampicillin resistance. Overnight cultures from single colonies were used for IPTG- and autoinduction of alginate lyase expression.
For IPTG induction, lysogenic broth (LB) supplemented with 50 μg mL−1 ampicillin, inoculated with an overnight culture to a starting OD600 of 0.1 was grown at 37 °C. When reaching an OD600 of 0.6, the temperature was reduced to 25 °C, and expression induced by addition of IPTG to a final concentration of 1 mM. Cells were grown overnight post induction before harvest. Expression with autoinduction was done as follows: an overnight culture was used to inoculate the autoinduction medium (6 g Na2HPO4, 3 g KH2PO4, 20 g tryptone, 5 g yeast extract, 5 g NaCl, 0.06% v/v glycerol, 0.05% w/v glucose and 0.04% w/v α-lactose, pH 7.2) to a starting OD600 of 0.1, cells were grown overnight at 25 °C, and harvested.
The expression of the lyases with the two expression strategies was evaluated by sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE, BioRad, CA, US). The most promising expression strategy was selected for scaled-up expression.
After the scaled-up expression, the cells were centrifuged and the pellets resuspended in Ni2+-column binding buffer (20 mM Na-phosphate buffer, 500 mM NaCl and 20 mM imidazole, pH 7), then sonicated to open the cells and centrifuged to remove cell debris. The supernatant containing the lyases was passed through a 0.45 μm filter before being loaded onto a 1 mL Ni2+-Sepharose HisTrap HP column (GE Healthcare, Uppsala, Sweden), equilibrated with binding buffer, on an ÄKTA purifier (GE Healthcare, Uppsala, Sweden). Unbound material was washed off the column with 5 column volumes (CV) of binding buffer. The alginate lyases were eluted with elution buffer (binding buffer with 500 mM imidazole) in a gradient from 0–100% elution buffer in 15 CV.
Protein purity was confirmed by SDS-PAGE and the protein concentration determined by the Lambert Beer law using baseline corrected absorption at 280 nm and extinction coefficients of 43![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 890 M−1 cm−1 (SALy) and 55
890 M−1 cm−1 (SALy) and 55![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 350 M−1 cm−1 (FALy), respectively.15 The benchmark alginate lyase, SigmALy, was purchased from Sigma-Aldrich (Steinheim, Germany) (undisclosed microbial origin, but previously Sigma-Aldrich presented this enzyme as being derived from Flavobacterium multivorum).
350 M−1 cm−1 (FALy), respectively.15 The benchmark alginate lyase, SigmALy, was purchased from Sigma-Aldrich (Steinheim, Germany) (undisclosed microbial origin, but previously Sigma-Aldrich presented this enzyme as being derived from Flavobacterium multivorum).
2.2 Seaweed
Laminaria digitata was harvested from the Danish North Sea coast end of August 2012. Prior to processing the material was washed successively four times with water to remove residual sand and salt. After washing, the seaweed biomass was stored at −20 °C until use. The dry matter content was determined after thawing. By weight, the dry biomass consisted of 57.1% glucose, 8% mannitol, 17.2% mannuronic and 5.7% guluronic acid, and ∼4.5% of other carbohydrate moieties (calculated as hydrated monomers).3 The seaweed was processed through a lab refiner mill, at a disc distance of 0.3 mm, as described earlier.5 Characterization of the milled Laminaria digitata gave (hydrated monomer) glucose and mannitol concentrations of 46.6 and 6.7% of dry weight. The contents of the other carbohydrates were the same as before milling.2.3 Alginate lyase characterization
To determine the optimum pH and temperature a randomized face-centered composite design was employed (Jump®9 program (SAS)). Reaction temperature was varied between 30 and 50 °C and pH from 4.5 to 8.5. For the thermal stability studies, enzyme solutions were incubated at different temperatures (40 to 60 °C) at pH 6 and 7 for 0, 15, 45, 90, 240, and 480 min before the activity was measured. Enzyme activity was measured on sodium alginate (Sigma-Aldrich, Steinheim, Germany) at a substrate concentration of 0.2% w/v at the particular pH in phosphate–citrate buffer at 40 °C (in the temperature optimization experiments the temperature was varied according to the experimental design). Activity was determined online over time in an Infinite 200 microplate reader (TECAN, Salzburg, Austria) with continuous data collection (Tecan i-control v 1.5.14.0, TECAN, Salzburg, Austria). Activity was quantified as formation of double bonds at an absorbance of 235 nm caused by lyase induced β-elimination.For the substrate specificity assessment, activity was measured on sodium alginate, poly-mannuronic acid (>5000 kDa and <5000 kDa) and guluronic acid, respectively, at pH 7. Pure substrates were purchased from Carbosynth Ltd., Berkshire, UK.
2.4 Enzymatic decomposition of brown seaweed
Enzymatic seaweed saccharification was conducted on 650 mg substrate by dry weight in 13 mL of slurry (5% substrate concentration). Temperature was 40 °C in a buffer system at pH 6 with 51 mM phosphate 14 mM citrate buffer. Treatment was performed with 1% E/S (Enzyme/Substrate level in % by weight) of the selected alginate lyase and 10% E/S (v/w) of the cellulase preparation Cellic®CTec2 (Novozymes A/S, Bagsværd, Denmark) in a horizontal roller mixer at 60 rpm. Samples of 250 μL were taken at 0, 0.5, 1, 2, 4, 6 and 8 hours during the enzymatic liquefaction.Viscosity assessment during enzymatic treatment was done on 1500 mg dry material (5% substrate concentration) in a Rapid Visco analyser RVA (Newport, UK) every 8 seconds for a total of 60 minutes at 60 rpm. Subsequently, samples were transferred to the roller mixer and the treatment was extended to a total of 24 hours. Samples were taken at 2, 4, 8, 14 and 24 hours. Reactions were stopped by addition of 5 M NaOH. After reaction the liquefied fraction was decanted from the insoluble pellet remaining after centrifugation for 30 min at 14![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 × g. Enzymatic treatments on pure laminarin (Sigma-Aldrich) and poly-mannuronic acid (>5000 kDa) were conducted in Eppendorf tubes in a thermomixer at 1400 rpm. For these sequential enzymatic treatments the poly-mannuronic acid was first enzymatically treated with the particular alginate lyase for one hour and the reaction stopped by heat (95 °C for 10 min), then the slurry was mixed with laminarin. Reaction conditions were set as those used for the enzymatic experiments described above. The substrate concentration ratio in the reactions corresponded to the available glucan to alginic acid as present in the fresh seaweed. Laminarin was deconstructed by treatment with Cellic®CTec2 for 30, 60 and 120 minutes and the reaction stopped by heat (95 °C for 10 min).
000 × g. Enzymatic treatments on pure laminarin (Sigma-Aldrich) and poly-mannuronic acid (>5000 kDa) were conducted in Eppendorf tubes in a thermomixer at 1400 rpm. For these sequential enzymatic treatments the poly-mannuronic acid was first enzymatically treated with the particular alginate lyase for one hour and the reaction stopped by heat (95 °C for 10 min), then the slurry was mixed with laminarin. Reaction conditions were set as those used for the enzymatic experiments described above. The substrate concentration ratio in the reactions corresponded to the available glucan to alginic acid as present in the fresh seaweed. Laminarin was deconstructed by treatment with Cellic®CTec2 for 30, 60 and 120 minutes and the reaction stopped by heat (95 °C for 10 min).
2.5 Sulfuric acid hydrolysis
After lyophilization a 2-step sulfuric acid hydrolysis described previously3 was applied on (i) the refiner milled slurry, (ii) the insoluble residues remaining after enzymatic treatment, and (iii) the enzymatically released sugar solutions, prior to carbohydrate analysis by high performance anion exchange chromatography with pulsed amperometric detection (HPAEC-PAD), see below.2.6 Carbohydrate analysis
2.7 Elemental analysis
C, H, N and S contents in the seaweed were measured using a Vario EL cube elemental analyzer (Elementar Hanau, Germany).2.8 Statistics
One-way analyses of variances (one-way ANOVA): 95% confidence intervals were compared as Tukey–Kramer intervals calculated from pooled standard deviations (Minitab Statistical Software, Addison-Wesley, Reading, MA).3 Results and discussion
3.1 Recombinant enzyme expression
Sphingomonas sp. strain A1 encodes three endotype alginate lyases (A1-I, A1-II [family PL7], and A1-III [family PL5]) and additionally harbors what appears to be a silent gene, the A1-II′ gene,12 which codes for a PL7 alginate lyase designated A1-II′. Based on the broad activity found for the recombinantly expressed A1-II′ enzyme on alginate molecules (poly-(G), poly-(M), and poly-(MG)) the application potential of the A1-II′ enzyme (SALy) in alginate processing has been proposed.13,17 Recently, another alginate lyase, derived from a Flavobacterium sp., was discovered and also proposed to have application potential for alginate degradation and for biochemicals and biofuels production from brown seaweeds.14 This Flavobacterium derived alginate lyase (FALy) was previously successfully overexpressed in E. coli BL21 with IPTG induction.14 However, we found this enzyme to express best in E. coli C41 with autoinduction (Fig. S1, ESI†). The Sphingomonas sp. enzyme (SALy) was successfully expressed at high yields (12.8 g L−1 cell extract) and purified from the cell extract of an E. coli BL21 transformant (Fig. S1, ESI†). In terms of mode of action both the SALy and the FALy were described previously as being endolytic and belonging to family 7 of the Polysaccharide Lyases (PL).12–14,17The FALy was moreover previously reported to mainly release tri-saccharides, but also to be able to release di-, tetra-, penta-, and hexa-oligo-saccharides in lesser amounts.14 The depolymerization pattern of alginate after treatment with SALy was reported to result in final products of unsaturated tri- and tetrasaccharide uronates.18
In order to assess the possible use of these alginate lyases in brown seaweed saccharification, the pH and temperature activity responses, thermal stabilities, and substrate specificities of SALy and FALy were investigated and compared to those of the commercially purchased alginate lyase (SigmALy).
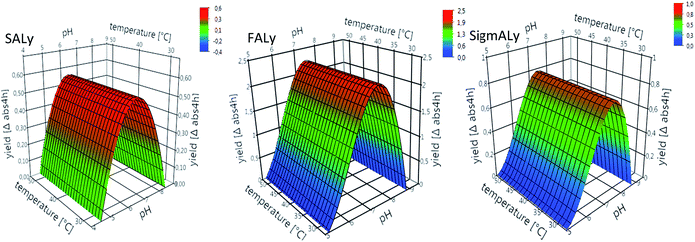
Alginate lyases from Flavobacterium sp. have previously been reported to have pH optimum of ∼7.5 and with the pH optimum being dependent on temperature in the range 20–35 °C.16,19 When comparing the activity response models for the FALy and SigmALy, the FALy showed higher activity towards higher pH (Fig. 1). Originally, the optimal pH for the FALy enzyme was reported as 8.5 (at 40–45 °C) with only about 40% activity at pH 7.5.14 In the activity response surface model ∼1/3 of the max activity of the FALy was achieved at pH 6 (Fig. 1). Other PL 7 lyases have been reported to have pH optimum from pH 7–8.5 and optimal reaction temperature at ∼50 °C.10,20 Due to its lower pH optimum, notably the SALy could be an appropriate candidate to combine with fungal cellulases to increase glucose release from brown seaweeds. The findings for SALy (Fig. 1) contrasted those reported by Miyake et al.12 They reported this enzyme to have highest activity at pH 7.5, and moreover found the enzyme to have temperature optimum at 40 °C and a fast decrease in activity above 45 °C.12 Yoon et al.18 expressed the three alginate lyases A1-I, A1-II, and A1-III from Sphingomonas sp. and found the A1-II enzyme to be most active at pH 8 and 70 °C, but also reported that 50% activity was lost after only 10 min at 50 °C.18 Recently, another alginate lyase derived from Sphingomonas sp. was characterized as having pH and temperature optima of pH 6.5 and 50 °C,21i.e. in accord with our data for the SALy.
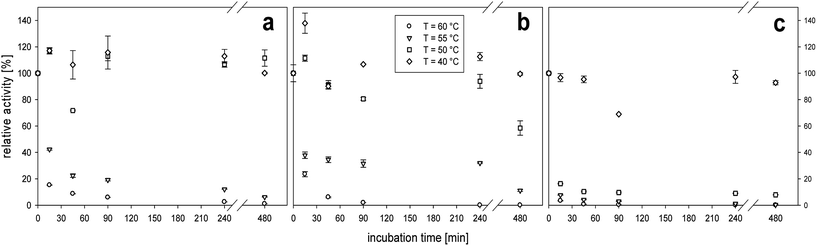
Likewise, in the present work, the FALy remained active at elevated temperatures, i.e. retained 30–40% activity at 55 °C for 4 hours but lost its activity drastically at 60 °C (Fig. 2b). This enzyme has been described previously to be stable for 1 h only up to 45 °C.14,20 Previously reported stability tests14 have employed incubation in 50 mM Tris–HCl buffer at pH 8.5, and we ascribe the stability differences to be due to differences in the methodologies used.
The purchased alginate lyase, SigmALy, was only stable at the lowest incubation temperature of 40 °C (Fig. 2c). Temperatures of ≥50 °C thus led to immediate activity loss (Fig. 2c). The thermal stability experiment was also conducted at pH 7, and enzyme activity readings revealed similar thermal stabilities as described for pH 6 (data not shown). The SALy was generally more active at pH 6 while it was the other way around for FALy and SigmALy. The data are in accordance with the determined optima for pH and temperature (see Section 3.1.1). As outlined below, in Section 3.2, the thermal stability of the alginate lyases was sufficient to boost cellulase action during extended saccharification at 40 °C. Higher thermal stability of the alginate lyases may nevertheless be required to allow processing at elevated temperatures, where e.g. microbial contamination risks are lower, and substrate solubility may be higher. Enzyme immobilization is not a first choice in alginate biomass applications as mass transfer constraints are high because of the insolubility/viscosity of the substrate. Instead, increased thermostability may be achievable via targeted enzyme protein engineering as proven successful for other bacterial lyases.
| Enzyme | Initial rate [% of total substrate/min] | |||
|---|---|---|---|---|
| Poly-M < 5000 kDa | Poly-M > 5000 kDa | Poly-G | Na-alginate | |
| SALy | 0.013 | 0.014 | 0.022 | 0.036 |
| FALy | 0.111 | 0.102 | 0.058 | 0.093 |
| SigmALy | (0.034) | (0.046) | 0.122 | 0.063 |
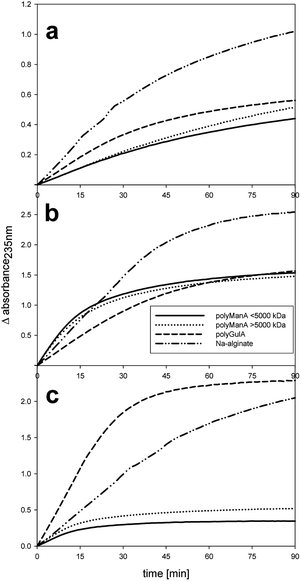
After an initial active cleavage period of about 5 min on poly-(M) the reaction of the SigmALy almost ceased (Fig. 3).
In contrast, the SigmALy had high activity on poly-(G) (Table 1, Fig. 3). It is likely that the initial cleavage by SigmALy of poly-(M) was an artifact, due to impurity of the substrate since approx. 15% by weight of the poly-(M) was analyzed as guluronic acid by HPAEC-PAD post sulfuric acid treatment (data not shown).
The SigmALy has previously been reported to be substrate specific towards guluronate components.14 This distinct substrate selectivity is rare as hitherto reported alginate lyases usually display at least a moderate processivity for the other heteropolymer.9,22
Overall, FALy had the highest decomposition ability, especially when taking into account that this enzyme was assessed at the lowest enzyme loading of 0.03% enzyme per substrate (E/S) (Fig. 3). After 30 min of reaction, i.e. the initial reaction, SALy created per release of unsaturated M-unit (>5000 kDa) 1.5 unsaturated G-units while the FALy catalyzed the release of 0.8 units of unsaturated G-units blocks per release of unsaturated M-unit (>5000 kDa) (Fig. 3).
3.2 Application on brown seaweed
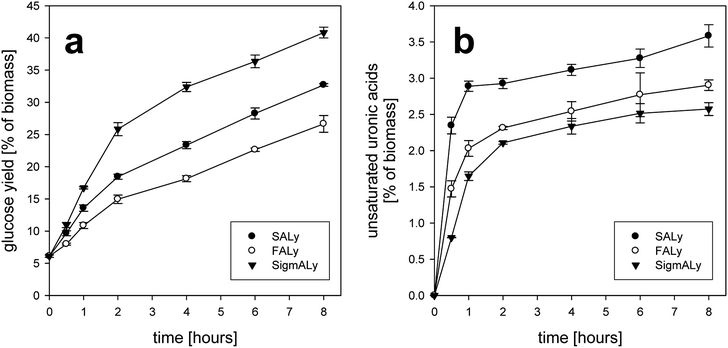
Combined application of alginate lyase and cellulase was found previously to be superior over cellulase application alone on the release of glucose from Laminaria digitata.5,6 The alginate lyase apparently catalyze the removal of alginate to render the laminarin and cellulose more susceptible to cellulase catalyzed degradation.5 This apparent synergistic effect of the alginate lyase–cellulase combination can be explained when considering that the matrix polysaccharides (alginate, glucans, and fucose-containing sulfated polysaccharides) are presumed to be tightly associated in the brown seaweed cell walls.23A glucose yield of 40.8% of dry weight milled seaweed corresponds to 87.6% of the potential available glucose after 8 hours of treatment (Fig. 4a). Application together with the lyases FALy and SALy was assumed to perform better. As expected, the lyases showed activity towards both homopolymers, poly-(M) and poly-(G), and the pH optimum for SALy suited better with application of pH 6 (Section 3.1). The release of glucose was significantly lower than expected. After 2 hours the FALy released 14.9% and SALy 18.4% of glucose from total seaweed by dry weight. This corresponded to 58%, respectively 71% compared to what has been released by the cellulase preparation applied together with SigmALy (Fig. 4a). The glucose yield for treatment with SigmALy started to bend off after 2 hours. Hence, the comparative relative yields of FALy and SALy rose to 65% (26.7% w/w biomass), respectively 80% (32.7% w/w biomass) after 8 hours of treatment.
Lower lyase dosis (of the purchased lyase from Sigma-Aldrich) but also lower substrate loading with maximum glucose recovery of 80% (∼25% after 8 hours) was achieved previously on dried and milled material after 24 hours of enzymatic treatment.6 Equally overall sugar recovery (glucose and mannitol) of over 90% was reached with a substrate concentration of 15% w/v but was 78% with increased solid loading (25%) after 29 hours with no change over treatment extension until 48 hours.24 Both investigations were conducted on dried material using the cellulase preparation Celluclast 1.5L and cellobiase 188 (Novozymes) at about pH 5. The preparation Celluclast 1.5L released less reducing sugars than another commercially available cellulase from L. digitata.25 Drying has been shown to hinder enzymatic glucose release, although from lignocellulosic material.26,27
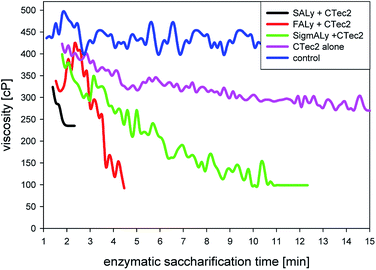
Celluclast has been found to retain 80% of the activity at pH 6 (optimum pH 5.2) on brown seaweed Macrocystis pyrifera. In contrast, for alginate lyases, including the endo-type lyase from Sigma-Aldrich, the activity was below 10% at pH 6 and about one third at pH 7 compared to at pH 7.5.28 With respect to temperature, glucose release could be enhanced by raising the temperature as yields were doubled by a temperature increase from 37 °C to 50 °C.28 However, to allow suitable conditions for all alginate lyases, the temperatures in the experiment of Fig. 5 were set to 40 °C. The purchased lyase from Sigma-Aldrich (SigmALy) was tested to have significant activity losses for temperatures ≥ 50 °C (Section 3.1.2).
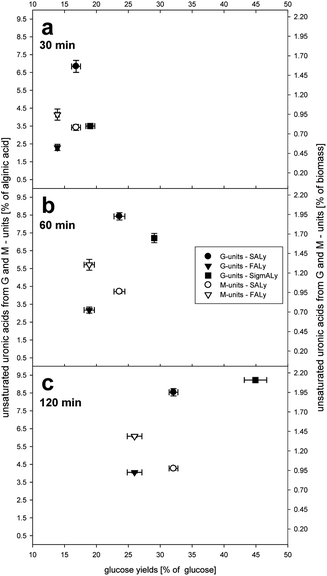 | ||
| Fig. 5 Yields of unsaturated uronic acids deriving from poly-guluronic acids (G-units, left y-axis) and poly-mannuronic acids (M-units, right y-axis) as % of potential total alginic acid of Laminaria over glucose yields (% of potential glucose); after 30 (a), 60 (b) and 120 min (c) of enzymatic saccharification with Cellic®CTec2 and alginate lyase (SALy, FALy and purchased lyase SigmALy). Potential total content of glucose and alginic acid monomers determined by HPAEC post sulfuric acid hydrolysis.3 Each data point represents the average value of independent duplicates, bi-dimensional bars indicate the standard deviation. | ||
For saccharification of the alginate within the seaweed only the first two hours of reaction appeared to be crucial as the uronic acids release leveled off at the end of reaction (Fig. 4b). This reaction pattern (leveling off) on alginate degradation with lyase concentration of ≥1% per substrate has been seen before.29 Moreover, Fig. 4b showed that the initial fast degradation of the alginate was achievable already within one hour with the application of SALy (2.9% unsaturated uronic acid residues per dry biomass). Potentially, this could be a result of a higher enzyme activity due to the more suitable pH conditions (pH optimum for SALy was 6.5; Section 3.2.1). Thomas et al.16 reported an intermediate initial degradation to larger oligosaccharides (DP 4 to 20) followed by a slower chopping of the alginate into dimeric uronates of DP of 2 over several hours.
Whereas nearly complete glucose release was achieved from just milled seaweed (Fig. 5a) a harsher pretreatment for decomposition of alginate from brown seaweed was proposed elsewhere.28 A 5-fold increase of uronic acids after 2 hour treatment with exo- and endo-alginate-lyases was reported post a sulfuric acid pretreatment compared to none.28 However, the seaweed Macrocystis pyrifera was dried and cut prior to use.28
In general, yields of unsaturated uronic acids were lower with FALy and SigmALy than with SALy (Fig. 4b). First, pH 6 was closer to the optimum of SALy. However, SigmALy was still efficient at pH 5.5 Second, SigmALy was not active on to poly-mannuronic acid unlike the other lyases. 4-Deoxy-4,5-unsaturated mono-uronates, such as the reaction products from exolytic alginate lyase action, will gradually convert non-enzymatically to the stable 5-keto structure, which does not absorb at 235 nm. Hence, a UV-absorbance product profile (A235) for exo-lyases producing unsaturated uronate monosaccharides will first provide an absorption increase, followed by a decrease.16 The A235-profiles in the present work did not show any such absorbance decrease after the absorption (Fig. 4b), confirming that the applied lyases were endo-acting, which appeared sufficient for the release of glucose (Fig. 5a).
Nonetheless, addition of exolytic oligoalginate lyase to produce monosaccharide units of alginate was recently demonstrated.21,29 Further, these monosaccharides were shown to be available for ethanol production by a newly discovered organism.30
Regardless of the mannuronic acid content an efficient disruption of alginate requires a lyase with high activity on G–G linkages.31 Based on the initial rates derived from the lyase activity on pure substrates (Table 1) ratios of G-cleavages to M-cleavages (G![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) M cleavage ratio) were calculated. G
M cleavage ratio) were calculated. G![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) M cleavage for SALy was 1.5
M cleavage for SALy was 1.5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 and for FALy 0.6
1 and for FALy 0.6![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1. SigmALy was not active on mannuronic acid. Subsequently, these ratios were transferred to calculate if the amount of unsaturated uronic acids derived from seaweed alginate saccharification (Fig. 4b) whether could be attributed to the cleavage of G-units or M-units (Fig. 5). The unsaturated M- and G-units were plotted over the glucose yields for the crucial first 2 hours (30, 60 and 120 min) of alginate degradation. Hence, the more unsaturated M-units were released the lower the glucose yields were obtained (Fig. 5).
1. SigmALy was not active on mannuronic acid. Subsequently, these ratios were transferred to calculate if the amount of unsaturated uronic acids derived from seaweed alginate saccharification (Fig. 4b) whether could be attributed to the cleavage of G-units or M-units (Fig. 5). The unsaturated M- and G-units were plotted over the glucose yields for the crucial first 2 hours (30, 60 and 120 min) of alginate degradation. Hence, the more unsaturated M-units were released the lower the glucose yields were obtained (Fig. 5).
The M/G ratio in the present L. digitata was 3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 with a total amount of guluronic acid of 5.7% (w/w).3 Taking into account the fact that the purchased lyase was almost only active on poly-(G) approx. 37% of all present guluronic acid did undergo a β-elimination leading to unsaturated uronic acid at the reducing end with an average DP of 2–3 (Fig. 5c). The mode of action of the purchased lyase from Sigma-Aldrich (SigmALy) was described as endolytic, releasing mainly trimers.14 In the same study FALy was found to release oligomers of DP 5–7 within the first 20 h of reaction. Hence, the presence of longer oligomers could describe the lower yields of unsaturated uronic acids deriving from G-units of FALy compared to the other two lyases (Fig. 5). For FALy the yield stabilized at 18% after 2 hours of reaction and would have produced oligomers of average DP 5–6 (Fig. 5c). SALy was described to release tri- and tetrasaccharides.14,18 This corresponded with the release of unsaturated (G)-units from seaweed using SALy of 8.6% of the total content of alginate (equal to 34% of guluronic acid). Hence, G-unit trimers were achieved already after 60 min of reaction (Fig. 5b). Further, a significant consumption from of guluronic acid after 120 min could not be observed (Fig. 5c). However, also for (M)-units no significant further increase over reaction time over 2 hours was achieved, both poly-(M) active lyases (SALy and FALy) similarly only released unsaturated M-units of 4–6% of the total mannuronic acid (Fig. 5c).
1 with a total amount of guluronic acid of 5.7% (w/w).3 Taking into account the fact that the purchased lyase was almost only active on poly-(G) approx. 37% of all present guluronic acid did undergo a β-elimination leading to unsaturated uronic acid at the reducing end with an average DP of 2–3 (Fig. 5c). The mode of action of the purchased lyase from Sigma-Aldrich (SigmALy) was described as endolytic, releasing mainly trimers.14 In the same study FALy was found to release oligomers of DP 5–7 within the first 20 h of reaction. Hence, the presence of longer oligomers could describe the lower yields of unsaturated uronic acids deriving from G-units of FALy compared to the other two lyases (Fig. 5). For FALy the yield stabilized at 18% after 2 hours of reaction and would have produced oligomers of average DP 5–6 (Fig. 5c). SALy was described to release tri- and tetrasaccharides.14,18 This corresponded with the release of unsaturated (G)-units from seaweed using SALy of 8.6% of the total content of alginate (equal to 34% of guluronic acid). Hence, G-unit trimers were achieved already after 60 min of reaction (Fig. 5b). Further, a significant consumption from of guluronic acid after 120 min could not be observed (Fig. 5c). However, also for (M)-units no significant further increase over reaction time over 2 hours was achieved, both poly-(M) active lyases (SALy and FALy) similarly only released unsaturated M-units of 4–6% of the total mannuronic acid (Fig. 5c).
Potentially, poly-(M) and poly-(G) interacted competitively with the lyases active on both substrates exhibiting increased binding affinity towards poly-(G). Iwamoto et al.22 indicated a strong reduced production of unsaturated mannuronates from poly-(M) by the presence of poly-(G), the higher the concentration of (M) the higher the reduction.
At a concentration of 0.1% for both of poly-(M) and poly-(G) the production of unsaturated UA was halved compared to (M) as the only product and only one third at increased concentration of poly-(M) to 0.2% S/V.22 Conclusively, as the total (M) in the present reaction volume of L. digitata was about 0.9% of (M) the presence of approx. 0.3% of poly-(G) might inhibited any further activity of the enzyme on the mannuronic acid blocks in the brown seaweed. Product inhibition was emphasized for the degradation of alginate by Sphingomonas sp. derived lyases, also when exo- and endolytic alginate lyases were acting together.29 This could also indicate that by addition of exolytic lyase a faster release of glucose is unlikely as an increase in enzyme loading did not enhance the decomposition of brown seaweed.5
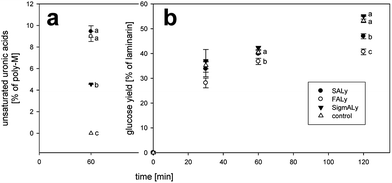
Nonetheless, degradation of poly-mannuronic acid led to an inhibition of glucose release from seaweed. The more activity towards M-blocks the more the glucose yields decreased (Fig. 5). Analogously, release of glucose from isolated commercially available laminarin mixed with pretreated poly-(M) decreased with respect to activity on poly-(M) (Fig. 6). Pretreatment with the lyases SALy and FALy inhibited the glucose release significantly after 2 hours (Fig. 6b). For FALy the inhibition was significant already after 1 hour of reaction (Fig. 6b). Treatment with the purchased lyase (SigmALy) did yield in similar amounts as the control containing no lyase (Fig. 6b). Hence, non-activity towards poly-(M) apparently protected the cellulase catalyzed glucan degradation.
A 24 hours enzymatic treatment with the FALy or SALy supplemented to the cellulase preparation Cellic®CTec2 left behind an insoluble residue constituting ∼20% by weight of the original seaweed substrate dry matter (Table 2). This residue could be separated from the liquid by centrifugation. Interestingly, treatment with the SigmALy, which appeared to have high affinity for poly-guluronic acid (Fig. 3c) catalyzed the liquefaction of more of the seaweed material and left behind 12.4% by weight only (Table 2). The achieved degrees of enzymatic saccharification were significantly higher than those achieved previously by 48 h-saccharification at pH 5 at 50 °C of 26% of the original L. digitata biomass.6
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 × g for 30 min
000 × g for 30 min
| Treatment | Residue | Liquefaction | ||
|---|---|---|---|---|
| Amount | N-recoverya | Mannitolb | Glucosec | |
| [% of original biomass] | [% of original biomass] | |||
| a After elemental analysis; raw material N = 0.73% (equal to 100%).3 b Hydrated monomers after HPAEC analysis. c Hydrated monomers after determination with enzyme assay. | ||||
| SALy + CTec2 | 19.3 | 80.1 | 3.4 | 51.8 |
| FALy + CTec2 | 20.3 | 83.5 | 5.9 | 48.2 |
| SigmALy + CTec2 | 12.4 | 71.4 | 5.0 | 52.7 |
| CTec2 alone | 51.8 | 68.4 | 3.9 | 46.0 |
| SigmALy alone | 28.9 | 78.5 | 2.3 | 17.6 |
Compared to the raw material 70–80% by weight of nitrogen was recovered in the solid residue indicating that the majority of the seaweed protein (constituting ∼3% by weight of the original seaweed biomass)3 was left in this fraction (Table 2).
The data (Table 2) are in accord with the recently published findings on the same seaweed material that a protein enriched residue having similar amino acid profile as the raw material remained insoluble after extensive saccharification.6
Beyond nitrogen, the residue also contained a mixture of carbohydrates at a level equivalent to 4% of the total seaweed carbohydrates by dry weight (data not shown). The different liquefied seaweed fractions contained glucose of 51.8, 48.2 and 52.7% by weight of the dry matter (Table 2), corroborating that nearly all the potential glucose was released.
Carbohydrate analysis HPAEC-PAD showed that besides glucose, the liquefied fraction also contained mannitol. The mannitol levels measured (3.4 to 5.9% of the original milled seaweed biomass; Table 2) made up about 80% of the original content of mannitol in the biomass. Sulfuric acid treatment of the liquefied fraction released fucose along with minor monosaccharides (data not shown). The presence of fucose after hydrolysis of the liquefied fraction indicated that fucoidan was released from the brown seaweed matrix during the enzymatic treatment. Such enzyme-assisted extraction of fucoidan polysaccharides may be a gentle extraction technique for obtaining bioactive fucoidans from brown seaweeds.32
In conclusion, the application of alginate lyases together with Cellic®CTec2 for 24 hours enabled almost complete release of the glucose and mannitol harbored in the L. digitata. New yeast strains have been shown to be able to convert mannitol into ethanol.33 8 h treatment was enough to release 90% of the glucose if the guluronic acid specific SigmALy along with the cellulase preparation was applied and 14 h were sufficient with the use of SALy whereas 24 hours with FALy and cellulase were required for complete glucose release (data not shown).
4 Conclusions
Expression of the endolytic bacterial alginate lyase from Sphingomonas sp. (SALy) was successful and high yields of 12.8 g L−1 cell extract were achieved. This enzyme had high activity at pH from 5.5 to 7 and thermal stability up to 50 °C, making it a promising candidate to support glucose release from brown seaweed catalyzed by the commercial, fungally derived cellulase preparation Cellic®CTec2. Like the endolytic lyase from Flavobacterium sp (FALy, optima pH 7.5), the SALy was active on both alginate epimers poly-mannuronic acid and poly-guluronic acid. In contrast, a purchased lyase (SigmALy) was only endolytically active towards poly-(G). The M/G ratio of the investigated brown seaweed Laminaria digitata was 3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1. The guluronic acid was presumably degraded to smaller oligomers of DP 3 by SALy, of DP 5–6 for FALy and DP 2–3 for SigmALy. However, only 4–6% of the mannuronic acid present in the seaweed was released enzymatically. Moreover, degradation of poly-(M) led to a decreased release rate of glucose from L. digitata by the cellulase preparation. In conclusion, not only the binding activity of the lyase towards poly-(G) was higher, the degradation of poly-(M) was apparently inhibiting the enzymatic glucose release.
1. The guluronic acid was presumably degraded to smaller oligomers of DP 3 by SALy, of DP 5–6 for FALy and DP 2–3 for SigmALy. However, only 4–6% of the mannuronic acid present in the seaweed was released enzymatically. Moreover, degradation of poly-(M) led to a decreased release rate of glucose from L. digitata by the cellulase preparation. In conclusion, not only the binding activity of the lyase towards poly-(G) was higher, the degradation of poly-(M) was apparently inhibiting the enzymatic glucose release.
Nevertheless, enzymatic treatment for 24 hours was sufficient to release all potential glucose from the glucan rich L. digitata (51% moieties) regardless the applied lyase. Viscosity reduction occurred primarily in the first minutes of the reaction. Since the enzymatic glucose release continued for several hours, the rapid viscosity data indicated that the alginate lyases were more required to decompose the alginate in the cell wall to enhance access for the cellulase to the glucan, rather than to decrease viscosity. In the solid pellet after enzymatic treatment 70–80% of the nitrogen was recoverable and fucose polymers, likely fucoidan, were present in the liquefied fraction. Hence combined treatment with cellulase and alginate lyase was in essence a biorefining treatment that helped solubilize the valuable sulfated fucose polysaccharides (fucoidan) in addition to mannitol and glucose from brown seaweeds, whilst leaving the proteins insoluble.
Acknowledgements
This work was supported by the Danish Council for Strategic Research via the MacroAlgaeBiorefinery (MAB3) project. The authors acknowledge Drs Annette Bruhn and Michael Bo Rasmussen (Aarhus University, Denmark) and Ditte B. Toerring and Kristian O. Nielsen (Danish Shellfish Centre, DTU Aqua) for providing the seaweed. Christiane Riegert, Hamburg University is acknowledged for assisting with the elemental analyses.References
- A. Brown and J. Tustin, Algae – The Future for Bioenergy? Summary and conclusions from the IEA Bioenergy ExCo64 Workshop. IEA Bioenergy: ExCo: 2010:02, 2010 Search PubMed.
- A. Dave, Y. Huang, S. Rezvani, D. McIlveen-Wright, M. Novaes and N. Hewitt, Bioresour. Technol., 2013, 135, 120–127 CrossRef CAS PubMed.
- D. Manns, A. L. Deutschle, B. Saake and A. S. Meyer, RSC Adv., 2014, 4, 25736–25746 RSC.
- H. Kim, C.-G. Lee and E. Lee, Biotechnol. Bioprocess Eng., 2011, 16, 843–851 CrossRef CAS.
- D. Manns, S. Andersen, B. Saake and A. Meyer, J. Appl. Phycol., 2016, 28, 1287–1294 CrossRef CAS.
- X. Hou, J. H. Hansen and A.-B. Bjerre, Bioresour. Technol., 2015, 197, 310–317 CrossRef CAS PubMed.
- E. Percival and R. H. McDowell, Chemistry and enzymology of marine algal polysaccharides, Academic Press Inc., London, 1967 Search PubMed.
- O. A. Aarstad, A. Tøndervik, H. Sletta and G. Skjåk-Bræk, Biomacromolecules, 2012, 13, 106–116 CrossRef CAS PubMed.
- B. Zhu and H. Yin, Bioengineered, 2015, 6, 125–131 CrossRef CAS PubMed.
- T. Y. Wong, L. A. Preston and N. L. Schiller, Annu. Rev. Microbiol., 2000, 54, 289–340 CrossRef CAS PubMed.
- O. Miyake, W. Hashimoto and K. Murata, Protein Expression Purif., 2003, 29, 33–41 CrossRef CAS PubMed.
- O. Miyake, A. Ochiai, W. Hashimoto and K. Murata, J. Bacteriol., 2004, 186, 2891–2896 CrossRef CAS PubMed.
- M. Yamasaki, K. Ogura, W. Hashimoto, B. Mikami and K. Murata, J. Mol. Biol., 2005, 352, 11–21 CrossRef CAS PubMed.
- L. Huang, J. Zhou, X. Li, Q. Peng, H. Lu and Y. Du, J. Ind. Microbiol. Biotechnol., 2013, 40, 113–122 CrossRef CAS PubMed.
- M. R. Wilkins, E. Gasteiger, A. Bairoch, J. C. Sanchez, K. L. Williams, R. D. Appel and D. F. Hochstrasser, Methods Mol. Biol., 1999, 112, 531–552 CAS.
- F. Thomas, L. C. E. Lundqvist, M. Jam, A. Jeudy, T. Barbeyron, C. Sandström, G. Michel and M. Czjzek, J. Biol. Chem., 2013, 288, 23021–23037 CrossRef CAS PubMed.
- K. Ogura, M. Yamasaki, B. Mikami, W. Hashimoto and K. Murata, J. Mol. Biol., 2008, 380, 373–385 CrossRef CAS PubMed.
- H.-J. Yoon, W. Hashimoto, O. Miyake, M. Okamoto, B. Mikami and K. Murata, Protein Expression Purif., 2000, 19, 84–90 CrossRef CAS PubMed.
- A. Inoue, K. Takadono, R. Nishiyama, K. Tajima, T. Kobayashi and T. Ojima, Mar. Drugs, 2014, 12, 4693–4712 CrossRef CAS PubMed.
- H. Kim, H.-J. Ko, N. Kim, D. Kim, D. Lee, I.-G. Choi, H. Woo, M. Kim and K. Kim, Biotechnol. Lett., 2012, 34, 1087–1092 CrossRef CAS PubMed.
- H. Park, N. Kam, E. Lee and H. Kim, Mar. Biotechnol., 2012, 14, 189–202 CrossRef CAS PubMed.
- Y. Iwamoto, R. Araki, K.-i. Iriyama, T. Oda, H. Fukuda, S. Hayashida and T. Muramatsu, Biosci., Biotechnol., Biochem., 2001, 65, 133–142 CrossRef CAS PubMed.
- E. Deniaud-Bouët, N. Kervarec, G. Michel, T. Tonon, B. Kloareg and C. Hervé, Ann. Bot., 2014, 114, 1203–1216 CrossRef PubMed.
- M. Alvarado-Morales, I. B. Gunnarsson, I. A. Fotidis, E. Vasilakou, G. Lyberatos and I. Angelidaki, Algal Res., 2015, 9, 126–132 CrossRef.
- C. H. Vanegas, A. Hernon and J. Bartlett, Int. J. Ambient Energy, 2015, 36, 2–7 CrossRef CAS.
- X. Luo and J. Y. Zhu, Enzyme Microb. Technol., 2011, 48, 92–99 CrossRef CAS PubMed.
- X. L. Luo, J. Y. Zhu, R. Gleisner and H. Y. Zhan, Cellulose, 2011, 18, 1055–1062 CrossRef CAS.
- M. C. Ravanal, R. Pezoa-Conte, S. von Schoultz, J. Hemming, O. Salazar, I. Anugwom, O. Jogunola, P. Mäki-Arvela, S. Willför, J.-P. Mikkola and M. E. Lienqueo, Algal Res., 2016, 13, 141–147 CrossRef.
- M. Ryu and E. Y. Lee, J. Ind. Eng. Chem., 2011, 17, 853–858 CrossRef CAS.
- A. J. Wargacki, E. Leonard, M. N. Win, D. D. Regitsky, C. N. S. Santos, P. B. Kim, S. R. Cooper, R. M. Raisner, A. Herman, A. B. Sivitz, A. Lakshmanaswamy, Y. Kashiyama, D. Baker and Y. Yoshikuni, Science, 2012, 335, 308–313 CrossRef CAS PubMed.
- K. Formo, O. A. Aarstad, G. Skjåk-Bræk and B. L. Strand, Carbohydr. Polym., 2014, 110, 100–106 CrossRef CAS PubMed.
- M. T. Ale and A. S. Meyer, RSC Adv., 2013, 3, 8131–8141 RSC.
- A. Ota, S. Kawai, H. Oda, K. Iohara and K. Murata, J. Biosci. Bioeng., 2013, 116, 327–332 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c6ra06669k |
| This journal is © The Royal Society of Chemistry 2016 |