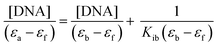Synthesis, crystal structure, DFT/TDDFT calculation, photophysical properties and DNA binding studies of morpholino moiety ligand based two Cu(II) complexes in combination with carboxylates†
Aparup Paula,
Soumen Mistria,
Apurba Bhuniaa,
Soumen Mannaa,
Horst Puschmannb and
Subal Chandra Manna*a
aDepartment of Chemistry and Chemical Technology, Vidyasagar University, Midnapore 721102, West Bengal, India. E-mail: scmanna@mail.vidyasagar.ac.in; Fax: +91 03222 275329
bDepartment of Chemistry, University of Durham, South Road, Durham DH1 3LE, UK
First published on 13th June 2016
Abstract
The complexes [Cu(L)(pa)] (1) and [Cu(L)(mb)] (2) (HL = o-{(3-morpholinopropylimino)methyl}phenol; pa = 3-phenylacrylate; mb = p-methylbenzoate) have been synthesized and characterized by elemental analysis, single crystal X-ray crystallography, FT-IR, UV-vis electronic absorption spectroscopic analysis. Complexes 1 and 2 are mononuclear with distorted square pyramidal geometries where the Schiff base coordinates to copper(II) in the tridentate (O, N, N) chelating mode. These three atoms plus one carboxylate oxygen define the equatorial plane of the square pyramid while the axial position is occupied by carboxylate oxygen at a relatively longer distance. Weak C–H⋯π interactions result the formation of 2D supramolecular structures for both complexes. At room temperature complexes 1 and 2, exhibit fluorescence with a quantum yield (Φs) of 0.129 and 0.117, respectively. The UV-vis electronic absorption and IR spectral data of complexes have been compared with the results obtained by employing DFT and time dependent density functional theory (TD-DFT) calculation using the B3LYP, B3PW91 and MPW1PW91 functionals, with LanL2DZ basis set. The results of these calculations are functional-dependent and among the functionals, B3LYP proved to better reproduce the experimental results. The interactions of complexes with the calf thymus DNA (CT-DNA) were investigated using electronic absorption and fluorescence spectroscopic techniques. The studies reveal that the binding affinities of complexes 1 and 2 with CT-DNA are in the order of 1.68 × 105 M−1 and 2.271 × 105 M−1, respectively. A study of the effect of various metal ions on the electronic absorption and fluorescence spectra of HL reveals that it selectively senses the Cu(II) ions.
Introduction
Coordination compounds of 3d metal ions are important due to their potential application in the areas of catalysis,1 magnetism,2 chemical sensor,3 medicinal chemistry,4 etc. Schiff bases are popular polydentate ligands in the chemistry of 3d metal ions.5 Many 3d metal Schiff base coordination compounds are reported with the aim to model biologically important molecules6 and these compounds also play important role in biology due to their antimicrobial,7 antifungal,8 antibacterial,9 antitumor,10 antiviral,11 antipyretic,8a and antidiabetic activities.12 These coordination compounds are also used for the treatment of several diseases like neurological disorders, diabetes, lymphomas, carcinomas etc. Among the 3d metals, vanadium, iron and zinc compounds show high potential for the development of metallopharmaceutics.13 Some copper(II) compounds are also reported as exhibiting interesting anticancer activity.14 Kinetic studies of the interaction between DNA and copper compounds under physiological condition are important for the development of copper metal based new metallo-pharmaceuticals.Density functional theory (DFT) and time dependent density functional theory (TD-DFT) are important techniques to explain molecular structure, electronic and spectroscopic properties of 3d metal based compounds. In the literature, there are limited numbers of report on the DFT computation to explore the electronic properties of copper(II) compounds.15 o-{(3-Morpholinopropylimino)methyl}phenol (HL) is an important polydentate Schiff base ligand in the chemistry of 3d metal ions. With this ligand the compounds [Zn(L)2], [Cu(L)2],16 and [Zn(HL)(Br)2]17 are reported in the literature, where it chelates to the metal center with η:η(O,N) coordination mode. Until now there are no reports of the Cu(II) compound of HL ligand in combination with aromatic carboxylates. In the present contribution we report synthesis, crystal structure, DFT calculation and fluorescence property of two mononuclear complexes, [Cu(L)(pa)] (1) and [Cu(L)(mb)] (2) (pa = 3-phenylacrylate; mb = p-methyl benzoate). The IR and electronic spectral properties of HL and complexes were explained by DFT computation. The effect of various metal ions on the electronic absorption and fluorescence spectra of HL shows that it selectively senses Cu(II) ions. The interaction of CT-DNA with complexes 1 and 2 was studied using electronic absorption and fluorescence spectroscopic techniques.
Experimental
Materials
High purity 3-(4-morpholinyl)propylamine (Alfa-Aesar) and salicylaldehyde (Merck-India) were purchased and used as received. All other chemicals used were of analytical grade and used without further purification. Solvents used for spectroscopic studies were purified and dried by standard procedures immediately before use.18Physical measurements
Elemental analyses (C, H and N) were performed using a Perkin-Elmer 240C elemental analyzer. IR spectra were recorded as KBr pellets on a Bruker Vector 22FT IR spectrophotometer operating from 400 to 4000 cm−1. NMR spectra of ligand recorded on Bruker 400 MHz instrument. Electronic absorption spectra were obtained in different solvents (acetonitrile, methanol, ethanol and dichloromethane) with a Shimadzu UV-1601 UV-vis spectrophotometer at room temperature. Quartz cuvettes with a 1 cm path length and a 3 cm3 volume were used for all measurements. Emission spectra were recorded on a Hitachi F-7000 spectrofluorimeter. Room temperature (300 K) spectra were obtained in different solvents (acetonitrile, methanol, ethanol and dichloromethane) using a quartz cell of 1 cm path length. The slit width was 2.5 nm for both excitation and emission. ESI-MS spectra of the compounds and HL in methanol were recorded on an Agilent Q-TOF 6500 mass spectrometer and the software used for mass analysis is Mass hunter. Electrochemical measurements performed under a dry argon atmosphere with a BAS Epsilon electrochemical system. A three-electrode assembly comprising a glassy carbon (for reduction) or Pt (for oxidation) working electrode, Pt auxiliary electrode, and an aqueous Ag/AgCl reference electrode were used. Cyclic voltammetric (CV) measurements were carried out at 298 K using methanolic solution of complexes (ca. 1 mM) and the concentration of supporting electrolyte tetraethylammonium perchlorate (TEAM) was maintained at 0.1 M. The potentials reported were referenced against the Ag/AgCl electrode, which under given experimental conditions gave a value of 0.36 V for ferrocene/ferrocenium couple.The fluorescence quantum yield was determined using phenol as a reference and methanol medium for both complexes and reference. Emission spectra were recorded by exciting the complex and the reference phenol at the same wavelength, maintaining nearly equal absorbance (∼0.1). The area of the emission spectrum was integrated using the software available in the instrument and the quantum yield calculated19 according to the following equation:
Synthesis of the ligand
A methanolic solution of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 mixture of 3-(4-morpholinyl)propylamine and salicylaldehyde was refluxed for 3 h. The resulting yellow color solution cooled to room temperature and solid yellow compound was obtained after evaporation of solvent. Re-crystallization of compound using methanol as solvent results yellow crystals of HL. Yield 0.198 g (80%). Anal. calc. for C14H20N2O2 (248): C, 67.80; H, 8.12; N, 11.29%. Found: C, 67.82; H, 8.09; N, 11.32 (%). 1H NMR (400 MHz, CDCl3, δ, ppm): 1.61–1.65 (2H, m, –CH2–), 2.35–2.43 (4H, m, –CH2–N–CH2–), 2.71–2.74 (2H, m, –CH2–N), 3.60–3.70 (4H, m, –CH2–O–CH2–; 2H, m,
1 mixture of 3-(4-morpholinyl)propylamine and salicylaldehyde was refluxed for 3 h. The resulting yellow color solution cooled to room temperature and solid yellow compound was obtained after evaporation of solvent. Re-crystallization of compound using methanol as solvent results yellow crystals of HL. Yield 0.198 g (80%). Anal. calc. for C14H20N2O2 (248): C, 67.80; H, 8.12; N, 11.29%. Found: C, 67.82; H, 8.09; N, 11.32 (%). 1H NMR (400 MHz, CDCl3, δ, ppm): 1.61–1.65 (2H, m, –CH2–), 2.35–2.43 (4H, m, –CH2–N–CH2–), 2.71–2.74 (2H, m, –CH2–N), 3.60–3.70 (4H, m, –CH2–O–CH2–; 2H, m, ![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N–CH2–), 4.86 (1H, s, Ar–OH), 6.83–7.31 (4H, m, Ar–H), 8.32 (H, s, –CH
N–CH2–), 4.86 (1H, s, Ar–OH), 6.83–7.31 (4H, m, Ar–H), 8.32 (H, s, –CH![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N–). 13C NMR (400 MHz, CDCl3, δ, ppm): 165.04 (–CH
N–). 13C NMR (400 MHz, CDCl3, δ, ppm): 165.04 (–CH![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N–), 161.25 (Ar–C–OH), 116.96–132.12 (Ar–C), 53.45–66.90 (morpholine–C and N–CH2–CH2–N), 27.54 (–CH2–).
N–), 161.25 (Ar–C–OH), 116.96–132.12 (Ar–C), 53.45–66.90 (morpholine–C and N–CH2–CH2–N), 27.54 (–CH2–).
Synthesis of complexes
Caution! Metal perchlorates in the presence of organic ligands are potentially explosive. Only a small amount of the material should be prepared and handled with care.The complexes have been synthesized by adopting the procedure schematically given in Scheme 1.
Crystallographic data collection and refinement
Data collections of complexes 1 and 2 were carried out at 120 K on an Oxford Diffraction Gemini Ultra diffractometer. Cell refinement, indexing and scaling of the data sets were done with CrysAlisPro package, Version 1.171.35.10.20 The structures were solved by using the olex2.solve solution program21 using the charge flipping algorithm and refined by the full matrix least-squares method based on F2 with all observed reflections.22 All the calculations were performed using the WinGX.23 Packing diagrams were done with graphical program Diamond.24 Crystal data and details of refinements are given in Table 1. CCDC reference numbers are 1405167 and 999890 for 1 and 2, respectively.| Complex | 1 | 2 |
|---|---|---|
| a R1 = ∑∣∣Fo∣ − ∣Fc∣∣/∑∣Fo∣, wR2 = [ ∑w(Fo2 − Fc2) 2/∑w(Fo2)2]1/2. | ||
| Empirical formula | C23H26CuN2O4 | C22H26CuN2O4 |
| Formula mass, g mol−1 | 458.00 | 445.99 |
| Crystal system | Monoclinic | Monoclinic |
| Space group | P21/c | P21/c |
| a, Å | 10.2427(3) | 10.7277(3) |
| b, Å | 24.3438(6) | 19.7595(4) |
| c, Å | 9.3982(4) | 10.9426(3) |
| α, deg | 90 | 90 |
| β, deg | 114.260(4) | 115.971(4) |
| γ, deg | 90 | 90 |
| V, Å3 | 2136.46(14) | 2085.31(12) |
| Z | 4 | 4 |
| T, K | 120 | 120 |
| D(calcd), g cm−3 | 1.424 | 1.421 |
| μ(Mo-Kα), mm−1 | 1.054 | 1.078 |
| F(000) | 956 | 932 |
| Theta range, deg | 2.2–28.5 | 2.1–28 |
| No. of collected data | 17![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 742 742 |
18![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 063 063 |
| No. of unique data | 5408 | 5043 |
| Rint | 0.032 | 0.040 |
| Observed reflections [I > 2σ(I)] | 4714 | 4158 |
| Goodness of fit (F2) | 1.023 | 1.058 |
| Parameters refined | 271 | 263 |
| R1 [I > 2σ(I)] a | 0.0343 | 0.0349 |
| wR2 [I > 2σ(I)] a | 0.0816 | 0.0850 |
| Residuals, e Å−3 | −0.41, 0.58 | −0.48, 0.49 |
Theory and computational methods
All computations were performed using the Gaussian 09 (G 09) software package25 by using the Becke's three-parameter hybrid exchange and the Lee–Yang–Parr non-local correlation functionals (B3LYP)26 the Becke's three-parameter Perdew–Wang 1991 functional (B3PW91)27 the Barone's Modified Perdew–Wang 1991 exchange and Perdew and Wang's 1991 correlation functionals (MPW1PW91).28 In the calculation 6-31G(d-p) basis set was assigned to all elements with the exception of copper for which the Los Alamos effective core potentials plus the Double Zeta (LanL2DZ)29 basis set were employed. The geometric structures of the complexes in the ground state (doublet) were fully optimized at the B3LYP, B3PW91 and MPW1PW91 levels. The vibration frequency calculations were performed to ensure that the optimized geometries represent local minima associated with positive eigen values only.Vertical electronic excitations based on B3LYP, B3PW91 and MPW1PW91 functionals were obtained with the time-dependent density functional theory (TD-DFT) formalism30 in different solvents (acetonitrile, methanol, ethanol and dichloromethane) using the conductor-like polarisable continuum model (CPCM).31 GaussSum32 was used to calculate the fractional contributions of various groups to each molecular orbital. Calculated coordination geometries of complexes are shown in Table 2.
| 1 | 2 | |||
|---|---|---|---|---|
| Exp | Calcd | Exp | Calcd | |
| a Using conductor-like polarizable continuum model (CPCM) in methanol; basis set, LanL2DZ. | ||||
| Bond distances | ||||
| Cu(1)–O(1) | 1.8931(15) | 1.9526 | 1.9048(15) | 1.9531 |
| Cu(1)–O(2) | 2.0013(13) | 2.0145 | 1.9714(14) | 2.0098 |
| Cu(1)–O(3) | 2.3538(14) | 2.6014 | 2.3606(17) | 2.6109 |
| Cu(1)–N(1) | 1.9319(16) | 1.9699 | 1.9236(17) | 1.9684 |
| Cu(1)–N(2) | 2.1184(16) | 2.1274 | 2.1605(17) | 2.1299 |
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) |
||||
| Bond angles | ||||
| O(1)–Cu(1)–O(2) | 91.30(6) | 93.31 | 92.46(6) | 93.10 |
| O(1)–Cu(1)–O(3) | 126.74(5) | 121.59 | 129.33(6) | 123.19 |
| O(1)–Cu(1)–N(1) | 94.65(7) | 93.36 | 94.24(7) | 93.33 |
| O(1)–Cu(1)–N(2) | 138.21(6) | 139.28 | 138.11(7) | 138.38 |
| O(2)–Cu(1)–O(3) | 60.20(5) | 57.23 | 60.29(6) | 56.96 |
| O(2)–Cu(1)–N(1) | 155.37(6) | 153.21 | 161.69(8) | 153.31 |
| O(2)–Cu(1)–N(2) | 99.05(6) | 98.08 | 97.07(6) | 98.38 |
| O(3)–Cu(1)–N(1) | 97.69(6) | 97.53 | 102.73(7) | 98.30 |
| O(3)–Cu(1)–N(2) | 92.83(5) | 97.09 | 90.02(6) | 96.12 |
| N(1)–Cu(1)–N(2) | 92.33(6) | 93.60 | 89.26(7) | 93.83 |
DNA binding experiments
where [DNA] is the concentration of CT-DNA, εa is the extinction co-efficient value of the complex at a given CT-DNA concentration, εf and εb are the extinction co-efficient of the complex, in free solution and when fully bound to CT-DNA, respectively. The plot of [DNA]/(εa – εf) vs. [DNA] gives a straight line with
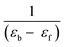 and
and 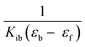 as slope and intercept, respectively. From the ratio of the slope to the intercept the value of Kib was calculated.
as slope and intercept, respectively. From the ratio of the slope to the intercept the value of Kib was calculated.| F0/F = 1 + Ksv[complex] |
Results and discussion
Synthetic aspects
The multisite coordinating ligand, HL, was prepared by a one pot synthesis employing condensation of the 3-(4-morpholinyl)propylamine and salicylaldehyde in methanol under reflux condition, and characterized by NMR (Fig. 1), ESI mass (Fig. 1S†), electronic absorption and emission spectra. Using HL, 3-phenylacrylate and p-methylbenzoate complexes 1 and 2 were synthesized at room temperature.Crystal structure description
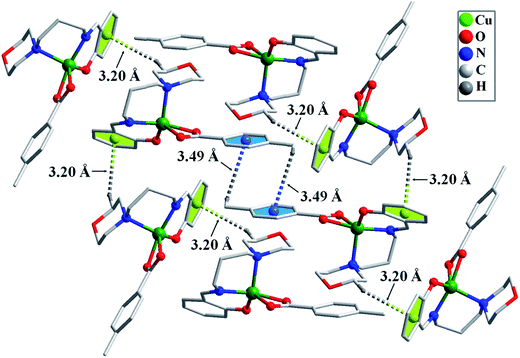
Crystal structure determination reveals that both the complexes 1 and 2 are mononuclear with five coordinated Cu(II) centers. The ORTEP drawing of the metal centers of 1 and 2 with the atom numbering scheme are shown in Fig. 2 and 3, while selected geometrical parameters are given in Table 2. The metal is coordinated by tridentate (O, N, N) chelating Schiff base, and completes the distorted square pyramidal geometry with two chelating carboxylate (3-phenylacrylate for 1, and p-methylbenzoate for 2) oxygen atoms, resulting in a CuN2O3 chromophore. The Schiff base O(1), N(1), N(2) donors and carboxylate oxygen atom O(2) define the equatorial plane around each pseudo square pyramidal copper ion [Cu(1)–O(1)1.8931(15), Cu(1)–O(2) 2.0013(13), Cu(1)–N(1) 1.9319(16), Cu(1)–N(2) 2.1184(16) Å, O(1)–Cu(1)–N(2) 138.21(6)°, O(2)–Cu(1)–N(1) 155.37(6)° for 1; [Cu(1)–O(1) 1.9048(15), Cu(1)–O(2) 1.9714(14), Cu(1)–N(1) 1.9236(17), Cu(1)–N(2) 2.1605(17) Å, O(1)–Cu(1)–N(2) 138.11(7)°, O(2)–Cu(1)–N(1) 161.69(8)° for 2]. The axial position is occupied by the oxygen atom from carboxylate oxygen O(3) [Cu(1)–O(3), 2.3538(14) and 2.3606(17) Å for 1 and 2, respectively]. In both the complexes the axial Cu–O bond length is longer than equatorial Cu–O bond length probably due to Jahn–Teller distortion. In complex 1, the bond angels between the successive coordinating equatorial atoms with central atom are 91.30(6), 94.65(7), 92.33(6), 99.05(6)° and the angel between axial atom, central atom and equatorial atoms are 126.74(5), 97.69(6), 92.83(5), 60.20(5)°. Whereas in 2, the bond angels between the successive coordinating equatorial atoms with central atom are 92.46(6), 94.24(7), 89.26(7), 97.07(6)° and the angel between axial atom, central atom and equatorial atoms are 129.33(6), 102.73(7), 90.02(6), 60.29(6)°.The trigonality τ parameter34 for complexes 1 and 2 were calculated as (α − β)/60, where α and β are the two largest coordination bond angles. For a regular trigonal bipyramidal structure with D3h symmetry has τ = 1 and for a regular C4v square pyramidal geometry τ = 0. Calculated values of τ were 0.286 and 0.393 for complexes 1 and 2, respectively, suggesting that the complexes posses distorted square pyramidal geometries. Both the complexes form 2D supramolecular structures (Fig. 4 and 5) with C–H⋯π interactions35 (C–H⋯Cg, 2.76 and 3.67 Å for 1; C–H⋯Cg, 3.20 and 3.49 Å, Table 1S†).
IR spectral studies
The most important absorption bands in IR spectroscopy (Fig. 2S and 3S†) of complexes 1 and 2 are summarized in experimental section. Both the complexes exhibit bands at 1629 cm−1 corresponding to νas(OCO), while the νs(OCO) vibration appears at 1406 cm−1 (for 1) and 1410 cm−1 (for 2). Aromatic ν (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C) and azomethine ν(C
C) and azomethine ν(C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N) stretching vibrations for compounds appear in the region 1413–1535 cm−1.
N) stretching vibrations for compounds appear in the region 1413–1535 cm−1.
ESI mass spectrometry
ESI mass spectra of HL, and complexes 1–2 were recorded in methanol. The ESI mass spectra of HL (Fig. 1S†) shows a peak at m/z = 249.125, corresponding to [C14H20N2O2]˙+ (calc. m/z = 248.322), which confirm the chemical composition of ligand. On the other hand the spectra of 1 (Fig. 6) shows three peaks at m/z = 458.060, 311.044 and 249.125, corresponding to [C23H26CuN2O4]˙+ (calc. m/z = 458.00), [C14H19CuN2O2]+ (calc. m/z = 310.86), (after removal of (pa)) and [C14H19N2O2 + H]+ (calc. m/z = 248.322), mono cations, respectively. Whereas for 2 (Fig. 4S†) the peaks are at 446.134, 310.037 and 249.125, corresponding to [C22H26CuN2O4]˙+ (calc. m/z = 445.99), [C14H19CuN2O2]+ (calc. m/z = 310.86), (after removal (mb)) and [C14H19N2O2 + H]+ (calc. m/z = 248.322) mono cations, respectively. This result evidences that the coordination environments for 1 and 2 in methanol are similar to that detected in the solid states.Electronic absorption spectra of HL, complexes 1 and 2
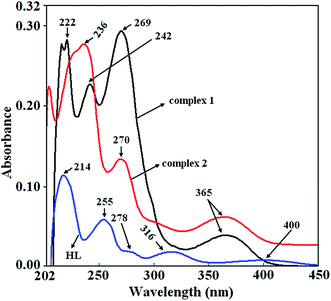
The electronic absorption spectra (Fig. 7 and 5S–7S†) of HL and complexes 1–2 have been studied in acetonitrile (ACN), methanol (MeOH), ethanol (EtOH) and dichloromethane (DCM), and the relevant data are summarized in Tables 3 and 4. The methanolic solution of HL shows (Fig. 7) significant transitions at 214 (2.56 × 105 L mol−1 cm−1), 254 (1.18 × 105 L mol−1 cm−1), 278 (3.6 × 104 L mol−1 cm−1), 316 (3.7 × 104 L mol−1 cm−1), and 400 (1.4 × 104 L mol−1 cm−1) nm. Tables 3 and 4 and Fig. 5S–7S† show the solvatochromic shift of the electronic absorption spectral bands. The electronic spectral bands at 214 and 278 nm are slightly blue shifted in more polar solvent acetonitrile, whereas red shifting observed in relatively less polar solvent like dichloromethane. The spectrum of the methanolic solution of complex 1 shows significant transitions at 222 (1.37 × 104 L mol−1 cm−1), 242 (1.09 × 104 L mol−1 cm−1), 269 (1.31 × 104 L mol−1 cm−1) and 365 (1.4 × 103 L mol−1 cm−1) nm. The spectral bands at 269 nm and 365 nm are assigned due to L−1 to copper and 3-phenylacrylate to copper charge transfer transitions, respectively. On the other hand complex 2 in methanol shows significant transitions at 204 (5.61 × 104 L mol−1 cm−1), 236 (7.09 × 104 L mol−1 cm−1), 270 (3.03 × 104 L mol−1 cm−1), 305 (7.7 × 104 L mol−1 cm−1) and 365 (9.9 × 103 L mol−1 cm−1). The spectral band at 270 nm is assigned due to the combination of L−1 to copper and p-methyl benzoate to copper charge transfer transitions. Whereas the band at 365 nm is due to p-methylbenzoate to copper charge transfer transition. The studies of the electronic absorption spectra in other solvents show solvatochromic shift of electronic absorption spectral bands (Tables 3 and 4 and Fig. 6S–7S†) for both the complexes.| Solvents | State | λcal (nm), εcal (M−1 cm−1), (eV) | Oscillator strength (f) | λexp (nm), εexp (M−1 cm−1), (eV) | Key transition | Charactera | |
|---|---|---|---|---|---|---|---|
| a LMCT = Schiff base to metal charge transfer; L1MCT = carboxylate to metal charge transfer; ILCT = intra Schiff base charge transfer; L1LCT = carboxylate to Schiff base charge transfer. | |||||||
| HL | ACN | S3 | 278.06, 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 255, (4.45) 255, (4.45) |
0.0826 | 276, 82![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 092, (4.52) 092, (4.52) |
HOMO−1 → LUMO (52%) | n → π* |
| S7 | 210.44, 30![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000, (5.89) 000, (5.89) |
0.3489 | 214, 582![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 854, (5.79) 854, (5.79) |
HOMO−1 → LUMO+1 (57%) | n → π* | ||
| MeOH | S3 | 278.02, 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 257, (4.45) 257, (4.45) |
0.0836 | 278, 36![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000, (4.45) 000, (4.45) |
HOMO−1 → LUMO (53%) | n → π* | |
| S7 | 210.38, 30![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000, (5.89) 000, (5.89) |
0.3433 | 214, 256![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000,(5.79) 000,(5.79) |
HOMO−1 → LUMO+1 (56%) | n → π* | ||
| EtOH | S3 | 278.15, 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 258, (4.45) 258, (4.45) |
0.0860 | 278, 34![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 532, (4.45) 532, (4.45) |
HOMO−1 → LUMO (53%) | n → π* | |
| S7 | 210.54, 30![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 067, (5.89) 067, (5.89) |
0.3502 | 215, 227![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 703, (5.76) 703, (5.76) |
HOMO−1 → LUMO+1 (57%) | n → π* | ||
| DCM | S3 | 278.45, 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 403, (4.45) 403, (4.45) |
0.1031 | 279, 77![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 987, (4.50) 987, (4.50) |
HOMO−1 → LUMO (59%) | n → π* | |
| S7 | 211.00, 31![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 002, (5.88) 002, (5.88) |
0.3440 | 219, 553![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 709, (5.66) 709, (5.66) |
HOMO−1 → LUMO+1 (56%) | n → π* | ||
| 1 | ACN | D13 | 373.78, 5007, (3.31) | 0.0302 | 375, 1257, (3.30) | SOMO−4(β) → LUMO(β) (34%) | LMCT |
| D36 | 270.35, 15![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 325, (4.58) 325, (4.58) |
0.0022 | 273, 9342, (4.54) | SOMO−4(α) → LUMO+1(α) (79%) | L1LCT | ||
| MeOH | D15 | 362.47,18![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 523, (3.42) 523, (3.42) |
0.0177 | 365, 1400, (3.39) | SOMO−7(β) → LUMO(β) (23%) | L1MCT | |
| D38 | 268.81, 14![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 867, (4.61) 867, (4.61) |
0.0010 | 269, 13![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 100, (4.60) 100, (4.60) |
SOMO−11(β) → LUMO(β) (85%) | LMCT | ||
| EtOH | D15 | 362.46, 18![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 525, (3.42) 525, (3.42) |
0.0181 | 368, 1342, (3.36) | SOMO−7(β) → LUMO(β) (23%) | L1MCT | |
| D38 | 268.82, 14![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 870, (4.61) 870, (4.61) |
0.001 | 270, 9887, (4.59) | SOMO−11(β) → LUMO(β) (85%) | LMCT | ||
| DCM | D13 | 375.05, 7048, (3.30) | 0.03 | 381, 1275, (3.25) | SOMO−4(β) → LUMO(β) (32%) | LMCT | |
| D35 | 274.82, 9836, (4.51) | 0.0132 | 277,8200, (4.47) | SOMO−5(β) → LUMO+1(β) (41%) | ILCT | ||
| 2 | ACN | D12 | 374.79, 3897, (3.30) | 0.0289 | 375![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 401, (3.30) 401, (3.30) |
SOMO−5(β) → LUMO(β) (24%) | LMCT |
| D30 | 270.83, 1257, (4.57) | 0.0099 | 272, 1310, (4.55) | SOMO−2(α) → LUMO(α) (47%) | L1LCT | ||
| MeOH | D13 | 366.00,13![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 789, (3.38) 789, (3.38) |
0.0108 | 365, 9900, (3.39) | SOMO−4(β) → LUMO(β) (33%) | L1MCT | |
| D31 | 269.61, 18![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 241, (4.59) 241, (4.59) |
0.001 | 270, 30![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 300, (4.59) 300, (4.59) |
SOMO−10(β) → LUMO(β) (46%) | LMCT | ||
| EtOH | D13 | 366.13, 13![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 802, (3.38) 802, (3.38) |
0.011 | 368, 3258, (3.39) | SOMO−4(β) → LUMO(β) (33%) | L1MCT | |
| D31 | 269.68, 18![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 246, (4.59) 246, (4.59) |
0.0011 | 271, 11![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 057, (4.59) 057, (4.59) |
SOMO−10(β) → LUMO(β) (47%) | LMCT | ||
| DCM | D12 | 376.46, 3345, (3.29) | 0.0288 | 381, 1457, (3.25) | SOMO−5(β) → LUMO(β) (22%) | LMCT | |
| D30 | 273.48, 1977, (4.53) | 0.013 | 274, 4340, (4.52) | SOMO−2(α) → LUMO(α) (45%) | L1LCT | ||
| UV-visa λmax,b (ε, M−1 cm−1) | λemission (nm) | Δνc (nm) | φd | Erede (V) | FT-IRf,g (cm−1) | ||
|---|---|---|---|---|---|---|---|
| a Wavelength in nanometer.b Molar extinction coefficient in M−1 cm−1 in methanol solvent.c Stoke shift.d Fluorescence quantum yield.e Methanol solution (supporting electrolyte NEt4ClO4, working electrode glassy carbon, reference electrode Ag/AgCl, scan rate 100 mV s−1).f In KBr pellet.g Wavenumber. | |||||||
| HL | ACN | 214 (5.82 × 105), 254 (1.62 × 105), 276 (2.92 × 104), 315 (8.87 × 104), 404 (4.22 × 103) | 365 | 111 | — | — | |
| MeOH | 214 (2.56 × 105), 254 (1.18 × 105), 278 (3.6 × 104), 316 (3.7 × 104), 400 (1.40 × 104) | 313, 359 | 59, 105 | 0.293 | |||
| EtOH | 215 (2.27 × 105), 254 (1.04 × 105), 278 (3.25 × 104), 315 (3.38 × 104), 400 (1.07 × 104) | 363 | 109 | ||||
| DCM | 219 (5.53 × 105), 254 (5.28 × 105), 279 (3.52 × 104) 316 (1.02 × 105), 407 (3.98 × 103) | 366 | 112 | ||||
| 1 | ACN | 221 (1.04 × 104), 245 (6.94 × 103), 273 (9.34 × 103), 375 (1.25 × 103) | 426, 457 | 51, 82 | 0.420 | 2843 [ν(C–H)], 1629 [νas(OCO)], 1535 [ν(C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N)] 1413 [νs(OCO)] N)] 1413 [νs(OCO)] |
|
| MeOH | 222 (1.37 × 104), 242 (1.09 × 104), 269 (1.31 × 104), 365 (1.4 × 103) | 412, 434 | 47, 69 | 0.129 | |||
| EtOH | 222 (1.04 × 104), 242 (8.08 × 103), 270 (9.88 × 103), 368 (1.34 × 103) | 417, 454 | 49, 86 | ||||
| DCM | 225 (8.82 × 103), 248 (5.97 × 103), 277 (8.20 × 103), 381 (1.27 × 103) | 436, 463 | 55, 82 | ||||
| 2 | ACN | 224 (3.20 × 103), 238 (3.28 × 103), 272 (1.31 × 103), 305 (5.42 × 102), 375 (4.01 × 102) | 426, 457 | 51, 82 | 0.436 | 2856 [ν(C–H)], 1629 [νas(OCO)], 1535 [ν(C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N)], 1410 [νs(OCO)] N)], 1410 [νs(OCO)] |
|
| MeOH | 204 (5.61 × 104), 236 (7.09 × 104), 270 (3.03 × 104), 305 (7.7 × 104), 365 (9.9 × 103) | 412, 434 | 47, 69 | 0.117 | |||
| EtOH | 204 (2.39 × 104), 237 (2.48 × 104), 271 (1.10 × 104), 300 (3.77 × 103), 368 (3.25 × 103) | 417, 455 | 49, 87 | ||||
| DCM | 204 (9.88 × 103), 243 (1.00 × 104), 274 (4.34 × 103), 307 (1.62 × 103), 381 (1.45 × 103) | 437, 463 | 56, 82 | ||||
Fluorescence spectra of complexes 1 and 2
The methanolic solution of HL shows emission at 359 nm (Fig. 8S†) when excited at 254 nm and the position of the emission band remain unchanged when λex is varied between 244 and 264 nm. On the other hand, on excitation at 365 nm, methanolic solution of both the complexes 1 and 2 exhibit luminescence bands (Fig. 9S and 10S†) at 412, 434 and 459 nm. These band positions remain unchanged when λex is varied between 355 and 375 nm. Both the complexes have identical excitation and emission spectral bands, which indicate that only change of carboxylate ligand (pa with mb) do not significantly effect in the electronic energy levels of the complexes. This is also supported by the results of DFT calculation of the complexes (vide infra). The energies and compositions of different molecular orbitals for both the complexes are very much closed. For complexes 1 and 2, the calculated fluorescence quantum yields (Φs) are 0.129 and 0.117, respectively. Results of the studies of the emission spectra of HL and complexes in different solvents are shown in Table 4 and Fig. 8S, 11S, 12S†. For HL, maximum blue shift of emission band observed in methanol, whereas maximum red shift in dichloromethane. Emission spectral bands for both the complexes show almost identical solvatochromic shift.Geometrical optimization and electronic structure
DFT and TD-DFT computations of optimized structures of HL and complexes 1 and 2 were performed to establish their electronic structure and spectral transitions. The geometric structures of the isolated ligand HL and complexes 1 and 2 were fully optimized at the B3LYP, B3PW91 and MPW1PW91 levels in the ground state (singlet for HL and doublet for complexes 1 and 2). The optimized structures of HL and complexes 1 and 2 along with their Mulliken charge distribution are depicted in Fig. 13S–15S.† TD-DFT calculations in methanol using conductor-like polarizable continuum model (CPCM) were performed and theoretically possible spin-allowed (singlet–singlet for HL and doublet–doublet for complexes) electronic transitions with their assignment are listed in Tables 3 and 2S–4S.† The TD-DFT results show that for HL, HOMO−18 → LUMO and HOMO → LUMO are the possible highest and lowest energy electronic transitions, respectively. The experimental electronic spectral bands (Fig. 7) at 214 and 278 nm in methanolic solution may be assigned as HOMO−1 → LUMO+1 and HOMO−1 → LUMO transitions, respectively. Both the bands at 214 and 278 nm corresponds to n → π* transitions (Table 3). It is interesting to note that the bands at 214 and 278 nm showed red shifting in relatively less polar solvent like dichloromethane, whereas blue shift resulted in relatively more polar solvent acetonitrile. This experimental observation also corroborate the nature of n → π* electronic transitions obtained from theoretical calculation. For n → π* electronic transitions, polar solvent stabilizes the ground electronic state more compared to corresponding excited state and hence with increasing solvent polarity blue shifting result. The results of TD-DFT calculation of HL using conductor-like polarizable continuum model (CPCM) in other solvents (ethanol/acetonitrile/dichloromethane) are summarized in Table 3. These results (Table 3 and Fig. 16S–18S†) evident the experimentally observed solvatochromic shifts of spectral bands.Calculated bond lengths and angles along with the experimentally measured values for complexes 1 and 2 are listed in Table 2. The comparison of bond lengths and angles between calculated and X-ray structure shows sufficient agreement.
The orbital diagram along with their energies and contributions from the ligands and metal for 1 and 2 are given in Fig. 19S and 20S,† and correlations of MOs are given in Fig. 21S–23S.† For α-MOs of 1, the energies of highest singly occupied molecular orbital (SOMO) and lowest unoccupied molecular orbital (LUMO) are −5.9 eV and −2.0 eV, respectively. Whereas for 2 corresponding values are −5.9 eV and −1.88 eV. For β-MOs of 1, the energies of SOMO and LUMO are −5.84 eV and −3.2 eV, respectively and for 2 the corresponding values are −5.85 eV and −3.22 eV, respectively. This results evidence that SOMO–LUMO energy difference of α-MOs are larger [ΔE, 3.9 eV (for 1); 4.02 eV (for 2)] compare to corresponding values of β-MOs [ΔE, 2.64 eV (for 1); 2.63 eV (for 2)].
Contribution of Schiff base (L), carboxylate and copper to SOMO and LUMO of α-MOs are [96% L, 1% pa, 3% Cu for 1; 97% L, 1% mb, 2% Cu for 2] and [1% L, 98% pa, 1% Cu for 1; 99% L, 0% mb, 1% Cu for 2], respectively. Whereas corresponding contributions to β-MOs are [95% L, 1% pa, 4% Cu for 1; 95% L, 1% mb, 4% Cu for 2] and [32% L, 7% pa, 61% Cu for 1; 32% L, 7% mb, 61% Cu for 2]. In summary for both complexes SOMO of α-MO and β-MO are characterized by the Schiff base ligand orbitals (≥95%). LUMO of α-MO and β-MO for 1 are characterized by the 3-phenylacrylate and copper orbitals, respectively, while for 2, LUMO of α-MO and β-MO are characterized by the Schiff base and copper orbitals, respectively. From TD-DFT calculations, the theoretically possible spin-allowed doublet–doublet electronic transitions of complexes with their assignment are listed in Tables 3 and 3S–4S.† For 1, the TD-DFT results show that SOMO−8(α) → LUMO(α) and SOMO(β) → LUMO(β) are the possible highest and lowest energy electronic transitions, respectively. On the other hand, for 2, SOMO−7(α) → LUMO(α) and SOMO(β) → LUMO(β) represent the possible highest and lowest energy electronic transitions, respectively. For both 1 and 2, the highest energy electronic transition in methanol is LL1CT (Schiff base to carboxylate inter ligand charge transfer) in nature, and the lowest energy electronic transition is LMCT (Schiff base to metal charge transfer) in nature. Experimental electronic transition in methanol for both the complexes at ∼270 nm may be assign as LMCT [SOMO−11(β) → LUMO(β) for 1; SOMO−10(β) → LUMO(β) for 2] transition, and the transition at ∼365 nm is L1MCT (carboxylate to metal charge transfer) [SOMO−7(β) → LUMO(β) for 1; SOMO−4(β) → LUMO(β) for 2] in nature. TD-DFT calculation for complexes using conductor-like polarizable continuum model (CPCM) in other solvents (ethanol/acetonitrile/dichloromethane) corroborate the experimentally observed solvatochromic shift of electronic absorption spectral bands (Table 3 and Fig. 24S–29S†).
Comparison of the results of DFT and TDDFT computations using different functionals reveals that B3LYP functional produce the structural and spectroscopic data which are more close to experimental results than B3PW91 and MPW1PW91 functionals (Tables 5S and 6S and Fig. 30S–32S†).
Redox properties of complexes
The electrochemical behavior of the complexes was investigated in methanol solution by cyclic voltammetry in the potential range between 0 and +0.6 V. Voltammetric data are collected in Table 4 and the voltammograms are displayed in Fig. 8. The cyclic voltammograms show the irreversible reduction processes at 0.42 and 0.44 V for complexes 1 and 2, respectively.DNA binding studies of complexes
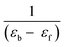 and
and  as slope and intercept, respectively. The values of intrinsic binding constants (Kib) were calculated from the ratio of slope to the intercept and calculated values of Kib for complexes 1 and 2 were 1.68 × 105 and 2.271 × 105 L mol−1, respectively.
as slope and intercept, respectively. The values of intrinsic binding constants (Kib) were calculated from the ratio of slope to the intercept and calculated values of Kib for complexes 1 and 2 were 1.68 × 105 and 2.271 × 105 L mol−1, respectively.
Photophysical properties of HL
 | ||
| Fig. 11 Change in UV-vis spectra of HL (3 mL 0.98 μM methanolic solution) upon gradual addition of 60 μL 7.69 μM methanolic solution of Cu(II) ion (nitrate salt). | ||
To explore UV-vis metric selectivity of HL towards the Cu(II) in the presence of other metal ions, we added 150 μL 25.66 μM methanolic solution of different metal ions [Na(I), K(I), Ca(II), Mg(II), Sr(II), Co(II), Ni(II), Cd(II), Pb(II) and Hg(II)] to the solution of adduct [3 mL 0.98 μM HL and 150 μL 25.66 μM Cu(NO3)2·3H2O] and found no noticeable change in UV-vis spectra of ensemble except slight increase in absorbance at 362 nm (Fig. 34S†), indicating excellent selectivity for Cu(II) over these competing metal ions. The reversible sensing ability of HL for Cu(II) was tested and the results are shown in Fig. 35S.† Addition of Cu(II) to HL results new bands at 239, 272 and 368 nm, which indicate the formation of Cu(II) and HL complex. But when EDTA was added as chelating ligand to the solution of Cu(II) and HL mixture, resulted the return of metal free HL spectrum, which evidences the reversible sensing ability of HL. Job's plot (at 316 nm, Fig. 12) analysis revealed the formation of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 stoichiometrical complex between Cu(II) and HL. To explain electronic spectral change of HL in presence of Cu(NO3)2·3H2O, we have calculated TD-DFT for all possible model compounds [6-31G(d-p) basis set for all atoms, except Cu for which LanL2DZ; CPCM model in methanol]. It is to note that TD-DFT results (Table 7S†) of five coordinated model compound [Cu(L)(NO3)] (Fig. 36S†) well fitted with the experimental electronic spectra of solution containing 1
1 stoichiometrical complex between Cu(II) and HL. To explain electronic spectral change of HL in presence of Cu(NO3)2·3H2O, we have calculated TD-DFT for all possible model compounds [6-31G(d-p) basis set for all atoms, except Cu for which LanL2DZ; CPCM model in methanol]. It is to note that TD-DFT results (Table 7S†) of five coordinated model compound [Cu(L)(NO3)] (Fig. 36S†) well fitted with the experimental electronic spectra of solution containing 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 mixture of HL and Cu(NO3)2·3H2O. This observation supports the formation of 1
1 mixture of HL and Cu(NO3)2·3H2O. This observation supports the formation of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 stoichiometrical complex between Cu(II) and HL with composition [Cu(L)(NO3)].
1 stoichiometrical complex between Cu(II) and HL with composition [Cu(L)(NO3)].
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 stoichiometry40
1 stoichiometry40 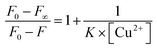 , where F0, F, F∞ are the emission intensities of HL considered in the absence of Cu(II), at an intermediate concentration of Cu(II) and at a concentration of complete interaction, respectively. Here K is the association constant and the plot of
, where F0, F, F∞ are the emission intensities of HL considered in the absence of Cu(II), at an intermediate concentration of Cu(II) and at a concentration of complete interaction, respectively. Here K is the association constant and the plot of 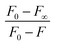 versus
versus  gives a straight line (Fig. 39S†) The rate constant (K) calculated from the slope of the line and the calculated value was 10.28 × 10−2 L mol−1.
gives a straight line (Fig. 39S†) The rate constant (K) calculated from the slope of the line and the calculated value was 10.28 × 10−2 L mol−1.Conclusion
Two copper(II) complexes were synthesized and characterized by spectroscopic methods, single crystal X-ray diffraction and electrochemical studies. Copper(II)–Schiff base in combination with aromatic carboxylate ligand pa (1) and mb (2) generates mononuclear distorted square pyramidal geometries and extends to 2D through C–H⋯π interactions. The DFT computations of the isolated complexes are in good agreement with the structural results obtained by X-ray diffraction. Results of TD-DFT calculations have been discussed in term of theoretically possible spin allowed electronic transitions along with their assignments. The influence of different functionals used in geometry optimization and excite state TD-DFT calculations of HL and complexes reveals that among the three, B3LYP, B3PW91 and MPW1PW91 functionals, B3LYP was able to best reproduce the experimental results. Solvatochromic shift of electronic absorption and emission spectral bands studied in acetonitrile, methanol, ethanol and dichloromethane, and results of TD-DFT calculation using conductor-like polarizable continuum model (CPCM) corroborate the experimentally observed solvatochromic shift of electronic spectral bands. Though the theoretical calculation for fluorescence emission spectra41 and DFT calculation based on long-range-corrected potentials (e.g. LC-BLYP)42 are beyond the scope of the present study, these calculations may be more significant for this type of work. Kinetics of interactions of 1 and 2 with CT-DNA was investigated and the binding affinities are in the order of 1.68 × 105 L mol−1 and 2.271 × 105 L mol−1, respectively. Study of the effect of various metal ions on the electronic absorption and fluorescence spectra of HL reveals that it selectively senses the Cu(II) ions.Acknowledgements
The authors gratefully acknowledge the financial assistance given by the CSIR, Government of India, to Dr Subal Chandra Manna (Project No. 01(2743)/13/EMR-II).References
- (a) L. Chen, J. Wang, Y.-Z. Liu, Y. Song, X.-T. Chen, Y.-Q. Zhang and Z.-L. Xue, Eur. J. Inorg. Chem., 2015, 271 CrossRef CAS; (b) A. V. Funes, L. Carrella, L. Sorace, E. Rentschler and P. Alborés, Dalton Trans., 2015, 44, 2390 RSC.
- (a) L. Chen, J. Wang, Y.-Z. Liu, Y. Song, X.-T. Chen, Y.-Q. Zhang and Z.-L. Xue, Eur. J. Inorg. Chem., 2015, 271 CrossRef CAS; (b) S. C. Manna, E. Zangrando, J. Ribas and N. Ray Chaudhuri, Dalton Trans., 2007, 1383 RSC; (c) S. Mistri, E. Zangrando, A. Figuerola, A. Adhikary, S. Konar, J. Cano and S. C. Manna, Cryst. Growth Des., 2014, 14, 3276 CrossRef CAS; (d) S. C. Manna, E. Zangrando, J. Ribas and N. Ray Chaudhuri, Eur. J. Inorg. Chem., 2008, 1400 CrossRef CAS.
- (a) K. B. Kim, H. Kim, E. J. Song, S. Kim, I. Noh and C. Kim, Dalton Trans., 2013, 42, 16569 RSC; (b) S. Mistri, E. Zangrando and S. C. Manna, Polyhedron, 2013, 49, 252 CrossRef CAS.
- (a) B. Duff, V. R. Thangella, B. S. Creaven, M. Walsh and D. A. Egan, Eur. J. Pharmacol., 2012, 689, 45 CrossRef CAS PubMed; (b) A. T. Chaviara, P. C. Christidis, A. Papageorgiou, E. Chrysogelou, D. J. Hadjipavlou-Litina and C. A. Bolos, J. Inorg. Biochem., 2005, 99, 2102 CrossRef CAS PubMed.
- (a) P. Mukherjee, M. G. B. Drew, M. Estrader and A. Ghosh, Inorg. Chem., 2008, 47, 7784 CrossRef CAS PubMed; (b) A. Bhunia, S. Manna, S. Mistri, A. Paul, R. K. Manne, M. K. Santra, V. Bertolasic and S. C. Manna, RSC Adv., 2015, 5, 67727 RSC.
- J. Adhikary, P. Chakraborty, S. Das, T. Chattopadhyay, A. Bauzá, S. K. Chattopadhyay, B. Ghosh, F. A. Mautner, A. Frontera and D. Das, Inorg. Chem., 2013, 52, 13442 CrossRef CAS PubMed.
- (a) M. S. Islas, J. J. M. Medina, L. L. L. Tévez, T. Rojo, L. Lezama, M. G. Merino, L. Calleros, M. A. Cortes, M. R. Puyol, G. A. Echeverría, O. E. Piro, E. G. Ferrer and P. A. M. Williams, Inorg. Chem., 2014, 53, 5724 CrossRef CAS PubMed; (b) A. Valent, M. Melnik, D. Hudecova, B. Dudova, R. Kivekas and M. R. Sundberg, Inorg. Chim. Acta, 2002, 340, 15 CrossRef CAS.
- (a) A. M. Abu-Dief and I. M. A. Mohamed, Beni-Suef University Journal of Basic and Applied Sciences, 2015, 4, 119 CrossRef; (b) F. Rahaman and B. H. M. Mruthyunjayaswamy, Complex Met., 2014, 1, 88 CrossRef.
- (a) S. Muche, I. Levacheva, O. Samsonova, L. Pham, G. Christou, U. Bakowsky and M. Hołyńska, Inorg. Chem., 2014, 53, 7642 CrossRef CAS PubMed; (b) A. Wojciechowska, A. Gągor, M. Duczmal, Z. Staszak and A. Ozarowski, Inorg. Chem., 2013, 52, 4360 CrossRef CAS PubMed; (c) P. K. Panchal, D. H. Patel and M. N. Patel, Synth. React. Inorg., Met.-Org., Nano-Met. Chem., 2004, 34, 1223 CrossRef CAS.
- (a) D.-D. Yin, Y.-L. Jiang and L. Shan, Chin. J. Chem., 2001, 19, 1136 CrossRef CAS; (b) X. Zhong, J. Yi, J. Sun, H.-L. Wei, W.-S. Liu and K.-B. Yu, Eur. J. Med. Chem., 2006, 41, 1090 CrossRef CAS PubMed.
- (a) J. Vančo, O. Švajlenová, E. Račanská, J. Muselik and J. Valentova, J. Trace Elem. Med. Biol., 2004, 18, 155 CrossRef; (b) A. Garoufis, S. K. Hadjikakou and N. Hadjiliadis, Coord. Chem. Rev., 2009, 253, 1384 CrossRef CAS.
- J. Vančo, J. Marek, Z. Trávníček, E. Račanská, J. Muselík and O. Švajlenová, J. Inorg. Biochem., 2008, 102, 595 CrossRef PubMed.
- Y. Yoshikawa and H. Yasui, Curr. Top. Med. Chem., 2012, 12, 210 CrossRef CAS PubMed.
- (a) B. Duff, V. R. Thangella, B. S. Creaven, M. Walsh and D. A. Egan, Eur. J. Pharmacol., 2012, 689, 45 CrossRef CAS PubMed; (b) A. T. Chaviara, P. C. Christidis, A. Papageorgiou, E. Chrysogelou, D. J. Hadjipavlou-Litina and C. A. Bolos, J. Inorg. Biochem., 2005, 99, 2102 CrossRef CAS PubMed; (c) A. Bhunia, S. Manna, S. Mistri, A. Paul, R. K. Manne, M. K. Santra, V. Bertolasic and S. C. Manna, RSC Adv., 2015, 56, 7727 Search PubMed; (d) S. Mistri, A. Paul, A. Bhunia, R. K. Manne, M. K. Santra, H. Puschmann and S. C. Manna, Polyhedron, 2016, 104, 63 CrossRef CAS.
- (a) S. Mistri, S. García-Granda, E. Zangrando and S. C. Manna, Polyhedron, 2013, 50, 333 CrossRef CAS; (b) S. Mistri, V. Bertolasi and S. C. Manna, Polyhedron, 2015, 88, 101 CrossRef CAS.
- N. A. I. Hisham, H. Khaledi, H. M. Ali and H. A. Hadi, J. Coord. Chem., 2012, 65, 3006 Search PubMed.
- S.-S. Qian, H.-H. Li, H. Zhu, Z.-M. Yang, Z.-L. You and H.-L. Zhu, Synth. React. Inorg., Met.-Org., Nano-Met. Chem., 2013, 43, 412 CrossRef CAS.
- D. D. Perrin, W. L. F. Armarego and D. R. Perrin, Purification of Laboratory Chemicals, Pergamon Press, Oxford, U.K., 1980 Search PubMed.
- J. R. Lakowicz, Principles of Fluorescence Spectroscopy, Springer, New York, USA, 3rd edn, 2006 Search PubMed.
- CrysAlis PRO, Agilent Technologies Ltd, Yarnton, 2010 Search PubMed.
- O. V. Dolomanov, L. J. Bourhis, R. J. Gildea, J. A. K. Howard and H. Puschmann, OLEX2: a complete structure solution, refinement and analysis program, J. Appl. Crystallogr., 2009, 42, 339 CrossRef CAS.
- G. M. Sheldrick, A short history of SHELX, Acta Crystallogr., 2008, A64, 112 CrossRef PubMed.
- L. J. Farrugia, J. Appl. Crystallogr., 1999, 32, 837 CrossRef CAS.
- K. Brandenburg, DIAMOND (Version 3.2i), Crystal Impact GbR, Bonn, Germany, 1999 Search PubMed.
- Gaussian 09, revision A.02, Gaussian, Inc., Wallingford, CT, 2009 Search PubMed.
- C. Lee, W. Yang and R. G. Parr, Phys. Rev., 1988, 37B, 785 Search PubMed.
- H. Ihee, J. Kua, W. A. Goddard III and A. H. Zewail, J. Phys. Chem. A, 2001, 105, 3623 CrossRef CAS.
- J. P. Safko, J. E. Kuperstock, S. M. McCullough, A. M. Noviello, X. Li, J. P. Killarney, C. Murphy, H. H. Patterson, C. A. Bayse and R. D. Pike, Dalton Trans., 2012, 41, 11663 RSC.
- P. J. Hay and W. R. Wadt, J. Chem. Phys., 1985, 82, 270 CrossRef CAS.
- (a) R. E. Stratmann, G. E. Scuseria and M. J. Frisch, J. Chem. Phys., 1998, 109, 8218 CrossRef CAS; (b) M. E. Casida, C. Jamorski, K. C. Casida and D. R. Salahub, J. Chem. Phys., 1998, 108, 4439 CrossRef CAS.
- M. Cossi, N. Rega, G. Scalmani and V. Barone, J. Comput. Chem., 2003, 24, 669 CrossRef CAS PubMed.
- N. M. O'Boyle, A. L. Tenderholt and K. M. Langner, J. Comput. Chem., 2008, 29, 839 CrossRef PubMed.
- A. Wolfe, G. H. Shimer and T. Mechan, Biochemistry, 1987, 26, 6392 CrossRef CAS PubMed.
- A. W. Addison, T. N. Rao, J. Reedijk, J. V. Rijn and G. C. Verschoor, J. Chem. Soc., Dalton Trans., 1984, 1349 RSC.
- (a) M. Nishio, M. Hirota and Y. Umezawa, The C–H⋯π Interaction: Evidence, Nature and Consequences, Wiley-VCH, New York, 1998 Search PubMed; (b) M. Nishio, CrystEngComm, 2004, 6, 130 RSC; (c) M. d. C. Fernandez-Alonso, F. J. Canada, J. Jimenez-Barbero and G. Cuevas, J. Am. Chem. Soc., 2005, 127, 7379 CrossRef CAS PubMed; (d) D. Braga, S. L. Giaffreda, F. Grepioni, L. Maini and M. Polito, Coord. Chem. Rev., 2006, 250, 1267 CrossRef CAS; (e) H. J. Schneider, Angew. Chem., Int. Ed., 2009, 48, 3924 CrossRef CAS PubMed.
- (a) Q.-L. Zhang, J.-G. Liu, H. Chao, G.-Q. Xue and L.-N. Ji, J. Inorg. Biochem., 2001, 83, 49 CrossRef CAS PubMed; (b) Z.-C. Liu, B.-D. Wang, B. Li, Q. Wang, Z.-Y. Yang, T.-R. Li and Y. Li, Eur. J. Med. Chem., 2010, 45, 5353 CrossRef CAS PubMed; (c) R. K. Gupta, G. Sharma, R. Pandey, A. Kumar, B. Koch, P.-Z. Li, Q. Xu and D. S. Pandey, Inorg. Chem., 2013, 52, 13984 CrossRef CAS PubMed; (d) E. Ramachandran, D. Senthil Raja, N. P. Rath and K. Natarajan, Inorg. Chem., 2013, 52, 1504 CrossRef CAS PubMed.
- (a) F. Mancin, P. Scrimin, P. Tecilla and U. Tonellato, Chem. Commun., 2005, 2540 RSC; (b) L. Tjioe, A. Meininger, T. Joshi, L. Spiccia and B. Graham, Inorg. Chem., 2011, 50, 4327 CrossRef CAS PubMed.
- (a) P. Kumar, S. Gorai, M. Kumar, B. Mondal and D. Manna, Dalton Trans., 2012, 41, 7573 RSC; (b) K. Jeyalakshmi, Y. Arun, N. S. P. Bhuvanesh, P. T. Perumal, A. Sreekantha and R. Karvembu, Inorg. Chem. Front., 2015, 2, 780 RSC.
- (a) J.-B. Lepecq and C. Paoletti, J. Mol. Biol., 1967, 27, 87 CrossRef CAS PubMed; (b) R. Palchaudhuri and P. J. Hergenrother, Curr. Opin. Biotechnol., 2007, 18, 497 CrossRef CAS PubMed; (c) A. J. Geall and I. S. Blagbrough, J. Pharm. Biomed. Anal., 2000, 22, 849 CrossRef CAS PubMed; (d) M. A. Kostiainen, J. G. Hardy and D. K. Smith, Angew. Chem., Int. Ed., 2005, 44, 2556 CrossRef CAS PubMed.
- (a) X. Peng, X. Tang, W. Qin, W. Dou, Y. Guo, J. Zheng, W. Liu and D. Wang, Dalton Trans., 2011, 40, 5271 RSC; (b) A. Mallick and N. Chahattopadhyay, Photochem. Photobiol., 2005, 81, 419 CrossRef CAS PubMed; (c) H. A. Benesi and J. H. Hildebrand, J. Am. Chem. Soc., 1949, 71, 2703 CrossRef CAS.
- (a) P. Gąsiorski, K. S. Danel, M. Matusiewicz, T. Uchacz, W. Kuźnik and A. V. Kityk, J. Fluoresc., 2012, 22, 81–91 CrossRef PubMed; (b) P. Gąsiorski, K. S. Danel, M. Matusiewicz, T. Uchacz, W. Kuźnik, Ł. Piątek and A. V. Kityk, Mater. Chem. Phys., 2012, 132, 330–338 CrossRef.
- A. V. Kityk, J. Phys. Chem. A, 2012, 116, 3048–3055 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. CCDC 1405167 and 999890. For ESI and crystallographic data in CIF or other electronic format see DOI: 10.1039/c6ra05570b |
| This journal is © The Royal Society of Chemistry 2016 |