Identification of modes of interactions between 9-aminoacridine hydrochloride hydrate and serum proteins by low and high resolution spectroscopy and molecular modeling†
Abstract
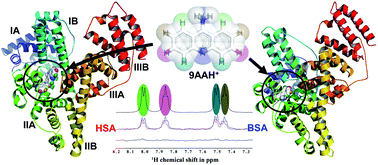
Photophysical studies on binding interactions of a therapeutically important drug, 9-aminoacridine hydrochloride hydrate (9AA-HCl) with serum proteins, bovine serum albumin (BSA) and human serum albumin (HSA), have been performed using several low and high resolution spectroscopic techniques in conjunction with molecular modeling, which disclose some salient features of such interactions in ground and excited sates. The studies reveal that although 9AA-HCl forms ground state complexes with both BSA and HSA, their individual modes of binding interaction are quite different. Its resonance energy transfer efficiency is 79% and 72% with BSA and HSA respectively. It undergoes photoinduced electron transfer (PET) with BSA, but both PET and excited state proton transfer simultaneously with HSA, which are confirmed further by laser flash photolysis studies coupled with a magnetic field. Thermodynamic analyses indicate that the binding of 9AA-HCl with BSA and HSA is controlled primarily by changes in enthalpy and entropy, respectively, with corresponding binding stoichiometries of 1 : 1 and 1 : 2, respectively. Circular dichroism spectra depict some structural changes of both the serum proteins while interacting with 9AA-HCl. Docking analyses unveil the crucial role of the disparity between the cavity sizes of the proteins which might be the foremost reason behind the differential behavior of the drug towards BSA and HSA. The binding site pocket of HSA to be docked with 9AA-HCl is 1.7 times larger in dimensions than that of BSA, which facilitates HSA to bind with 9AA-HCl with higher stoichiometry compared to BSA. These differences in binding modes as well as affinities have been further confirmed by saturation transfer difference (STD) NMR experiments which show the ligand 9AA-HCl binds to BSA with higher affinity compared to HSA. In addition, unlike BSA, HSA can accommodate more than one ligand, which corroborates well with docking and fluorescence studies.


 Please wait while we load your content...
Please wait while we load your content...