Hollow silver alginate microspheres for drug delivery and surface enhanced Raman scattering detection†
Abstract
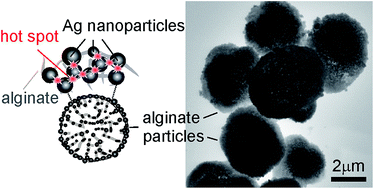
Multifunctional silver alginate hydrogel microspheres are assembled via a template assisted approach using calcium carbonate cores. Sodium alginate is immobilized into the highly porous structure of calcium carbonate microspheres followed by cross-linking in the presence of silver ions. The simultaneous processes of the growth of silver nanoparticles in the alginate matrix and the removal of the calcium carbonate template are triggered by ascorbic acid. The abundance of silver nanoparticles and their interparticular junctions in the alginate network allow for the detection of solutes using Raman spectroscopy using the surface of the plasmonic microspheres. Rhodamine B was used to illustrate the potential applications of such multifunctional plasmonic alginate hydrogel microspheres for sensing at low concentrations. A proof of principle for using such particles for the quick identification of microorganisms is then demonstrated using the Escherichia coli bacterium.
- This article is part of the themed collection: Surface enhanced Raman Spectroscopy: Editors collection for RSC Advances

 Please wait while we load your content...
Please wait while we load your content...