HSAF-induced antifungal effects in Candida albicans through ROS-mediated apoptosis
Abstract
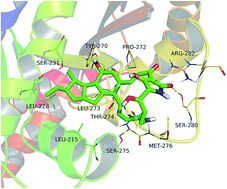
Heat-stable antifungal factor (HSAF) belongs to polycyclic tetramate macrolactams (PTMs), which inhibits many fungal pathogens and is effective in inhibiting Candida albicans (C. albicans). In this study, we found that HSAF induced the apoptosis of C. albicans SC5314 through inducing the production of reactive oxygen species (ROS). Nevertheless, we validated the efficacy of HSAF against candidiasis caused by C. albicans in a murine model in vivo, and HSAF significantly improved survival and reduced fungal burden compared to vehicles. A molecular dynamics (MD) simulation was also investigated, revealing the theoretical binding mode of HSAF to the β-tubulin of C. albicans. This study first found PTMs-induced fungal apoptosis through ROS accumulation in C. albicans and its potential as a novel agent for fungicides.

 Please wait while we load your content...
Please wait while we load your content...