 Open Access Article
Open Access ArticleGeometric effects of nano-hole arrays for label free bio-detection†
Seunguk
Kim
a,
Jeong Hee
Shin
a,
Samhwan
Kim
b,
Seung-Jun
Yoo
b,
Byoung Ok
Jun
a,
Cheil
Moon
b and
Jae Eun
Jang
*a
aDepartment of Information and Communication Engineering, Daegu Gyeongbuk Institute of Science and Technology (DGIST), 333, Techno jungang-daero, Hyeonpung-myeon, Dalseong-gun, Daegu, 711-873, Korea. E-mail: jang1@dgist.ac.kr
bDepartment of Brain & Cognitive Sciences, Daegu Gyeongbuk Institute of Science and Technology (DGIST), 333, Techno jungang-daero, Hyeonpung-myeon, Dalseong-gun, Daegu, 711-873, Korea
First published on 15th January 2016
Abstract
Geometric effects of nano-hole arrays were investigated for label free bio-detection. Because the nano-hole arrays were designed to present a filtered peak wavelength in the visible light region, filtered color changes caused by different biomolecules were easily observed with a microscope or even by the naked eye. Generally, many biomolecules are transparent or colorless in the visible range, so that it is hard to distinguish among them using visible observation. However, their molecular structure and composition induce some differences in the dielectric constant or refractive index causing a filtered color shift in the nano-hole array structure. Here, the contribution of geometric parameters such as the hole diameter and the spacing between nano-holes for bio-detection was evaluated to maximize the change in color among different biomolecules. A larger hole size and space between the holes enabled the biomolecules to be easily distinguished. Even if the change in color was not distinctive enough by eye in some cases, it was possible to distinguish the change by simple analysis of the ‘Hue’ values or by the ‘Lab’ color coordinates obtained from the photo images. Therefore, this technique can have high probability of realization for real-time detection of cells without the use of bio-markers.
The most powerful analysis tool for real-time observation of cells so far is the microscope in bio-related fields. However, the resolution limit is a barrier to bio-analysis because many bio-objects are smaller than 1 μm. Various new optical approaches have been suggested with improvements in the quality of optical components to solve the resolution limit. The other important issue without much progress yet is poor selectivity among various biomolecules such as proteins due to their transparent or colorless optical characteristic in the visible range. To provide selectivity, bio-markers, generally fluorescent materials, are used for ‘in vivo’ or ‘in vitro’ bio-detection;1–3 however, these detected molecules are not native to the human body. Therefore, they can lead to undesirable results sometimes.4–6 One of the promising solutions for marker-free detection is surface plasmon resonance (SPR), which is the resonant oscillation of conduction electrons at the interface between a negative and positive permittivity material stimulated by incident light. SPR reflectivity measurements can be used to detect molecular adsorption, such as polymers, organic molecules or proteins.7–10 The mechanism of detection is as follows: the adsorbing molecules cause changes in the local index of refraction, changing the resonance conditions of the surface plasmon waves. However, technically, a detector system such as a precise reflection angle detector or spectrum analyzer is essential.11 In addition, an anti-body coating is required to enhance the sensitivity for certain conditions. The phase contrast microscopy, usually preferred in biology, is an optical microscopy which converts phase shifts of the light passing through a transparent target to brightness changes in the image. They are able to be visualized when shown as brightness variations although phase shifts are invisible. However, it just enhances the contrast factor of the image and easily causes Halo effect.12 Due to the reasons as described above, more appropriate and simpler technique for real-time detection is required.
A structural color filter (SCF) based on nano-hole arrays which originate from localized surface plasmon resonance (LSPR) could be an important solution for marker-free detection. Once the nano-holes are periodically perforated on metallic thin films, the incident light into these arrays is filtered out, and specific wavelengths are transmitted.13–15 If we can select the appropriate design factors for SCFs, the filtered peak wavelength can be in the visible light region. For a general color filter (CF) using color pigments, if a transparent biomolecule is dropped onto the CF, it just changes the brightness of the filtered light not the color coordinates (Fig. 1a and c). However, in the case of a SCF, the filtered wavelength can be selected according to the optical characteristics of a biomolecule such as the refractive index or dielectric constant as well as the structural dimension of the SCF. This means that even though two transparent biomolecules are dropped onto a SCF, the SCF has distinguishable transmission results depending on its dielectric constant or its refractive index, i.e., the transmissive colors are different in the same SCF. Fig. 1 is a schematic description of such a difference. Therefore, we can distinguish the transparent biomolecules with a microscope or even by the naked eye when using the SCF without any bio-markers.
Herein, we investigated the role of the geometric factors of SCFs such as the hole diameter and spacing between the holes in the bio-detection mechanism, especially to maximize the color shift for high material sensitivity. In addition, by using fabricated filters as a cell-detector, we confirmed that they are suitable for classifying individual biomolecules even if their size is extremely small.
To investigate the geometric effects on the spectral properties, SCFs were fabricated with varying hole diameters (120–220 nm) and spacing conditions (260–400 nm). To form the metallic layer, 150 nm thick Al films were deposited onto a glass substrate by thermal evaporation. A SiO2 layer was also deposited by a RF sputtering system at a thickness of 50 nm as a mask in the Al etching process. Periodic nano-hole arrays were formed with an electron beam lithography system (JEOL JBX-9300FS) with an accelerating voltage of 100 kV. The hole arrays consisted of a quadrate arrangement with the same spacing for both the x- and y-axes. This square structure makes the SCFs independent of the polarization effect from the incident light. After formation of the PMMA mask pattern, the SiO2 and Al films were etched by an inductively coupled plasma and reactive ion etching (ICP-RIE) system. The SiO2 layer used as an etch mask remained on the Al films to enhance the resonance modes of upper and bottom interfaces of patterned Al films and to avoid reactions with water, air, or biological solutions. More details of experimental procedure are described in Fig. S1.†Fig. 2a shows an optical microscope (OM, transmission mode) image and scanning electron microscope (SEM) images of the SCFs. As shown in Fig. 2a, each filter has a different transmission color and intensity from the different geometric factors (hole diameters and spacing). The transmission spectral properties of the fabricated SCFs were measured with a CCD based spectrometer (DALSA PRO-5200) which had an available wavelength between 400 nm and 780 nm with a 1 nm resolution. The light source was set vertical to the filters when the spectral measurements were performed. Fig. 2b shows the measured transmission spectra of the SCFs which consist of increasing spacing between the holes from 260 to 400 nm with a 20 nm step while the diameter of the holes was kept at 120 nm. Although the increase in spacing led to a decrease in the transmission intensity, it was clearly confirmed that it induces a red-shift in the resonance wavelengths, i.e., the filtering colors are changed from bluish green to red. When we chose a spacing smaller than 260 nm, we also got a blue color filtering result. Fig. 2c shows the effect of the hole diameters on the transmission spectra of the filters. The spacing was fixed at 400 nm while the diameter was changed from 120 to 200 nm with a 20 nm step. In contrast to the dynamic spectral shift of the SCFs with variable spacing (Fig. 2b), the change in the hole diameters mainly affected the transmission intensity of the filters. The increase in the hole diameter induced a higher transmittance and broadened the half width at full maximum (HWFM) of the spectra. In addition to this main tendency, a small red-shift occurred when the diameter of the holes increased, as well. This shift is due to the cut-off behavior of the filters, i.e., longer wavelength lights are more sensitive and reactive to changes in the diameter.16,17Fig. 2d and e show the peak points of the resonance wavelengths according to the spacing and the diameter. Both (1,0)- and (1,1)-resonance wavelengths were red-shifted when the spacing and the diameter were increased. The integer pair (i,j) such as (1,0) and (1,1) is related with reciprocal lattice vectors in square structural arrays. In addition, it plays an important role to determine resonance modes in SCFs. Therefore, we can conclude that even though the spacing of nano-hole arrays primarily determines the position of the resonance wavelengths, the diameter of the holes also has a partial role in the positioning regime.
A change in the surface environment such as the dielectric constant enables the SCFs to detect biomolecules. The resonance peak of the filtered wavelengths through the SCFs is approximately expressed as follows:
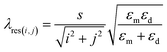 | (1) |
As shown in Fig. 3b–f, dramatic shifts of spectra occurred when the deionized water was dropped on filters. This visible color change comes from a difference between the dielectric constants of air (εd,air ≈ 1) and water (εd,water ≈ 1.33). Therefore, the filtered spectra moved toward longer wavelengths. In the case of the number ‘2’ filter in Fig. 3a and b, even though it looks like a blue-shift occurred (orange to yellow), both the (1,0)- and (1,1)-resonance peaks moved to longer wavelengths (red-shift). The (1,0)-peak just shifted into the near-infrared (NIR) region while the (1,1)-peak still remained in the visible light region as shown in Fig. 3e. As a result, the reddish color diminishes and only the yellow color becomes visible. For the number ‘1’ filter, the two main peaks shifted toward the NIR region and this resulted in a grey color (Fig. 3d). Eqn (1) also supports this spectral shift tendency. However, these color changes are not normal phenomenon for general pigment-based CFs. The water drop simply causes a change in the brightness. From this perspective, SCFs are advantageous in the detection of transparent biomolecules such as proteins. However, the difference in the dielectric constants of biomolecules sometimes can be small because they are diluted with a base solution with a low concentration, or they have a similar molecule structure. For these reasons, optimization of the geometric factors such as the spacing and diameter of the holes is needed to maximize the color difference among biomolecules and to increase the resolution of detection of the SCFs. According to eqn (1), it is clear that the spacing and the dielectric constant have a crucial role in determining the position of the resonance peaks of the filtered wavelengths.
Here, the dielectric constants of metal εm is fixed. The change in εd induced by biomolecules diluted with a base solution is assumed to be quite small. Therefore, we need to consider the spacing effect on the positioning regime of the resonance wavelengths. Additionally, the hole diameter is also an important geometry to enhance the selectivity of the SCFs to biomolecules because it can induce a filtering color change based on the difference in optical characteristics of the biomolecules, as well. For this reason, we fabricated two groups of SCFs. One group was organized with variable spacing (280, 320, and 360 nm), and the other consisted of variable diameters (140, 160, and 180 nm). Then, collagen and bovine serum albumin (BSA) solutions diluted to 20 μg ml−1 with phosphate buffered saline (PBS) were dropped onto the surface of the filters. Collagen is the main structural protein of the connective tissue in the form of elongated fibrils, whereas BSA is a globular protein abundant in the plasma.19–21 Finally, their optical property was measured by a spectrometer. Fig. 4a and b show the transmission spectra of the SCFs with the collagen and BSA solutions. From the results, several enhancement factors for improving the resolution of bio-detection are revealed from the measured transmission spectral property. At first, the BSA solution showed a higher transmittance than that of the collagen for all cases. However, this tendency was not what we expected because a similar result can be obtained with general CFs. Crucially, such a difference in transmission intensity also occurs according to the concentration of the same biomolecule.22 Therefore, it is not proper to use this detecting mechanism. Meanwhile, like in Fig. 3, the filtered resonance peaks shifted toward the red color direction with a drop of the solutions. The resonance wavelengths of the BSA solution were positioned slightly to the right of that of collagen for all the SCFs. This is due to the larger dielectric constant and refractive index of BSA compared to collagen.23,24 In this regime, another enhancing factor is available. By increasing the spacing between the holes, the difference in the main resonance wavelengths for both solutions increased shown in Fig. 4c. This can be explained easily by eqn (1). The spacing is the slop of eqn (1) such that a larger one can induce a larger difference between each resonance peak with the two dielectric constants of εd,BSA and εd,col. Furthermore, the increase in the diameter of the holes caused more red-shifted resonance wavelengths. At the same spacing condition (360 nm), a larger hole diameter resulted in much red-shift for both solutions shown in Fig. 4b. Thus, the increase in the diameter of the holes, which limits the transmission of longer wavelengths, makes the cut-off point move toward the right. In this mechanism, the shift rate of each spectrum also depends on the dielectric constant, and a more considerable red-shift occurs with a higher dielectric constant. Therefore, the increase in the spacing and diameter enhances the separation of the resonance peaks induced by each of the biomolecules.
Fig. 4c and d shows the difference between the resonance wavelengths of the collagen and BSA solutions according to various spacing and diameter conditions. Even though the molecules had a low concentration, a deviation of 2 nm was achieved by changing the spacing. In addition to this, we observed that the diameter of the holes also has a large role. The difference in the main resonance wavelength for collagen and BSA was enhanced to 5 nm with a hole diameter of 180 nm under a fixed spacing of 360 nm (Fig. 4d). In the case of a 1–2 nm peak shift, it is not easy to detect a color difference by eye or by a simple color analysis program, such that we may need a good spectrum analyzer. However, as mentioned previously, a spectrum analyzer is not appropriate for real time detection. Whereas, a 5 nm difference is detected by eyes and clearly detected by commercialized programs as like ‘Photoshop’ in ‘Hue’ value or ‘Lab’ color coordinates. Thus, SCFs consisting of larger geometric factors which are the spacing and hole diameter guarantee the higher detecting resolution. However, much larger geometric factors leading to a NIR shift of the resonance wavelengths and resulting in a failure of our designed intent that the transmission spectra are positioned in the visible light region.
To confirm the possibility of the SCFs to be utilized as a bio-detector, detection of live cells was performed with a microscope. In general, a cell is composed of many and diverse biomolecules such as proteins and nucleic acids. Thus, it is expected that transmission colors will change according to the type of biomolecules because each has its own dielectric constant. This property enables easier distinction among biomolecules. For this experiment, human embryonic kidney (HEK)-293 cells were dropped onto the surface of a general CF using a red pigment and onto our fabricated SCF. The SCF consisted of a spacing of 360 nm and a hole diameter of 180 nm. For low magnification, the image captured from the SCF is much clearer than that of the general CF due to the higher contrast which comes from both the brightness and color changes shown in Fig. 5a and b. As expected, just the difference in brightness was detected in the cell structure on the general CF due to its mechanistic principle. Most of the compositions constituting a cell is colorless or transparent so that different biomolecules induce only a brightness change. However, the SCF with cells showed different colors as well as a change in brightness depending on the molecules as long as their dielectric constants were not the same with each other. Therefore, with the SCF, it is much clearer by microscope observation. In highly magnified images, this tendency became stronger. The image from the SCF with the HEK-239 cell had various color pixels such as greenish yellow, yellow, red and base magenta, whereas, all the pixel colors were almost red for the general CF. The color change can be quantified with a simple commercialized image program using the ‘Hue’ value. Hue is one of the main properties of a color, which is part of the color appearance parameters, and technically defined as the degree of similarity or difference between stimuli which are described as red, orange, yellow, green, blue, and violet. In general, Hue is usually referred to as ‘colors’. The different Hues have different wavelengths in the spectrum. Therefore, we can quantify the spectrum difference from the Hue values.25 Here, we used ‘Adobe Photoshop CS6’ to analyze the results. There were two comparison points, the nucleus enriched with nucleic acids (white circle) and the cytosol abundant with proteins (yellow circle) (Fig. 5a and b). For the general CF, the Hue values were same as 0° for both the nucleus and cytosol areas, even though they are enriched with distinct biomolecules. In contrast, a big difference in the Hue values was induced by the SCFs. The cytosol area showed a reddish magenta color, which corresponds to a value of 345° for the Hue, whereas, the nucleus area had a value of 12°. Inside the cell, most of the pixel colors was slightly different because each point was occupied by different biomolecule compositions which are well-matched with the physiological analysis of the cell. When we used a ‘Lab’ space based on nonlinearly compressed CIE XYZ coordinates consisting of dimension L and ab for the lightness and opposition colors respectively, the matching with biomolecules was being more precise. As a result, the Lab values of the general color filter were measured as (L,a,b) = (54,72,51) and (57,70,45) for the nucleus and cytosol areas. This result indicates that the difference in both lightness and color is too small. However, the SCF induced values of (L,a,b) = (42,34,29) and (49,57,13) for the two points, such that these clearly show the distinct color difference with high resolution which enables easier matching with biomolecules (Fig. 5e and f). Because both parameters, ‘Hue’ and ‘Lab’, can be simply obtained from the photo images by commercialized image programs, we are able to trace the kind of biomolecules in this manner without need of any complicated spectrum analyzer. Therefore, we believe that SCFs can be utilized as a solution for real-time detection of cell configurations with some standardization.
In summary, we fabricated SCFs with varying geometric factors including the spacing between holes and the diameters of the holes and explained their effects on the detection resolution of biomolecules. The role of spacing is one of the important parameters. Moreover, in this study, we found that the hole diameter also has a large contribution in enhancing the resolution. An increase in both the spacing and diameter of nano-hole arrays induced a red-shift in the resonance wavelengths, and this tendency can maximize the difference between characteristic transmission spectra of biomolecules. When the geometric factors mentioned above are properly adjusted and optimized for each filter, further enhancement in the detecting ability can be obtained. Additionally, we showed that the SCFs can be used to distinguish the type of biomolecules in cells with general color spaces such as ‘Hue’ and ‘Lab’ coordinates. This is expected to have great potential in label-free and real-time ‘in vitro’ bio-detection with easy handling.
Acknowledgements
This research was supported by Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the MSIT (2014M3A9D7070668/2015R1A2A2 A01005043) and DGIST R&D Program of MSIT (15-BD-0404/15-BD-0402).Notes and references
- H. J. Avens and C. N. Bowman, Acta Biomater., 2010, 6, 83–89 CrossRef CAS PubMed.
- I. Lee, W.-K. Oh and J. Jang, J. Hazard. Mater., 2013, 252, 186–191 CrossRef PubMed.
- Y.-D. Zhu, J. Peng, L.-P. Jiang and J.-J. Zhu, Analyst, 2014, 139, 649–655 RSC.
- H. Ozaki and L. W. McLaughlin, Nucleic Acids Res., 1992, 20, 5205–5214 CrossRef CAS PubMed.
- M. L. Larramendy, W. El-Rifai and S. Knuutila, Cytometry, 1998, 31, 174–179 CrossRef CAS PubMed.
- M. Nagel, F. Richter, P. Haring-Bolivar and H. Kurz, Phys. Med. Biol., 2003, 48, 3625–3636 CrossRef CAS PubMed.
- A. G. Brolo, R. Gordon, B. Leathem and K. L. Kavanagh, Langmuir, 2004, 20, 4813–4815 CrossRef CAS PubMed.
- A. Kausaite-Minkstimiene, A. Ramanaviciene and A. Ramanavicius, Analyst, 2009, 134, 2051–2057 RSC.
- H. K. Hunt and A. M. Armani, Nanoscale, 2010, 2, 1544–1559 RSC.
- E. Kazuma and T. Tatsuma, Nanoscale, 2014, 6, 2397–2405 RSC.
- F. C. Chien and S. J. Chen, Biosens. Bioelectron., 2004, 20, 633–642 CrossRef CAS PubMed.
- T. Wilson and C. Sheppard, Theory and practice of scanning optical microscopy, Academic Press London, 1984 Search PubMed.
- C. Genet and T. Ebbesen, Nature, 2007, 445, 39–46 CrossRef CAS PubMed.
- Y. S. Do, J. H. Park, B. Y. Hwang, S. M. Lee, B. K. Ju and K. C. Choi, Adv. Opt. Mater., 2013, 1, 133–138 CrossRef.
- Y. Gu, L. Zhang, J. K. Yang, S. P. Yeo and C.-W. Qiu, Nanoscale, 2015, 7, 6409–6419 RSC.
- H. Bethe, Phys. Rev., 1944, 66, 163 CrossRef.
- K. L. van der Molen, F. B. Segerink, N. F. van Hulst and L. Kuipers, Appl. Phys. Lett., 2004, 85, 4316–4318 CrossRef CAS.
- M. R. Gartia, A. Hsiao, A. Pokhriyal, S. Seo, G. Kulsharova, B. T. Cunningham, T. C. Bond and G. L. Liu, Adv. Opt. Mater., 2013, 1, 68–76 CrossRef.
- M. J. Buehler, Proc. Natl. Acad. Sci. U. S. A., 2006, 103, 12285–12290 CrossRef CAS PubMed.
- P. Fratzl, Collagen: structure and mechanics, Springer Science & Business Media, 2008 Search PubMed.
- X. M. He and D. C. Carter, Nature, 1992, 358, 209–215 CrossRef CAS PubMed.
- D. Zhang, Y. Lu, J. Jiang, Q. Zhang, Y. Yao, P. Wang, B. Chen, Q. Cheng, G. L. Liu and Q. Liu, Biosens. Bioelectron., 2015, 67, 237–242 CrossRef CAS PubMed.
- H. Schwick and H. Haupt, The plasma proteins: structure, function, and genetic control, New York, 1985 Search PubMed.
- V. V. Tuchin, I. L. Maksimova, D. A. Zimnyakov, I. L. Kon, A. H. Mavlyutov and A. A. Mishin, J. Biomed. Opt., 1997, 2, 401–417 CrossRef CAS PubMed.
- M. D. Fairchild, Color appearance models, John Wiley & Sons, 2013 Search PubMed.
Footnote |
| † Electronic supplementary information (ESI) available: Optical microscope (OM) images, transmission spectra, and CIE 1931 chromaticity diagrams of additionally fabricated SCFs with various diameter to spacing ratios. See DOI: 10.1039/c5ra25797b |
| This journal is © The Royal Society of Chemistry 2016 |





