Cellulose as an efficient matrix for lipase and transaminase immobilization†
Stefânia P. de Souzaa,
Ivaldo I. Juniorb,
Guilherme M. A. Silvaa,
Leandro S. M. Mirandaa,
Marcelo F. Santiagoc,
Frank Leung-Yuk Lamd,
Ayad Dawoode,
Uwe T. Bornscheuer*e and
Rodrigo O. M. A. de Souza*a
aBiocatalysis and Organic Synthesis Group, Chemistry Institute, Federal University of Rio de Janeiro, Rio de Janeiro, Brazil. E-mail: rodrigosouza@iq.ufrj.br
bSchool of Chemistry, University of Rio de Janeiro, Rio de Janeiro, Brazil
cInstitute of Biophysics Carlos Chagas Filho (IBCCF), Federal University of Rio de Janeiro (UFRJ), Rio de Janeiro, Brazil
dDepartment of Chemical and Biomolecular Engineering, The Hong Kong University of Science and Technology, Hong Kong SAR, China
eInstitute of Biochemistry, Dept. of Biotechnology & Enzyme Catalysis, Greifswald University, Greifswald, Germany
First published on 17th December 2015
Abstract
Immobilization of enzymes is important to improve their stability and to facilitate their recyclability, aiming to make biocatalytic processes more efficient. One of the important aspects is the utilization of cheap, abundant, and environmentally friendly carriers for enzyme immobilization. Here we report the use of functionalized cellulose for lipase and transaminase immobilization. High immobilization efficiencies (up to 90%) could be achieved for the transaminase from Vibrio fluvialis. For immobilized lipase CAL-B as well as the transaminase, good conversions and recyclability could be demonstrated in kinetic resolutions to afford chiral alcohols or amines. Moreover, such application of the immobilized transaminase enabled very high conversions in a continuous-flow process in the asymmetric synthesis of (S)-phenylethylamine (80% conversion, >99% ee).
Introduction
As already pointed out by Sheldon & van Pelt,1 enzymes are Nature's sustainable catalysts. This statement is based on the mild conditions required for performing the requested transformation in addition to biocompatibility and biodegradability characteristics.2 Producing large quantities of an efficient and stable biocatalyst is still an important challenge even with advances in the 90s in biotechnology and protein engineering which have provided researchers with better tools for the discovery and improvement of enzymes by means of genetic manipulations.3The commercial large scale application of enzymes is not hampered by a lack of immobilization protocols, since it is easy to find different methods for enzyme immobilization in the literature,4–11 but rather because most of them use expensive carrier materials with prices being prohibitive for large-scale operations. Hence, finding a cheap and readily available support for enzyme immobilizations is crucial for the development of new immobilized biocatalysts. Cellulose is the most abundant biopolymer consisting of a linear polysaccharide chain composed of D-glucose units linked by β(1 → 4) glycosidic bonds, fully decorated with hydroxyl groups. Since the discovery of cellulose in 1839 by Anselme Payen, this natural biopolymer has been used for many different applications ranging from thermoplastic polymers to cellulose nanofibers.12–15
The use of cellulose as a matrix for enzyme immobilization dates from the beginning of the seventies when Kennedy and co-workers have modified cellulose for chymotrypsin A immobilization.16–18 A few years later lipases were also immobilized on cellulose by different methods and successfully used for hydrolytic and esterification reaction.19 As a cheap and abundant biopolymer, cellulose occurs as a very interesting matrix for enzyme immobilization representing an affordable immobilized biocatalyst for large-scale operations.
In continuation of our efforts on the development of carriers and protocols for enzyme immobilization,20–26 herein we report the functionalization of cellulose followed by enzyme immobilization (exemplified for lipase and transaminase) via covalent bonds and its application in the production of chiral alcohols and amines.
Materials and methods
Materials
Cellulose, 3-aminopropyltriethoxysilane (APTES), (3-glycidyloxypropyl)trimethoxysilane (GLYMO) and glutaraldehyde were purchased from Sigma-Aldrich. Immobilized Candida antarctica lipase B (Novozyme 435, NZ435 – 30 mg g−1 of support) was purchased from Novozymes as well as free Candida antarctica lipase B on phosphate buffer (Cal-B, 5.7 mg mL−1). GDH-105 (Lot# D12010) and LDH-101 (Lot# A10036) was purchased from Codexis Inc (Redwood City, CA). All other materials were at least reagent-grade.Preparation of APTES functionalized cellulose
(3-Aminopropyl)trimethoxysilane, APTES (5 mL) and tetrahydrofuran (50 mL) were mixed and 500 mg of cellulose were added. This mixture was stirred at room temperature for 3 h. Then the solvent was evaporated and 10 mL phosphate buffer pH 7 (50 mM) was added to the product, followed by 2 mL glutaraldehyde. This mixture was stirred at room temperature for 24 h. The product was filtered and washed repeatedly with distilled water to eliminate unreacted residues.Preparation of Glymo functionalized cellulose
(3-Glycidyloxypropyl)trimethoxysilane, GLYMO (5 mL) and tetrahydrofuran (50 mL) were mixed and 500 mg cellulose were added. This mixture was stirred at room temperature for 5 h. Then the solvent was evaporated and the product was filtered and washed repeatedly with distilled water to eliminate unreacted residues.Vibrius fluvialis transaminase production
Overnight cultures of Vfl-TA-WT (in pET24b vector with kanamycin resistance) were used for inoculation of 150 mL TB media containing 50 μg mL−1 kanamycin. When OD600 of 0.6 was reached IPTG was added for induction in a final concentration of 0.2 mM. The expression occurs at 20 °C for 18–20 h. The harvesting of cells was performed at 4000 × g for 15 min and 4 °C. French Press at 1700 psi was used for cell disruption after re-suspension in 50 mM NaPP pH 7.5, 0.1 mM PLP and 300 mM NaCl. The subsequent centrifugation step was done at 37![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 × g for 1 h and 4 °C. The crude lysate was filtered with a 0.2 μm filter and stored at 4 °C.
000 × g for 1 h and 4 °C. The crude lysate was filtered with a 0.2 μm filter and stored at 4 °C.
Immobilization conditions
The support (functionalized cellulose) was subjected to the following immobilization process:Lipase immobilization protocol: 2 mL lipase B from Candida antarctica enzyme solution (5.7 mg mL−1) was dissolved in 10 mL 0.025 mM phosphate buffer pH 7.0 and added to support (2 g). The mixture was stirred for 24 h at 40 °C using a flask shaker or thermomixer, before being filtered and dried under vacuum followed by drying over night at ambient temperature. Immobilization efficiency was evaluated by the difference between initial amount of units added and that in the supernatant after filtration of the immobilized enzyme.
Transaminase immobilization protocol: 5 mL Vibrius fluvialis transaminase solution (8.8 mg mL−1) was dissolved in 30 mL 50 mM phosphate buffer pH 7.5 and added to the support (1 g). The mixture was stirred for 24 h at 30 °C using a flask shaker or thermomixer, before being filtered and dried under vacuum followed by drying over night at ambient temperature. Immobilization efficiency was evaluated by the difference between the initial amount of units added and that in the supernatant after filtration of the immobilized enzyme.
Esterification reactions
The immobilized lipase (10 mg of support in 1 mL reaction media) was evaluated in an esterification reaction between oleic acid and ethanol (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, 100 mM in n-heptane) at different temperatures. The reactions were performed in cryotubes at 200 rpm agitation on a shaker. Samples (10 μL) were collected after 1 h. For calculation of the initial reaction velocity, the reaction times were varied from 5, 10, 15, 20, and 30 min. For thermal stability, the reaction was investigated at 50–70 °C. Quantification of esters formed was performed by GC-MS analysis.
1, 100 mM in n-heptane) at different temperatures. The reactions were performed in cryotubes at 200 rpm agitation on a shaker. Samples (10 μL) were collected after 1 h. For calculation of the initial reaction velocity, the reaction times were varied from 5, 10, 15, 20, and 30 min. For thermal stability, the reaction was investigated at 50–70 °C. Quantification of esters formed was performed by GC-MS analysis.
Kinetic resolution using immobilized lipase
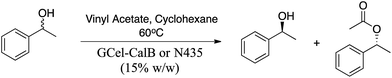
Rac-1-Phenylethanol (1 mmol, 122 mg), vinyl acetate (1 mol per eq.) as acyl donor, and 18 mg (15% w/w) of the corresponding immobilized enzyme were reacted in cyclohexane (3 mL) for 2, 4 and 5 h at 60 °C. Enantiomeric excess values (ee) were determined by chiral GC analysis (chiral column Betadex-325, for details as shown in the section of “GC analysis”).Discontinuous acetophenone assay
For immobilized transaminases a 24 deep-well plate was heated in a thermomixer to 30 °C. A total of 50 mg wet immobilized transaminase was used for each measurement. The reaction was started by adding 5 mL of pre-warmed reaction solution (HEPES buffer 50 mM, pH 7.5, 2.5 mM (±)-α-phenylethylamine ((±)-α-PEA), 5 mM pyruvate, and 0.5% DMSO). Then, 200 μL samples were taken and the absorbance was measured at 245 nm. The slope was determined and conversions calculated.27Asymmetric synthesis using immobilized transaminase
Asymmetric synthesis of (S)-phenylethylamine was performed using the GDH/LDH system for shifting the equilibrium and cofactor recycling.28,29 Reactions were performed in a 5 mL mixture using 50 mM HEPES buffer (pH 7.5), 10 mM acetophenone, 250 mM alanine, 10% DMSO, LDH-GDH (88 U mL−1 and 15 U mL−1, respectively), 1 mM NADH and 150 mM glucose, at 30 °C. For batch experiments, 100 mg immobilized enzyme was used in a total reaction volume of 5 mL on a thermomixer at 800 rpm. The starting mixture was stirred for 5 min while the instrument Asia Flow Reactor was equipped with Omnifit (6.6 mm × 10 mm with 0.3421 cm2 base and height 10 cm column) containing 2 g of the immobilized catalysts. The reaction solution was pumped using an Asia syringe pump at different flow rates in order to achieve the desired residence time. Samples were collected and analysed by HPLC.32GC analysis
All GC-MS measurements were carried out in duplicate using a DB 5 (Agilent, J&W Scientific®, USA) capillary column (30 m × 0.25 mm × 0.25 μm). The GC-MS samples were prepared by dissolving 10 μL of the sample in 98 mL of heptane and 10 μL of MSTFA (N-methyl-N-(trimethylsilyl) trifluoroacetamide). 1 μL of this sample was then injected into the Shimadzu CG2010 GC. The injector and detector temperatures were 250 °C and the oven temperature was constant at 60 °C for 1 min, and then increased by 10 °C min−1 to 250 °C, where it was held constant for 3 min. The conversion and the selectivity were analyzed from the peak areas in the chromatograms. GC-FID: chiral column Betadex-325 capillary column; 1 μL samples were injected at 100 °C. The oven was heated at 15 °C min−1 to 150 °C, at 8 °C min−1 to 200 °C, at 2 °C min−1 to 240 °C and then maintained for 4 min. After this, the oven was heated at 15 °C min−1 to 300 °C. Chiral GC analysis was performed on a Shimadzu GC-2010 chromatograph equipped with a FID, an AOC-20i autosampler and a chiral CP-Chirasil-Dex CB (25 m × 0.25 mm ID) or a chiral β-Dex325 (30 m × 0.24 mm ID) column using hydrogen as carrier gas. Injector and detector temperatures were set at 220 °C. GC-FID temperature program for 1-phenylethylamine (PEA) using β-Dex325 column: 90 °C|30 min → 180 °C, 40 °C min−1|10 min.HPLC analysis
For quantitative analysis 150 μL samples of the reaction mixture were derivatized with 15 μL trifluoroacetic acid (TFA), centrifuged (13![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 g, 5 min) and 15 μL of the supernatant were directly analyzed by HPLC (Hitachi LaChrom) using a reversed-phase C18 column (LiChrospher®). Detection took place with a UV detector set at 245 nm.
000 g, 5 min) and 15 μL of the supernatant were directly analyzed by HPLC (Hitachi LaChrom) using a reversed-phase C18 column (LiChrospher®). Detection took place with a UV detector set at 245 nm.
Thermogravimetric (TG) analysis
The TG curves were obtained in a thermogravimetric module, coupled to a thermal analyzer, both manufactured by Netzsch®. Thermogravimetric measurements were performed using a platinum sample holder containing about 10 mg of each immobilized enzyme. Each sample was heated from 35 to 600 °C at 10 °C min−1, under atmosphere of synthetic air and N2, both at a flow rate of 60 mL min−1.X-Ray photoelectron spectroscopy (XPS)
The XPS was conducted using a Kratos Axis Ultra DLD spectrometer. A monochromatic Al source was used as X-ray source. The soft X-rays emitted from excitation of Al source generate photoelectrons from the surface layers of atoms in a solid sample. The electrons thus emitted are analyzed according to their kinetic energy and the spectrum produced is used to identify the elements present and their chemical states.Infrared analysis
Analysis by infrared spectroscopy used a Shimadzu 8300 FTIR spectrophotometer. The spectrum was obtained with 32 scans and with 4 cm−1 resolution. For the analysis, 10 mg of a sample was placed in the sample collector to form tablets of approximately 2 mm thickness and 5 mm diameter without KBr addition.Scanning electron microscopy (SEM) analysis
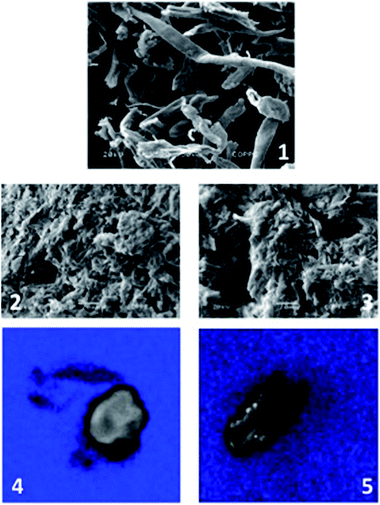
Cellulose, functionalized cellulose and supported enzyme had their structure analyzed by scanning electron microscopy (SEM) using a Zeiss EVO® 50H microscope. All micrographs were obtained using a Shimadzu® sputter equipment. For this, each sample was placed in a sample holder on carbon tape and then they were metallized under vacuum. After preparation, the samples were placed under the microscope and bombarded by an electron beam interacting with the sample's atoms. From the interaction between the electron beam and the sample, the radiation was possible to form a magnified image of the sample.Laser confocal microscopy analysis (LCMS)
Prior to being analyzed, the immobilized enzyme samples were mixed with fluorescamine solution (50 mg mL−1, in acetone) for 3 min to form a highly fluorescent product through the reaction between the primary amines in proteins and the fluorescamine. Fluorescence intensity quantification of both biocatalyst containing fluorescent lipase (GCel-CalB) and transaminase (ECel-VF) were analyzed by confocal microscopy (Zeiss, LSM 510 META). For this purpose, slides were mounted with VectaShield (Vector Laboratories) and covered with a glass lamina. Stack images were analyzed using Image J software (NIH).Results and discussion
First, cellulose was modified in order to introduce the epoxy (GLYMO) and amino (APTES) groups for covalent bond immobilization protocols. Cellulose was successively mixed with APTES and glutaraldehyde in order to deliver the glutaraldehyde-functionalized cellulose (GCel), after 24 h. The epoxy-functionalized cellulose (ECel), was also produced by the reaction between GLYMO and cellulose for 5 h, using THF as solvent (Scheme 1).After drying the functionalized cellulose (GCel and ECel) at ambient temperature for 48 h, the enzyme immobilization was performed. For the lipase immobilization, 2 mL lipase B from Candida antarctica solution was dissolved in 10 mL 0.025 mM phosphate buffer pH 7.0. The functionalized cellulose (GCel and ECel) was added (2 g) and allowed to react for 24 hours at 40 °C. Immobilization efficiency was evaluated by the difference between initial amount of enzyme added and that in the supernatant after filtration of the immobilized enzyme (Table 1). For the transaminase immobilization procedure, 5 mL Vibrius fluviaris transaminase solution was dissolved in 30 mL 50 mM HEPES buffer pH 7.5. The functionalized cellulose (GCel and ECel) was added (1 g) and underwent the reaction for 24 hours at 30 °C. No optimization was done in order to first verify the feasibility of using the functionalized cellulose as a matrix for enzyme immobilization. All supports were characterized by infrared (IR), thermogravimetry analysis (TG), scanning electron microscopy (SEM) and X-ray Photoelectron Spectroscopy (XPS) (see ESI† for further details).
| Immob. biocatalyst | Immob. efficiencya (%) | Amount of protein (mg g−1 of support) |
|---|---|---|
| a Immobilization efficiency was evaluated by the difference between initial amount of units added and that in the supernatant after filtration of the immobilized enzyme. | ||
| ECel-CalB | 3.5 | 0.2 |
| GCel-CalB | 10.5 | 0.6 |
| ECel-VF | 90.1 | 40.5 |
| GCel-VF | 27.7 | 12.4 |
The immobilization efficiency obtained for Cal-B on epoxy-cellulose (ECel-CalB), Cal-B on glutaraldehyde–cellulose (GCel-CalB), VF transaminase on epoxy–cellulose (ECel-VF) and VF transaminase on glutaraldehyde–cellulose (GCel-VF) are shown on Table 1.
As shown on Table 1, the immobilization protocol developed did not present efficient results for the immobilization of lipase B from Candida antarctica, leading to very low protein incorporation. The VF transaminase leads to good to excellent immobilization efficiencies on both epoxy (up to 90.1%, Table 1) and glutaraldehyde functionalized cellulose. In order to prove the concept of using functionalized cellulose as a renewable support for enzyme immobilization, the two supports were attempted where the immobilization efficiency was higher, C–B on glutaraldehyde–cellulose (GCel-CalB) and VF transaminase on epoxy–cellulose (ECel-VF).
The solid state 13C NMR spectroscopy of the functionalized cellulose clearly demonstrates the incorporation of the APTES and glutaraldehyde in the cellulosic material. The presence of the imino as well as the silyl propyl groups can be inferred by the presence of the signals in 163–164 ppm and the signals at higher field 10–50 ppm, respectively. When the same analysis was performed after the incorporation of the enzyme into the cellulosic support, an increase in the number of signals in the range of 164–180 ppm can be observed, which correlates to the presence of imino and amide functions (see ESI† for further details). This result corroborates the XPS analysis where a peak at the binding energy of 400 eV is observed for the glutaraldehyde-functionalized cellulose (GCel) being an indicator of nitrogen atoms in the sample. For the sample GCel-CalB there is a shoulder peak shown at the binding energy of 402 eV. This shoulder peak is the main difference compared to the GCel sample, meaning that the nitrogen atom is in a higher oxidation state, which can be related to the protein binding to the support (see ESI† for further details). In order to verify the presence of protein on the functionalized cellulose, we decided to perform a confocal laser scanning microscopy (CLSM) experiment to visualize the dispersion of protein into the support (Fig. 1).29,30 As displayed in Fig. 1a (ECel-VF) the white color reveals the presence of well dispersed protein on the surface of cellulose granules. In addition, the CLSM analysis of GCel-VF (Fig. 1b) shows a punctual localization of protein, which reflects the low immobilization efficiency compared to the epoxy-functionalized pattern (Table 1).
With the characterized results, catalytic performances of the immobilized enzymes at different reaction temperatures were evaluated in the designed reactions. The immobilized lipase GCel-CalB was evaluated by the esterification reaction between oleic acid and ethanol (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, 100 mM in heptane and 200 rpm), at different temperatures (50–70 °C) for 1 h. The behavior presented by GCel-CalB was compared to the commercial immobilized lipase Novozyme 435 and the results are presented, as shown in Table 2. A similar evaluation was made for ECel-VF immobilized enzyme, in this case the kinetic resolution of (+/-)-phenylethylamine (PEA) was used as standard reaction at temperatures ranging from 30 to 60 °C. Unfortunately, there is no commercial transaminase that can be used as a positive control for comparison purpose.
1, 100 mM in heptane and 200 rpm), at different temperatures (50–70 °C) for 1 h. The behavior presented by GCel-CalB was compared to the commercial immobilized lipase Novozyme 435 and the results are presented, as shown in Table 2. A similar evaluation was made for ECel-VF immobilized enzyme, in this case the kinetic resolution of (+/-)-phenylethylamine (PEA) was used as standard reaction at temperatures ranging from 30 to 60 °C. Unfortunately, there is no commercial transaminase that can be used as a positive control for comparison purpose.
| Entry | Immob. enzyme | Temperature (°C) | Conversion (%) |
|---|---|---|---|
a Reaction conditions: GCel-CalB or Novozyme 435 (10 mg immobilized enzyme in 1 mL reaction media) was evaluated in the esterification reaction between oleic acid and ethanol (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 – 100 mM in n-heptane) at different temperatures. 1 – 100 mM in n-heptane) at different temperatures. |
|||
| 1 | GCel-CalB | 50 | 85 |
| 2 | 60 | 97 | |
| 3 | 70 | 84 | |
| 4 | Novozyme 435 | 50 | 84 |
| 5 | 60 | 80 | |
| 6 | 70 | 81 | |
A first look at the results presented on Table 2, can take the reader to the wrong assumption that GCel-CalB and Novozyme 435 have the same behavior at different temperatures, but it is worthy to note that Novozyme 435 has 50× more protein attached to the support compared to GCel-CalB, making this one much more efficient than the commercial immobilized enzyme. GCel-CalB was also evaluated at higher temperatures (80 °C) and the results obtained show a similar behavior, leading to the desired product on 75% of conversion (see ESI† for further details). The results for temperature profile of ECel-VF (Table 3) show good results at temperatures ranging from 30 to 50 °C and a slight decrease is observed at 60 °C. Especially for the transaminase, the pH profile was also evaluated in the range between 7.5 and 9.5, where the case at pH 7.5 offered the best result.
| Entry | Immob. enzyme | Temp. (°C) | Conv. (%) |
|---|---|---|---|
| a Reaction conditions: ECel-VF (50 mg immobilized enzyme in 5 mL reaction media) was evaluated using the discontinuous acetophenone assay (5 mL 50 mM HEPES buffer, (±)-PEA 2.5 mM, 5 mM pyruvate, 0.5% DMSO and 800 rpm) at different temperatures. | |||
| 1 | ECel-VF | 30 | 38 |
| 2 | 40 | 42 | |
| 3 | 50 | 43 | |
| 4 | 60 | 35 | |
At this stage we also evaluated the recyclability of the immobilized biocatalysts GCel-CalB and ECel-VF. After each reaction the immobilized biocatalyst was washed three times with buffer and dried at ambient temperature for 24 h and this was repeated five times (Table 4).
| Entry | Immob. enzyme | Conversion (%) | ||||
|---|---|---|---|---|---|---|
| Cycle 1 | Cycle 2 | Cycle 3 | Cycle 4 | Cycle 5 | ||
a Reaction conditions: GCel-CalB or Novozyme 435 (10 mg immobilized enzyme in 1 mL reaction media) was evaluated in the esterification reaction between oleic acid and ethanol (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 – 100 mM in n-heptane) at 50 °C.b Reaction conditions: ECel-VF (50 mg immobilized enzyme in 5 mL reaction media) was evaluated using the discontinuous acetophenone assay (5 mL 50 mM HEPES buffer, (±)-PEA 2.5 mM, 5 mM pyruvate, 0.5% DMSO and 800 rpm) at 30 °C. 1 – 100 mM in n-heptane) at 50 °C.b Reaction conditions: ECel-VF (50 mg immobilized enzyme in 5 mL reaction media) was evaluated using the discontinuous acetophenone assay (5 mL 50 mM HEPES buffer, (±)-PEA 2.5 mM, 5 mM pyruvate, 0.5% DMSO and 800 rpm) at 30 °C. |
||||||
| 1 | Novozyme 435a | 84 | 71 | 56 | 26 | 20 |
| 2 | GCel-CalBa | 85 | 60 | 54 | 51 | 43 |
| 3 | ECel-VFb | 43 | 39 | 39 | 40 | 35 |
The findings of the recyclability study show that using the GCel-CalB immobilized lipase a decrease of the conversion was observed in the second cycle compared to the commercial enzyme while Novozyme 435 continues to lose its activity for four consecutive cycles to only 20% conversion. The immobilized transaminase ECel-VF presented a better stability affording the same conversion in fours cycles and only a slight decrease in the last one.
Finally, the newly developed immobilized biocatalysts GCel-CalB and ECel-VF were used for the synthesis of chiral products. The immobilized biocatalysts GCel-CalB was applied in the kinetic resolution of (R,S)-α-phenylethanol using vinyl acetate as acyl donor at 60 °C for 5 h (Table 5).
| Entry | Immob. enzyme | Reaction time | Conv.(%) | E | Productivity (g of product per h per mg protein) |
|---|---|---|---|---|---|
| a Reaction conditions: (R,S)-α-phenylethanol (1 mmol), vinyl acetate (1 mol per eq.) as acyl donor, and 18 mg (15% w/w) immobilized enzyme were reacted in cyclohexane (3 mL) for 2, 4, and 5 h at 60 °C. Enantiomeric excess values (ee) were determined by chiral GC analysis. | |||||
| 1 | Novozyme 435 | 2 hours | 49 | >200 | 0.053 |
| 2 | GCel-CalB | 2 hours | 35 | >200 | 1.80 |
| 3 | 4 hours | 40 | >200 | 1.08 | |
| 4 | 5 hours | 42 | >200 | 0.91 | |
Comparatively, GCel-CalB immobilized enzyme could not mimic the efficiency shown by Novozyme 435, arriving to 49% of conversion after 2 h. However a promising result could be obtained by the use of GCel-CalB with conversions reaching 40% after 4 h. The most important feature of this immobilized biocatalyst is the fact that even a low protein loading can lead to very high productivities (up to 1.8 g of product per h per mg immob. enzyme), which could never be reached by the commercial immobilized enzyme.
The immobilized transaminase ECel-VF was subject to an asymmetric synthesis protocol using acetophenone as starting material. At this stage – since the development of immobilized transaminases is an emerging area – the performance of ECel-VF was evaluated in both, batch and continuous-flow reactors (Table 6).
| Entry | Reaction/residence time | Conv. (%) | ee (%) |
|---|---|---|---|
| a Reaction conditions: 50 mM HEPES buffer (pH 7.5), 10 mM acetophenone, 250 mM alanine, 10% DMSO, LDH-GDH, 1 mM NADH and 150 mM glucose. For the batch experiments, 100 mg immobilized enzyme was used in a total reaction volume of 5 mL on a shaker at 800 rpm at 30 °C. For continuous-flow experiments, 2 g immobilized enzyme were packed arriving on a packed bed with a total volume of 10 mL. | |||
| 1 | 48 h (batch) | 31 | >99 |
| 2 | 96 h (batch) | 33 | >99 |
| 3 | 15 min (cont. flow) | 14 | >99 |
| 4 | 30 min (cont. flow) | 37 | >99 |
| 5 | 60 min (cont. flow) | 72 | >99 |
| 6 | 90 min (cont. flow) | 80 | >99 |
As observed in Table 6, the asymmetric synthesis protocol under continuous-flow conditions is better than in batch mode since lower reaction/residence times are needed and higher conversions could be obtained without compromising the enantiomeric excess. Probably, the continuous-flow system enables to enhance mass transfer and consequently leads to better conversions. The packed bed, after washing for 3 h with HEPES buffer, was stored and re-used with the same behaviour presented before.
Conclusion
In conclusion, we have developed one lipase (GCel-CalB) and one transaminase (ECel-VF) immobilized onto functionalized cellulose for different applications. Effects of temperature, pH and recyclability profiles were evaluated and satisfactory results were presented. The GCel-CalB immobilized enzyme has shown very good results in the kinetic resolution of (R,S)-α-phenylethanol enabling high productivities. ECel-VF immobilized enzyme demonstrates its excellent catalytic activity in the kinetic resolution as well as in asymmetric synthesis where batch and continuous flow protocols have been applied, reducing the reaction time from 48 h to 90 min and increasing the conversion from 30% to 80%.Acknowledgements
The authors thank CAPES, CNPq, FAPERJ and DLR (Grant No: 01DN14016) and the Alexander von Humboldt Foundation (Germany) for financial support.References
- R. A. Sheldon and S. van Pelt, Chem. Soc. Rev., 2013, 42, 6223–6235 RSC.
- U. T. Bornscheuer, Angew. Chem., Int. Ed., 2003, 42, 3336–3337 CrossRef CAS PubMed.
- U. T. Bornscheuer, G. W. Huisman, R. J. Kazlauskas, S. Lutz, J. C. Moore and K. Robins, Nature, 2012, 485, 185–194 CrossRef CAS PubMed.
- L. Bayne, R. V. Ulijn and P. J. Halling, Chem. Soc. Rev., 2013, 42, 9000–9010 RSC.
- S. Cantone, V. Ferrario, L. Corici, C. Ebert, D. Fattor, P. Spizzo and L. Gardossi, Chem. Soc. Rev., 2013, 42, 6262–6276 RSC.
- U. Hanefeld, L. Cao and E. Magner, Chem. Soc. Rev., 2013, 42, 6211–6212 RSC.
- M. Hartmann and X. Kostrov, Chem. Soc. Rev., 2013, 42, 6277–6289 RSC.
- A. Liese and L. Hilterhaus, Chem. Soc. Rev., 2013, 42, 6236–6249 RSC.
- E. Magner, Chem. Soc. Rev., 2013, 42, 6213–6222 RSC.
- R. C. Rodrigues, C. Ortiz, A. Berenguer-Murcia, R. Torres and R. Fernandez-Lafuente, Chem. Soc. Rev., 2013, 42, 6290–6307 RSC.
- F. Secundo, Chem. Soc. Rev., 2013, 42, 6250–6261 RSC.
- S. Alila, A. M. Ferraria, A. M. Botelho do Rego and S. Boufi, Carbohydr. Polym., 2009, 77, 553–562 CrossRef CAS.
- S. J. Eichhorn, A. Dufresne, M. Aranguren, N. E. Marcovich, J. R. Capadona, S. J. Rowan, C. Weder, W. Thielemans, M. Roman, S. Renneckar, W. Gindl, S. Veigel, J. Keckes, H. Yano, K. Abe, M. Nogi, A. N. Nakagaito, A. Mangalam, J. Simonsen, A. S. Benight, A. Bismarck, L. A. Berglund and T. Peijs, J. Mater. Sci., 2010, 45, 1–33 CrossRef CAS.
- Y. Habibi, L. A. Lucia and O. J. Rojas, Chem. Rev., 2010, 110, 3479–3500 CrossRef CAS PubMed.
- N. Lavoine, I. Desloges, A. Dufresne and J. Bras, Carbohydr. Polym., 2012, 90, 735–764 CrossRef CAS PubMed.
- A. S. Karsakevich, R. A. Zhagat and V. Y. Son, Khim. Prir. Soedin., 1976, 639–643 CAS.
- J. F. Kennedy and A. Rosevear, J. Chem. Soc., Perkin Trans. 1, 1974, 757–762 RSC.
- J. Kucera, Collect. Czech. Chem. Commun., 1979, 44, 804–807 CrossRef CAS.
- M. Rucka, B. Turkiewicz and J. S. Zuk, J. Am. Oil Chem. Soc., 1990, 67, 887–889 CrossRef CAS.
- A. S. de Miranda, L. S. M. Miranda and R. O. M. A. de Souza, Biotechnol. Adv., 2015, 33, 372–393 CrossRef CAS PubMed.
- A. S. de Miranda, R. C. Simon, B. Grischek, G. C. de Paula, B. A. C. Horta, L. S. M. de Miranda, W. Kroutil, C. O. Kappe and R. O. M. A. de Souza, ChemCatChem, 2015, 7, 984–992 CrossRef CAS.
- C. Garcia, I. I. Junior, R. O. M. A. de Souza and R. Luque, Catal. Sci. Technol., 2015, 5, 1840–1846 CAS.
- R. O. Lopes, S. Grimm, J. B. Ribeiro, I. C. R. Leal, L. S. M. Miranda and R. O. M. A. de Souza, J. Braz. Chem. Soc., 2015, 26, 550–554 CAS.
- S. P. de Souza, J. Bassut, H. V. Marquez, I. I. Junior, L. S. M. Miranda, Y. Huang, G. Mackenzie, A. N. Boa and R. O. M. A. de Souza, Catal. Sci. Technol., 2015, 5, 3130–3136 CAS.
- I. Itabaiana Jr, K. M. Goncalves, M. Zoumpanioti, I. C. R. Leal, L. S. M. E. Miranda, A. Xenakis and R. O. M. A. de Souza, Org. Process Res. Dev., 2014, 18, 1372–1376 CrossRef.
- F. K. Sutili, D. d. O. Nogueira, S. G. F. Leite, L. S. M. Miranda and R. O. M. A. de Souza, RSC Adv., 2015, 5, 37287–37291 RSC.
- H. Mallin, U. Menyes, T. Vorhaben, M. Höhne and U. T. Bornscheuer, ChemCatChem, 2013, 5, 588–593 CrossRef CAS.
- N. J. Turner, M. D. Truppo, D. Rozzell and J. C. Moore, Org. Biomol. Chem., 2009, 7, 395–398 Search PubMed.
- W. Kroutil, D. Rozzell, D. Clay, I. Lavandera and D. Koszelewski, Adv. Synth. Catal., 2008, 350, 2761–2766 CrossRef.
- D. J. Cui, L. L. Li and M. Y. Zhao, Ind. Eng. Chem. Res., 2014, 53, 16176–16182 CrossRef.
- Y. Li, G. Fei, W. Wei, B. Jian, H. M. Guang and Q. Z. Wei, J. Mol. Catal. B: Enzym., 2010, 66, 182 CrossRef CAS.
- S. Schätzle, M. Höhne, E. Redestad, K. Robins and U. T. Bornscheuer, Anal. Chem., 2009, 81, 8244–8248 CrossRef PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c5ra24976g |
| This journal is © The Royal Society of Chemistry 2016 |