Reconstruction of a helical trimer by the second transmembrane domain of human copper transporter 2 in micelles and the binding of the trimer to silver
Zhe Donga,
Liping Guana,
Chunyu Wanga,
Haoran Xub,
Zhengqiang Lib and
Fei Li*a
aState Key Laboratory of Supramolecular Structure and Materials, Jilin University, 2699 Qianjin Avenue, Changchun 130012, P. R. China. E-mail: feili@jlu.edu.cn; Fax: +86-431-85193421; Tel: +86-431-85168548
bKey Laboratory for Molecular Enzymology & Engineering, The Ministry of Education, Jilin University, 2699 Qianjin Avenue, Changchun 130012, P. R. China
First published on 28th December 2015
Abstract
Copper transporter 2 (Ctr2) is identified as a homologue of copper transporter 1 (Ctr1) and believed to transport copper and silver ions by a similar mechanism to Ctr1. Of three pore-forming transmembrane domains, the second transmembrane domain (TMD2) is highly conserved by the two proteins and the MXXXM motif in TMD2 is significant for the transport function of the proteins. However, whereas Ctr1 is a high-affinity transporter, Ctr2 is a low-affinity transporter. Herein, we studied the structure and assembly of the peptide corresponding to TMD2 of human Ctr2 (hCtr2) in sodium dodecyl sulfate (SDS) micelles and the binding of the micelle-bound peptide to silver ion using nuclear magnetic resonance, circular dichroism, isothermal titration calorimetry and electrophoresis. The peptide hCtr2-TMD2 formed an α-helix in SDS micelles, and could assemble mainly as a dimer at a lower molar ratio of peptide![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) SDS or a trimer at a higher peptide
SDS or a trimer at a higher peptide![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) SDS ratio. The peptide trimer could bind two silver ions by the coordination of the C-terminal part and two methionine residues in the MXXXM motif played an important role in the binding. Our results showed that the hCtr2-TMD2 trimer has weaker intermolecular interactions and a lower binding affinity for silver ion compared with the results of our previous study on hCtr1-TMD2. This finding suggests that the intermolecular interactions between the second transmembrane domains may play a significant role in the pore formation of the two transporters and the binding affinity for Ag(I).
SDS ratio. The peptide trimer could bind two silver ions by the coordination of the C-terminal part and two methionine residues in the MXXXM motif played an important role in the binding. Our results showed that the hCtr2-TMD2 trimer has weaker intermolecular interactions and a lower binding affinity for silver ion compared with the results of our previous study on hCtr1-TMD2. This finding suggests that the intermolecular interactions between the second transmembrane domains may play a significant role in the pore formation of the two transporters and the binding affinity for Ag(I).
Introduction
Copper transporter 1 and 2 (Ctr1 and Ctr2) belong to a Ctr family in mammalian cells. Ctr1 is a high affinity transporter that plays a key role in mediating copper entry from blood to cells across the plasma membrane.1–3 Ctr2 is a low affinity transporter with a majority located in late endosomes and lysosomes where it releases copper from the intracellular compartments to the cytosol.4 Ctr2 is also distributed at the plasma membrane with a small proportion and transports extracellular copper into cells.5Human Ctr1 (hCtr1) consists of 190 amino acids that are separated into an extracellular N-terminal domain, three transmembrane domains (TMD1–TMD3), a large intracellular loop between TMD1 and TMD2, and a cytosolic C-terminal tail. In the past years, Ctr1 has been widely studied and the structure and function of the protein have been well documented.3,6–12 The structure of Ctr1 has been characterized as a homotrimer that forms a pseudo-cone-shaped pore by an assembly of the transmembrane domains. Of the three transmembrane domains, TMD2 lines the pore and plays a crucial role in the function of Ctr1. A mutation of the methionine residues in the MXXXM motif of TMD2 to Leu is found to decrease the uptake of copper by Ctr1 dramatically and a histidine to arginine mutation at position 139 in TMD2 of hCtr1 is shown to accelerate the transport of copper by the protein.2 We previously detected the formation of a trimer by the isolated hCtr1-TMD2 segment in SDS micelles and the binding of the TMD2 trimer to Ag(I) by a chemical stoichiometry of 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3 of Ag(I):peptide, suggesting that the intrinsic interaction between the TMD2 helices may be closely associated with the formation of hCtr1 pore in cellular membranes.13 In addition, Ctr1 has multiple methionine and histidine residues in the N-terminal domain and a HCH motif in the C-terminal tail, which has also been proved to be important for the function of the protein.14–16 Although the actual mechanism underlying the function of Ctr1 has not been fully understood, a generally recognized model of mechanism has been proposed based on these studies. In this model, the N-terminal domain containing multiple copper binding sites (methionine and histidine residues) is responsible for the local concentration of copper at the border of plasma membrane, the MXXXM motif in the extracellular boundary of TMD2 forms rings at the entry of the pore and mediates the translocation of Cu(I) from the extracellular portion down to the intracellular portion, where the ion is taken over by the HCH motif in the intracellular C-terminal tail before it is recruited by the chaperones or other chelators.
3 of Ag(I):peptide, suggesting that the intrinsic interaction between the TMD2 helices may be closely associated with the formation of hCtr1 pore in cellular membranes.13 In addition, Ctr1 has multiple methionine and histidine residues in the N-terminal domain and a HCH motif in the C-terminal tail, which has also been proved to be important for the function of the protein.14–16 Although the actual mechanism underlying the function of Ctr1 has not been fully understood, a generally recognized model of mechanism has been proposed based on these studies. In this model, the N-terminal domain containing multiple copper binding sites (methionine and histidine residues) is responsible for the local concentration of copper at the border of plasma membrane, the MXXXM motif in the extracellular boundary of TMD2 forms rings at the entry of the pore and mediates the translocation of Cu(I) from the extracellular portion down to the intracellular portion, where the ion is taken over by the HCH motif in the intracellular C-terminal tail before it is recruited by the chaperones or other chelators.
In contrast, the structural and functional details that exist for Ctr1 are currently lacking for Ctr2 due to less investigations on Ctr2 to date. Therefore, the precise role of Ctr2 in copper transport in mammalian cells remains uncertain. Much of structural and functional information of Ctr2 is deduced based on the study results of Ctr1. Although hCtr2 shares only 41% amino acid homology with hCtr1 and lacks the N- and C-terminal domain characteristics of hCtr1, the residues in the transmembrane domains that are known to be essential for the transport function of Ctr1 are conserved in Ctr2.17 In particular, a MXXXM motif within TMD2 of Ctr1 that is known to be crucial for the transport function of Ctr1 is also conserved in TMD2 of Ctr2 and is confirmed to play a significant role in the function of Ctr2.4,17 Human Ctr2 is believed to have a topological and structural homology to hCtr1 and likely forms a trimer with TMD2 lining the channel-like pore. However, no experimental evidences are available to support the assumptions of the structure and assembly of Ctr2 to date.
It is well-established that Ctr1 and Ctr2 are also involved in the trafficking of Ag(I), an isoelectric ion to Cu(I).1,5,18 Several studies have provided evidences that the metabolism of silver is undertaken by proteins maintaining the copper homeostasis,19–21 including Ctr class of copper transporters (Ctr1 and Ctr2), copper chaperones and the copper efflux transporters. Because of wide applications of silver and its products in hygiene or as antimicrobial and anticancer agents,22–26 a full understanding of how the metal is absorbed into and accumulated in the mammalian cells and tissues is significant.
In light of the function of Ctr2 in transporting Cu(I) and Ag(I) and the potentially significant role of TMD2 in the architecture of Ctr2 pore, we examined in the current study the assembly structure of TMD2 of hCtr2 in SDS micelles and the binding to silver ion using circular dichroism (CD), SDS-PAGE, solution NMR spectroscopy and isothermal titration calorimetry (ITC) techniques. We observed the formation of a trimer with a head-to-head parallel pattern by hCtr2-TMD2 segment itself and a preferential binding of the peptide trimer to Ag(I) by the MXXXM motif with a binding affinity smaller than that of hCtr1-TMD2 trimer. This study provides an insight into the structure of TMD2 and its possible role in cellular uptake of Cu(I)/Ag(I) by Ctr2.
Experimental section
Materials
A 29-residue peptide with a sequence KHFGQSLIHVIQVVIGYFIMLAVMSYNKK was synthesized and purified by Apeptide Co., Ltd (Shanghai, China). The purity of the peptide greater than 95% was confirmed by mass spectroscopy and high-performance liquid chromatography. The peptide sequence H2-N27 corresponds to hCtr2 93-118 (the second transmembrane domain of the protein selected based on the BLAST analysis of hCtr1 and hCtr2).11 Three lysine (K) residues, one in the N-terminus and two in the C-terminus, were added to improve purification of the peptide. The detergent sodium dodecyl sulfate (SDS) and the organic solvent 1,1,1,3,3,3-hexafluoro-2-isopropanol (HFIP) were obtained from Sigma-Aldrich (St. Louis, MO) and Acros Organics (Morris Plains, NJ), respectively. SDS-d25 (98%) and D2O (99.8%) were purchased from Cambridge Isotope Laboratories, Inc. (Cambridge, MA). All reagents and solvents were utilized without further purification.Sample preparation
Samples of the peptide incorporated with SDS micelles were prepared using the method reported previously.13 Briefly, the peptide was dissolved in 200 μL HFIP solution and vortexed for 3–5 s, and then sonicated using a bath-type sonicator for 30 min. SDS (or SDS-d25 for the NMR experiments) was solubilized in 1 mL deionized water separately. The HFIP solution of the peptide was added into SDS (or SDS-d25) aqueous solution. The mixture was vortexed for 3–5 s and sonicated for 30 min. The resulting solution was further diluted with 1.6 mL deionized water and lyophilized overnight. The resulting powder was dissolved in deionized water or in H2O/D2O (9/1 v/v) to a desired volume in terms of the experiments performed. According to this method, we prepared following samples: 1 mM peptide in 120 mM and 20 mM SDS-d25 micelles (the molar ratio of peptide![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) SDS was 1
SDS was 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 120 and 1
120 and 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 20, respectively) for the NMR experiments, 50 μM and 500 μM peptide in 10 mM SDS micelles (the molar ratio of peptide
20, respectively) for the NMR experiments, 50 μM and 500 μM peptide in 10 mM SDS micelles (the molar ratio of peptide![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) SDS was 1
SDS was 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 200 and 1
200 and 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 20, respectively) for the ITC experiments, 50 μM peptide in 10 mM SDS micelles (the peptide
20, respectively) for the ITC experiments, 50 μM peptide in 10 mM SDS micelles (the peptide![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) SDS was 1
SDS was 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 200) for the SDS-PAGE experiment, and 20 μM peptide in 10 mM SDS micelles (the peptide
200) for the SDS-PAGE experiment, and 20 μM peptide in 10 mM SDS micelles (the peptide![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) SDS was 1
SDS was 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 500) for the CD experiments. The pH values of all sample solutions were adjusted to 6. In the titration experiments, the AgNO3 stock solution (200 mM) was prepared with deionized water and added in the peptide/detergent solutions at the molar ratios of Ag(I):peptide ranging from 0 to 4. The pH changes in the sample solutions induced by AgNO3 were no more than 0.2. The effects of such small pH variations on the results of the experiments were insignificant.
500) for the CD experiments. The pH values of all sample solutions were adjusted to 6. In the titration experiments, the AgNO3 stock solution (200 mM) was prepared with deionized water and added in the peptide/detergent solutions at the molar ratios of Ag(I):peptide ranging from 0 to 4. The pH changes in the sample solutions induced by AgNO3 were no more than 0.2. The effects of such small pH variations on the results of the experiments were insignificant.
Far-UV circular dichroism experiments
Far-UV CD spectra were recorded on a PMS-450 spectropolarimeter (Biologic, France) using 0.05 cm path-length quartz cell under a constant flow of N2 at room temperature. The spectra were scanned in a range of 190–260 nm at a scan speed of 0.1 nm s−1. The absorbance at a given wavelength was expressed as molar ellipticity (θ) in a unit of deg cm2 dmol−1. The CD signals of three scans were averaged and the reference spectrum was subtracted.SDS-PAGE experiments
Gel electrophoresis was carried out in a tricine buffer system with 0.3% SDS according to the method previously reported.27 The sample of 50 μM peptide incorporated into 10 mM SDS micelles in aqueous solution was prepared first. The sample was diluted with 4× SDS loading buffer and loaded to a 16.5%-polyacrylamide/tricine/SDS gel. The ultra low molecular weight marker in the range from 3.3 to 20.1 kDa was used. The experiment was run at 4 °C. A voltage of 30 V was used initially and the voltage was changed to 120 V once the sample entered the spacer gel. Immediately after the run, the gel was placed in a solution containing 0.5% gluteraldehyde and 30% ethanol for 30 min and stained with Coomassie Brilliant Blue R-250.NMR experiments and structural calculation
A standard set of two-dimensional NMR spectra including TOCSY (mixing time 100 ms) and NOESY (mixing time 100 ms) were recorded at 25 °C using a Bruker Avance 600 MHz spectrometer equipped with a cryoprobe. The WATERGATE technique was used to suppress the water signal. DSS (2,2-dimethyl-2-silapentane-5-sulfonate-d6) was used as an internal standard. NMR data were processed using Topspin 3.1 (Bruker) and analyzed with Sparky (http://www.cgl.ucsf.edu/home/sparky/). Distance restraints were obtained from the NOE intensities and the structures were calculated based on these restraints using software CYANA (version 1.0.6).28 The 20 structures with the lowest target functions were assessed using the program PROCHECK-NMR29 and displayed by MOLMOL (Version 2.6).30Isothermal titration calorimetry measurements
Calorimetric measurements were performed on an ITC200 system (GE Healthcare, USA) at a temperature of 25 °C using the method reported previously.31 Briefly, 1 and 10 mM AgNO3 solution containing 10 mM SDS in the syringe cell was titrated into the sample cell filled with the solution of 10 mM SDS containing 50 and 500 μM peptide, respectively, at a fixed stirring speed of 1000 rpm. A total of 27 injections were performed with an interval of 240 s between each injection. Control was performed with SDS micellar solution to measure the dilution heat of the titrant and was subtracted from the sample data.The ITC data were processed with the ORIGIN software package (version 7.0) supplied by the manufacturer and the parameters including the enthalpy change (ΔH), binding constant (Ka) and number of binding site (n) were obtained by a non-linear least-square fitting of the experimental data. The Gibbs free energy change (ΔG) and the entropy change (ΔS) were calculated based on Ka and (ΔH).31
Results and discussion
Structure and assembly of hCtr2-TMD2 in SDS micelles
The secondary structure of the hCtr2-TMD2 peptide incorporated with SDS micelles was analyzed by the CD spectrum (Fig. 1A). The spectrum displayed two negative minima at ca. 208 and 222 nm and a positive maximum at ca.194 nm, indicating the formation of an α-helical conformation. The self-assembly state of the peptide in SDS micelles was tested by the SDS-PAGE experiment at a molar ratio of peptide![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) SDS of 1
SDS of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 200. As shown in Fig. 1B, a clearly visible band at a molecular weight between 5.8 and 7.8 kDa was observed, suggesting the formation of a dimer (the molecular mass of monomer is 3.3 kDa).
200. As shown in Fig. 1B, a clearly visible band at a molecular weight between 5.8 and 7.8 kDa was observed, suggesting the formation of a dimer (the molecular mass of monomer is 3.3 kDa).
 | ||
| Fig. 1 (A) CD spectra of 20 μM peptide in 10 mM SDS micelles at pH 6 in the absence and presence of 50 μM AgNO3. (B) SDS-PAGE result of 50 μM peptide in 10 mM SDS at pH 6. | ||
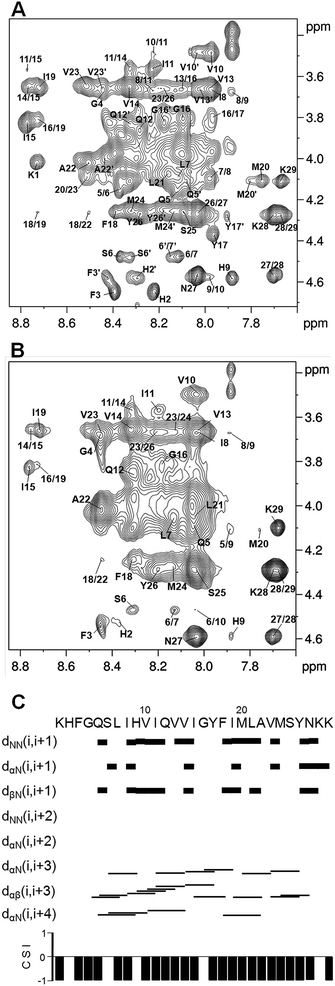
Two-dimensional TOCSY and NOESY spectra of 1 mM hCtr2-TMD2 in 120 mM SDS-d25 micelles (peptide![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) SDS = 1
SDS = 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 120) were recorded to obtain the structure of the peptide at an atomic level. The spectra demonstrated two groups of amino proton (HN) chemical shift for some residues and the cross peaks involved in the two groups of HN resonances were different in intensity (Fig. 2A). Two groups of resonances from the side-chain protons were also observed for certain residues, e.g., the Hδ proton on the iminazole ring of His2 and the aromatic protons of Tyr17. However, the residues with two groups of HN resonances were little different in the resonances of Hα protons. This suggests that the peptide is incorporated with SDS micelles as two states which are different mainly in topology or assembly, instead of the secondary structure.
120) were recorded to obtain the structure of the peptide at an atomic level. The spectra demonstrated two groups of amino proton (HN) chemical shift for some residues and the cross peaks involved in the two groups of HN resonances were different in intensity (Fig. 2A). Two groups of resonances from the side-chain protons were also observed for certain residues, e.g., the Hδ proton on the iminazole ring of His2 and the aromatic protons of Tyr17. However, the residues with two groups of HN resonances were little different in the resonances of Hα protons. This suggests that the peptide is incorporated with SDS micelles as two states which are different mainly in topology or assembly, instead of the secondary structure.
On the basis of the SDS-PAGE result that indicated the formation of a dimer at a peptide![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) SDS ratio of 1
SDS ratio of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 200, we inferred that a mixture of dimer and trimer could be obtained for the peptide in SDS micelles at a higher molar ratio of peptide
200, we inferred that a mixture of dimer and trimer could be obtained for the peptide in SDS micelles at a higher molar ratio of peptide![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) SDS (such as 1
SDS (such as 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 120 in the NMR experiment). In order to determine the oligomeric state of the peptide in the NMR sample with the molar ratio of peptide
120 in the NMR experiment). In order to determine the oligomeric state of the peptide in the NMR sample with the molar ratio of peptide![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) SDS-d25 of 1
SDS-d25 of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 120, we prepared a sample with the molar ratio of peptide
120, we prepared a sample with the molar ratio of peptide![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) SDS-d25 of 1
SDS-d25 of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 20. This peptide
20. This peptide![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) SDS ratio corresponds to the ratio of three peptide molecules to one SDS micelle given that one SDS micelle consists of 60 SDS molecules.32 Interestingly, the NOESY spectrum of the 1
SDS ratio corresponds to the ratio of three peptide molecules to one SDS micelle given that one SDS micelle consists of 60 SDS molecules.32 Interestingly, the NOESY spectrum of the 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 20 peptide/SDS-d25 sample displayed only one group of resonances that overlapped well with the group of signals with a weaker intensity in the NOESY spectrum of the 1
20 peptide/SDS-d25 sample displayed only one group of resonances that overlapped well with the group of signals with a weaker intensity in the NOESY spectrum of the 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 120 sample (Fig. 2B). This confirmed that the resonances with a stronger intensity in the NOESY spectrum of the 1
120 sample (Fig. 2B). This confirmed that the resonances with a stronger intensity in the NOESY spectrum of the 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 120 sample arise from the dimer of the peptide, while the resonances with a weaker intensity originate from the trimer of the peptide. The increase in the ratio of peptide
120 sample arise from the dimer of the peptide, while the resonances with a weaker intensity originate from the trimer of the peptide. The increase in the ratio of peptide![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) SDS-d25 from 1
SDS-d25 from 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 120 to 1
120 to 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 20 or in the ratio of peptide
20 or in the ratio of peptide![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) micelle from 1
micelle from 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2 to 3
2 to 3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 converted the assembly of the peptide in the micelles from a dimer-trimer mixed state to a pure trimeric state. Only one group of resonances obtained in the NOESY spectrum of the 1
1 converted the assembly of the peptide in the micelles from a dimer-trimer mixed state to a pure trimeric state. Only one group of resonances obtained in the NOESY spectrum of the 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 20 sample also implied that the peptide molecules in a trimer were arranged homogeneously by a parallel (head-to-head) pattern in SDS-d25 micelles.
20 sample also implied that the peptide molecules in a trimer were arranged homogeneously by a parallel (head-to-head) pattern in SDS-d25 micelles.
The NOESY spectrum of 1 mM peptide in 20 mM SDS-d25 micelles at pH 6 was used in the calculation of peptide structure. A series of sequential HN(i)–HN(i + 1) NOE connectivities and a series of medium-range connectivities including Hα(i)–HN(i + 3), Hα(i)–Hβ(i + 3) and Hα(i)–HN(i + 4) in the range of residues Gln5–Tyr26 were obtained from the NOESY spectrum (Fig. 2B and C). On the basis of the NOE cross-peaks, three dimensional structure of the peptide in trimer was calculated by CYANA and an α-helix spanning from Ser6 to Tyr26 was obtained by an average of the 20 structures with the lowest target functions (Fig. 3A). Ramachandran analyses for the 20 structures indicated that the dihedral angles φ and ψ in the helical span of the mean structure were distributed totally in the allowed region of an α-helix. The NMR restraints used in calculations and structural statistics extracted from calculation results are listed in Table 1.
| −Ag | +Ag | |
|---|---|---|
| Average target functions (Å2) | 0.04 ± 0.01 | 0.02 ± 0.00 |
| Number of nonredundant distance restraints | 174 | 207 |
| Intraresidual (|i − j| = 0) | 107 | 115 |
| Sequential (|i − j| = 1) | 36 | 49 |
| Medium (|i − j| ≤ 4) | 31 | 43 |
| Long range (|i − j| > 4) | 0 | 0 |
| Average sum of distance restraint violations (Å) | 0.40 ± 0.00 | 0.30 ± 0.00 |
| Average max. distance restraint violation (Å) | 0.10 ± 0.00 | 0.11 ± 0.01 |
| Average sum of torsion angle restraint violations (°) | 0.00 ± 0.00 | 0.0 ± 0.00 |
| Average max. of torsion angle restraint violation (°) | 0.00 ± 0.00 | 0.0 ± 0.00 |
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) |
||
| RMS deviation from the mean structure (Å) | ||
| All residues | ||
| Backbone heavy atoms | 2.95 ± 0.73 | 3.28 ± 0.79 |
| All heavy atoms | 4.35 ± 0.87 | 4.81 ± 0.91 |
| Residues in helix region | 6–26 | 6–26 |
| Backbone heavy atoms | 1.64 ± 0.59 | 2.00 ± 0.70 |
| All heavy atoms | 2.61 ± 0.69 | 3.13 ± 0.93 |
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) |
||
| Ramachandran plot statistics (for each helix span) | ||
| Residues in most favored region (%) | 83.8 | 93.2 |
| Residues in additionally allowed region (%) | 15.0 | 6.4 |
| Residues in generously allowed region (%) | 1.2 | 0.4 |
| Residues in disallowed region (%) | 0.0 | 0.0 |
Binding of hCtr2-TMD2 to silver ion
The binding behavior of hCtr2-TMD2 to Ag(I) was explored by the ITC experiments (Fig. 4). At a peptide![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) SDS of 1
SDS of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 200, an isotherm profile of the peptide titrated by Ag(I) displayed a transition at a Ag(I)
200, an isotherm profile of the peptide titrated by Ag(I) displayed a transition at a Ag(I)![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) peptide of ca. 1. The binding constant (Ka) of 1.09 × 105 M−1 was obtained by the fitting of the isotherm using a one-site binding mode (Table 2). Interestingly, an isotherm profile consisting of two transitions was observed at a peptide
peptide of ca. 1. The binding constant (Ka) of 1.09 × 105 M−1 was obtained by the fitting of the isotherm using a one-site binding mode (Table 2). Interestingly, an isotherm profile consisting of two transitions was observed at a peptide![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) SDS of 1
SDS of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 20. The first isothermal transition occurred at a molar ratio of Ag(I)
20. The first isothermal transition occurred at a molar ratio of Ag(I)![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) peptide of ca. 0.76 and the second at ca. 2.53. The binding constants of 5.87 × 105 M−1 and 1.34 × 105 M−1 were obtained for the first and the second transition, respectively, by the fitting of the isotherm using a two-site binding mode. This indicates that there are either two binding sites in the peptide trimer or two binding processes, the binding corresponding to the first isothermal transition is a higher affinity binding with a chemical stoichiometry of 2
peptide of ca. 0.76 and the second at ca. 2.53. The binding constants of 5.87 × 105 M−1 and 1.34 × 105 M−1 were obtained for the first and the second transition, respectively, by the fitting of the isotherm using a two-site binding mode. This indicates that there are either two binding sites in the peptide trimer or two binding processes, the binding corresponding to the first isothermal transition is a higher affinity binding with a chemical stoichiometry of 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3 for Ag(I)
3 for Ag(I)![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) peptide, and the binding corresponding to the second isothermal transition has a lower affinity but a much higher chemical stoichiometry of Ag(I)
peptide, and the binding corresponding to the second isothermal transition has a lower affinity but a much higher chemical stoichiometry of Ag(I)![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) peptide.
peptide.
 | ||
| Fig. 4 ITC profiles of 50 μM peptide (left) and 500 μM peptide (right) in 10 mM SDS micelles at pH 6 titrated with AgNO3. | ||
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) SDS 1
SDS 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 200) and 500 μM peptide in 10 mM SDS micellar solution (peptide
200) and 500 μM peptide in 10 mM SDS micellar solution (peptide![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) SDS 1
SDS 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 20) titrated with AgNO3 at pH 6, 25 °C
20) titrated with AgNO3 at pH 6, 25 °C
| Peptide/SDS | 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 200 200 |
1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 20 20 |
1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 20 20 |
|---|---|---|---|
| Transition | 1 | 1 | 2 |
| n | 1.16 ± 0.03 | 0.76 ± 0.02 | 2.53 ± 0.02 |
| Ka × 10−5 (M−1) | 1.09 ± 0.16 | 5.87 ± 0.14 | 1.34 ± 0.12 |
| ΔH (kcal mol−1) | −9.02 ± 0.32 | −14.45 ± 0.71 | −22.75 ± 0.68 |
| ΔG (kcal mol−1) | −6.91 ± 0.02 | −7.85 ± 0.01 | −7.01 ± 0.05 |
| ΔS (cal mol−1) | −7.11 ± 1.17 | −44.19 ± 2.62 | −24.43 ± 2.45 |
The thermodynamic parameters were also obtained by the ITC curve fitting (Table 2). The feature of the data (ΔH < 0 and ΔS < 0) indicates that the hydrogen bonding and van der Waals interactions are important in driving either one-site binding of the peptide dimer (a predominant oligomeric state of the peptide at a peptide![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) SDS of 1
SDS of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 200) or the two-site binding/two-process binding of the peptide trimer (an oligomeric state of the peptide at a peptide
200) or the two-site binding/two-process binding of the peptide trimer (an oligomeric state of the peptide at a peptide![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) SDS of 1
SDS of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 20) with silver ion, and the binding occurred spontaneously and exothermically.33
20) with silver ion, and the binding occurred spontaneously and exothermically.33
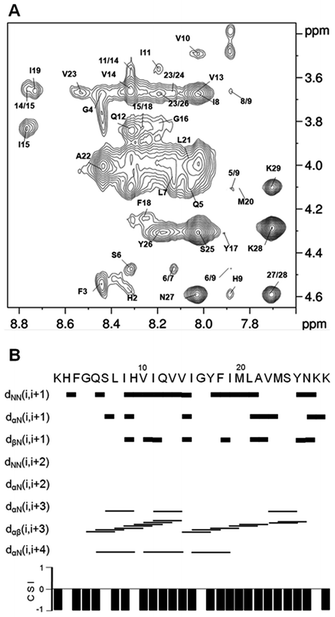
Based on the ITC results, we performed the NMR measurements for the samples of 1 mM peptide in 20 mM SDS-d25 solutions containing 1.5 mM and 3 mM AgNO3. Upon the addition of 1.5 mM Ag(I), the HN proton chemical shifts of some residues in the C-terminal half of the peptide (F18-Met24) were affected significantly, while those in the N-terminal half were little affected (Table 3). This indicates that the peptide binds Ag(I) through the residues of the C-terminal part. Of the residues with larger chemical shift changes, Met20 and Met24 displayed a dramatic downfield shift in Hγ (–CH2–S–), Hε (–S–CH3) and Hβ resonances as well as HN resonance (only Met20), and Val23 showed a dramatic downfield shift in HN resonance, while Phe18 and Leu21 displayed an evident upfield shift in HN and Hα resonance, respectively (Table 3). This implies that the sulfur atoms in the side-chains of Met24 and Met20 were involved in the binding to Ag(I), and the backbone nitrogen atoms of Met20 and Val23 could also participate in the coordination. The upfield shift of Phe18 HN resonance and Leu21 Hα resonance may be induced by the structural regulation of the C-terminal region.
| residue | HN | Hα | Hβ | Hγ | Hδ | Hε |
|---|---|---|---|---|---|---|
| a The data larger than 0.02 ppm are marked by bold. | ||||||
| LYS1 | −0.002 | −0.009 | 0 | 0 | −0.001 | |
| HIS2 | 0.011 | 0.001 | −0.006 | −0.004 | ||
| PHE3 | 0.001 | −0.004 | −0.001 | |||
| GLY4 | −0.004 | −0.006 | ||||
| GLN5 | 0.01 | 0.001 | 0 | −0.001 | −0.003 | |
| SER6 | 0.005 | −0.002 | 0.002 | |||
| LEU7 | −0.002 | −0.003 | −0.008 | −0.001 | −0.003 | |
| ILE8 | −0.006 | −0.001 | 0.002 | −0.002 | −0.012 | |
| HIS9 | −0.001 | −0.001 | 0.005 | 0 | ||
| VAL10 | −0.007 | −0.002 | 0.001 | −0.002 | ||
| ILE11 | 0.004 | 0.001 | 0.008 | 0.001 | 0.001 | |
| GLN12 | −0.001 | −0.009 | 0.001 | −0.006 | ||
| VAL13 | 0.008 | 0.006 | −0.004 | −0.001 | ||
| VAL14 | 0 | −0.002 | −0.001 | −0.003 | ||
| ILE15 | 0.002 | 0.006 | −0.002 | −0.003 | −0.001 | |
| GLY16 | −0.007 | −0.012 | ||||
| TYR17 | 0 | −0.002 | −0.003 | −0.011 | −0.011 | |
| PHE18 | −0.058 | 0 | −0.011 | 0.006 | ||
| ILE19 | 0.017 | 0.002 | −0.002 | −0.002 | −0.031 | |
| MET20 | 0.068 | −0.001 | 0.068 | 0.192 | 0.121 | |
| LEU21 | 0.008 | −0.084 | −0.004 | 0.001 | −0.007 | |
| ALA22 | −0.006 | −0.004 | 0.009 | |||
| VAL23 | 0.088 | 0 | 0.001 | 0.001 | ||
| MET24 | 0.004 | −0.001 | 0.059 | 0.152 | 0.127 | |
| SER25 | −0.013 | 0.023 | −0.001 | |||
| TYR26 | −0.005 | 0.005 | 0.001 | −0.001 | 0.003 | |
| ASN27 | −0.003 | −0.001 | −0.011 | −0.016 | ||
| LYS28 | 0.006 | −0.003 | −0.005 | −0.004 | 0 | 0 |
| LYS29 | 0.035 | 0.003 | −0.003 | −0.005 | 0.001 | 0.001 |
However, when the concentration of AgNO3 was increased from 1.5 mM to 3 mM, only the chemical shifts of Hγ (–CH2–S–) and Hε (–S–CH3) in Met20 and Met24 increased evidently, while the chemical shifts of all other residues were almost not changed in the NOESY spectrum. This means that there is only one binding site, i.e., the MXXXM motif, in the peptide trimer for silver. It is most likely that the peptide trimer binds two silver ions by the MXXXM motif first (the first isothermal transition), then two univalent silver clusters that may consist of three silver atoms replace the binding sites of two Ag(I) ions (the second isothermal transition) when the concentration of AgNO3 is increased. A weaker peptide–peptide interaction in the hCtr2-TMD2 trimer could provide a larger space of pore in the binding site to accommodate the silver clusters without changing the topology of peptide monomers in the micelles. As a result, the chemical shifts of all the residues except for those of two Met residues were not changed. Currently, the speculation of multi–silver binding has not been confirmed by the experimental data. Other possibilities of binding patterns could not be excluded either.
Although the presence of Ag(I) resulted in a distinct shifting of the proton resonances of the residues in the C-terminal region and a decrease in the intensity of some NMR signals, it affected the structure of the peptide slightly. An α-helix spanning from Ser6 to Tyr26 was also obtained by the structure calculation based on the NOE data from the NOESY spectrum of 1 mM peptide in 20 mM SDS-d25 micelles in the presence of 1.5 mM AgNO3 (Fig. 5 and 3B). Whereas a similar helicity was obtained for the peptide in SDS micelles at a peptide![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) SDS of 1
SDS of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 20 by the NMR experiments in the absence and presence of AgNO3, a small difference in the helicity (characterized by two absorbance minima) was observed in the CD spectra of the peptide in SDS micellar solutions with and without AgNO3 (Fig. 1). On the basis of the SDS-PAGE and NMR results that the peptide formed a dimer in SDS micelles at a peptide
20 by the NMR experiments in the absence and presence of AgNO3, a small difference in the helicity (characterized by two absorbance minima) was observed in the CD spectra of the peptide in SDS micellar solutions with and without AgNO3 (Fig. 1). On the basis of the SDS-PAGE and NMR results that the peptide formed a dimer in SDS micelles at a peptide![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) SDS of 1
SDS of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 200 and a trimer at a peptide
200 and a trimer at a peptide![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) SDS of 1
SDS of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 20, we inferred that the oligomeric state of the peptide in SDS micelles at a peptide
20, we inferred that the oligomeric state of the peptide in SDS micelles at a peptide![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) SDS of 1
SDS of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 500 as used in the CD experiment should be dimer and/or monomer. Slightly different effect of Ag(I) on the helicity of the peptide observed in the CD and NMR spectra may be attributed to the difference in the oligomeric degree of the peptide formed in different experimental conditions.
500 as used in the CD experiment should be dimer and/or monomer. Slightly different effect of Ag(I) on the helicity of the peptide observed in the CD and NMR spectra may be attributed to the difference in the oligomeric degree of the peptide formed in different experimental conditions.
Comparison with the results of hCtr1-TMD2
Although hCtr1 and hCtr2 share only 41% homology in the entire amino acid sequence, they are highly conserved in the transmembrane domains, especially in the second transmembrane domains. Therefore, the two transporters are believed to share similar topological features and similar oligomeric manner in membranes.34,35 Ctr1 has been revealed to form a pore in lipid membrane by the trimerization and the second transmembrane domain lines the pore of the trimer.6,12,36 Whether this is true for Ctr2 is currently unknown, but it has been known that the MXXXM motif that is highly conserved in TMD2 of both proteins is equally significant for the functions of Ctr1 and Ctr2,1,2,5,7,18 implying that TMD2 may play an important role in the functions of the two homological proteins. The functional studies have demonstrated that both Cu(I) and Ag(I) are the substrates of the two transporters. However, the fact that Ctr1 is a high affinity copper transporter but Ctr2 is a low affinity transporter indicates a difference between Ctr1 and Ctr2 in the structures and/or functions.Because the significance of TMD2 in the pore formation and transport function of Ctr1, we have previously studied the assembly structure of hCtr1-TMD2 in SDS micelles and the binding of the peptide to silver ion, and showed that this transmembrane segment itself has a strong propensity to assemble as a trimer in SDS micelles at a peptide![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) SDS of 1
SDS of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 120 and the MXXXM motif is involved in the binding to Ag(I) with an affinity (Kd) of ∼0.5 μM.13 In the present work, we undertook similar study on hCtr2-TMD2. The results indicated that the majority of hCtr2-TMD2 molecules form dimer at the experimental condition of 1 mM peptide, 120 mM SDS, pH 6, 25 °C and one-time HFIP treatment, whereas total amount of hCtr1-TMD2 form trimer at the same condition. The peptide hCtr2-TMD2 formed trimer entirely only when the molar ratio of peptide
120 and the MXXXM motif is involved in the binding to Ag(I) with an affinity (Kd) of ∼0.5 μM.13 In the present work, we undertook similar study on hCtr2-TMD2. The results indicated that the majority of hCtr2-TMD2 molecules form dimer at the experimental condition of 1 mM peptide, 120 mM SDS, pH 6, 25 °C and one-time HFIP treatment, whereas total amount of hCtr1-TMD2 form trimer at the same condition. The peptide hCtr2-TMD2 formed trimer entirely only when the molar ratio of peptide![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) SDS was increased to 1
SDS was increased to 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 20. This suggests that hCtr2-TMD2 has a less propensity to assemble as a trimer than hCtr1-TMD2. Notably, the ITC results showed that the hCtr2-TMD2 trimer binds Ag(I) with an affinity (Kd) of ∼1.7 μM, lower than that of hCtr1-TMD2 trimer (∼0.5 μM).13 This is also confirmed by the NMR spectra. Comparing with the 2D NMR spectra of hCtr1-TMD2 in SDS-d25 micelles, one could find that the amount of signals that are affected by Ag(I) is decreased in the NMR spectra of hCtr2-TMD2 (Table 3), indicating a weaker binding of Ag(I) by hCtr2-TMD2 than the binding by hCtr1-TMD2. Although no affinity data are available for silver transport by the two proteins, previous studies have shown that the affinity of hCtr1 for copper is higher than that of hCtr2 in terms of the Km values of ∼1.7 μM (ref. 1) and ∼7–11 μM (ref. 34) for hCtr1 and hCtr2, respectively. It is possible that hCtr1 and hCtr2 also have different affinities for Ag(I) uptake, like they do for Cu(I), considering that hCtr2 has a shorter N-terminal domain and less methionine and histidine residues in the domain than hCtr1, and the binding of hCtr2 to Ag(I) by the MXXXM motif of TMD2 may be less stronger than that of hCtr1, as observed in our studies. The difference in the affinity between hCtr1-TMD2 and hCtr2-TMD2 for Ag(I) may be associated with the difference in the intermolecular interactions in the transmembrane domains. However, why there is such a difference between hCtr1-TMD2 and hCtr2-TMD2 in the intermolecular interaction is not clear currently because most of the residues in the two peptide sequences are either same or very similar in property. Therefore, the two peptides should be similar rather than different in the aggregation ability. Further study should be done for answering this question. Despite having different intermolecular interactions, the trimers reconstructed by hCtr1-TMD2 and hCtr2-TMD2 in SDS micelles showed the same binding stoichiometry of 2
20. This suggests that hCtr2-TMD2 has a less propensity to assemble as a trimer than hCtr1-TMD2. Notably, the ITC results showed that the hCtr2-TMD2 trimer binds Ag(I) with an affinity (Kd) of ∼1.7 μM, lower than that of hCtr1-TMD2 trimer (∼0.5 μM).13 This is also confirmed by the NMR spectra. Comparing with the 2D NMR spectra of hCtr1-TMD2 in SDS-d25 micelles, one could find that the amount of signals that are affected by Ag(I) is decreased in the NMR spectra of hCtr2-TMD2 (Table 3), indicating a weaker binding of Ag(I) by hCtr2-TMD2 than the binding by hCtr1-TMD2. Although no affinity data are available for silver transport by the two proteins, previous studies have shown that the affinity of hCtr1 for copper is higher than that of hCtr2 in terms of the Km values of ∼1.7 μM (ref. 1) and ∼7–11 μM (ref. 34) for hCtr1 and hCtr2, respectively. It is possible that hCtr1 and hCtr2 also have different affinities for Ag(I) uptake, like they do for Cu(I), considering that hCtr2 has a shorter N-terminal domain and less methionine and histidine residues in the domain than hCtr1, and the binding of hCtr2 to Ag(I) by the MXXXM motif of TMD2 may be less stronger than that of hCtr1, as observed in our studies. The difference in the affinity between hCtr1-TMD2 and hCtr2-TMD2 for Ag(I) may be associated with the difference in the intermolecular interactions in the transmembrane domains. However, why there is such a difference between hCtr1-TMD2 and hCtr2-TMD2 in the intermolecular interaction is not clear currently because most of the residues in the two peptide sequences are either same or very similar in property. Therefore, the two peptides should be similar rather than different in the aggregation ability. Further study should be done for answering this question. Despite having different intermolecular interactions, the trimers reconstructed by hCtr1-TMD2 and hCtr2-TMD2 in SDS micelles showed the same binding stoichiometry of 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3 for Ag(I):peptide, suggesting that the two peptides bind Ag(I) by a similar manner, i.e., silver ions are chelated by two Met triads from Met20 and Met24. The binding of Ag(I) by methionine residues has been previously found either in the copper transporters or in the peptide segments.18,31,37–39
3 for Ag(I):peptide, suggesting that the two peptides bind Ag(I) by a similar manner, i.e., silver ions are chelated by two Met triads from Met20 and Met24. The binding of Ag(I) by methionine residues has been previously found either in the copper transporters or in the peptide segments.18,31,37–39
Conclusions
A trimeric structure of hCtr2-TMD2 can be reconstructed in SDS micelles at a molar ratio of peptide![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) SDS of 1
SDS of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 20. Similar to the hCtr1-TMD2 trimer, the hCtr2-TMD2 trimer binds two Ag(I) by a predominant coordination of two methionine triads from the MXXXM motif. However, the hCtr2-TMD2 trimer has a weaker intermolecular interaction and a lower affinity for Ag(I) than the hCtr1-TMD2 trimer. Our results suggest that the intermolecular interactions between the second transmembrane domains may play a significant role in the formation of oligomeric structure of hCtr1 and hCtr2 and in the binding affinity of the proteins for Ag(I)/Cu(I).
20. Similar to the hCtr1-TMD2 trimer, the hCtr2-TMD2 trimer binds two Ag(I) by a predominant coordination of two methionine triads from the MXXXM motif. However, the hCtr2-TMD2 trimer has a weaker intermolecular interaction and a lower affinity for Ag(I) than the hCtr1-TMD2 trimer. Our results suggest that the intermolecular interactions between the second transmembrane domains may play a significant role in the formation of oligomeric structure of hCtr1 and hCtr2 and in the binding affinity of the proteins for Ag(I)/Cu(I).
Acknowledgements
This work was financially supported by the NSFC (20934002).References
- J. Lee, M. M. O. Peña, Y. Nose and D. J. Thiele, J. Biol. Chem., 2002, 277, 4380–4387 CrossRef CAS PubMed.
- S. Puig, J. Lee, M. Lau and D. J. Thiele, J. Biol. Chem., 2002, 277, 26021–26030 CrossRef CAS PubMed.
- J. F. Eisses and J. H. Kaplan, J. Biol. Chem., 2005, 280, 37159–37168 CrossRef CAS PubMed.
- P. van den Berghe, D. Folmer, H. Malingre, E. van Beurden, A. Klomp, B. van de Sluis, M. Merkx, R. Berger and L. Klomp, Biochem. J., 2007, 407, 49–59 CrossRef CAS PubMed.
- J. Bertinato, E. Swist, L. Plouffe, S. Brooks and M. L'Abbe, Biochem. J., 2008, 409, 731–740 CrossRef CAS PubMed.
- S. G. Aller and V. M. Unger, Proc. Natl. Acad. Sci. U. S. A., 2006, 103, 3627–3632 CrossRef CAS PubMed.
- I. F. Tsigelny, Y. Sharikov, J. P. Greenberg, M. A. Miller, V. L. Kouznetsova, C. A. Larson and S. B. Howell, Cell Biochem. Biophys., 2012, 63, 223–234 CrossRef CAS PubMed.
- J. F. Quail, C.-Y. Tsai and S. B. Howell, J. Trace Elem. Med. Biol., 2014, 28, 151–159 CAS.
- H. Ohrvik and D. J. Thiele, Ann. N. Y. Acad. Sci., 2014, 1314, 32–41 CrossRef CAS PubMed.
- V. Martins, E. Bassil, M. Hanana, E. Blumwald and H. Gerós, Planta, 2014, 240, 91–101 CrossRef CAS PubMed.
- N. K. Wee, D. C. Weinstein, S. T. Fraser and S. J. Assinder, Int. J. Biochem. Cell Biol., 2013, 45, 960–963 CrossRef CAS PubMed.
- C. J. de Feo, S. G. Aller, G. S. Siluvai, N. J. Blackburn and V. M. Unger, Proc. Natl. Acad. Sci. U. S. A., 2009, 106, 4237–4242 CrossRef CAS PubMed.
- Z. Dong, Y. Wang, C. Wang, H. Xu, L. Guan, Z. Li and F. Li, J. Phys. Chem. B, 2015, 119, 8302–8312 CrossRef CAS PubMed.
- K. L. Haas, A. B. Putterman, D. R. White, D. J. Thiele and K. J. Franz, J. Am. Chem. Soc., 2011, 133, 4427–4437 CrossRef CAS PubMed.
- X. B. Du, H. Y. Li, X. H. Wang, Q. Liu, J. Z. Ni and H. Z. Sun, Chem. Commun., 2013, 49, 9134–9136 RSC.
- E. B. Maryon, S. A. Molloy, K. Ivy, H. Yu and J. H. Kaplan, J. Biol. Chem., 2013, 288, 18035–18046 CrossRef CAS PubMed.
- E. M. Rees, J. Lee and D. J. Thiele, J. Biol. Chem., 2004, 279, 54221–54229 CrossRef CAS PubMed.
- J. Bertinato, L. Cheung, R. Hoque and L. J. Plouffe, J. Trace Elem. Med. Biol., 2010, 24, 178–184 CAS.
- S. R. Hanson, S. A. Donley and M. C. Linder, J. Trace Elem. Med. Biol., 2001, 15, 243–253 CAS.
- A. Odermatt, R. Krapf and M. Solioz, Biochem. Biophys. Res. Commun., 1994, 202, 44–48 CrossRef CAS PubMed.
- M. Solioz and A. Odermatt, J. Biol. Chem., 1995, 270, 9217–9221 CrossRef CAS PubMed.
- P. L. Drake and K. J. Hazelwood, Ann. Occup. Hyg., 2005, 49, 575–585 CrossRef CAS PubMed.
- B. S. Atiyeh, M. Costagliola, S. N. Hayek and S. A. Dibo, Burns, 2007, 33, 139–148 CrossRef PubMed.
- J. J. Liu, P. Galettis, A. Farr, L. Maharaj, H. Samarasinha, A. C. McGechan, B. C. Baguley, R. J. Bowen, S. J. Berners-Price and M. J. McKeage, J. Inorg. Biochem., 2008, 102, 303–310 CrossRef CAS PubMed.
- K. I. Batarseh, Curr. Med. Chem., 2013, 20, 2363–2373 CrossRef CAS PubMed.
- S. Ray, R. Mohan, J. K. Singh, M. K. Samantaray, M. M. Shaikh, D. Panda and P. Ghosh, J. Am. Chem. Soc., 2007, 129, 15042–15053 CrossRef CAS PubMed.
- H. Li, F. Li, H. Sun and Z. Qian, Biochem. J., 2003, 372, 757–766 CrossRef CAS PubMed.
- P. Guntert, C. Mumenthaler and K. Wuthrich, J. Mol. Biol., 1997, 273, 283–298 CrossRef CAS PubMed.
- R. A. Laskowski, J. A. Rullmannn, M. W. MacArthur, R. Kaptein and J. M. Thornton, J. Biomol. NMR, 1996, 8, 477–486 CrossRef CAS PubMed.
- R. Koradi, M. Billeter and K. Wüthrich, J. Mol. Graphics, 1996, 14, 51–55 CrossRef CAS PubMed.
- Y. R. Wang, L. L. Wang and F. Li, RSC Adv., 2013, 3, 15245–15253 RSC.
- A. Öhman, P.-O. Lycksell, A. Juréus, Ü. Langel, T. Bartfai and A. Gräslund, Biochemistry, 1998, 37, 9169–9178 CrossRef PubMed.
- T. S. Olsson, M. A. Williams, W. R. Pitt and J. E. Ladbury, J. Mol. Biol., 2008, 384, 1002–1017 CrossRef CAS PubMed.
- C. P. Huang, M. Fofana, J. Chan, C. J. Chang and S. B. Howell, Metallomics, 2014, 6, 654–661 RSC.
- P. Abada and S. B. Howell, Met.-Based Drugs, 2010, 2010, 1–9 CrossRef PubMed.
- M. Schushan, Y. Barkan, T. Haliloglu and N. Ben-Tal, Proc. Natl. Acad. Sci. U. S. A., 2010, 107, 10908–10913 CrossRef CAS PubMed.
- F. Long, C.-C. Su, M. T. Zimmermann, S. E. Boyken, K. R. Rajashankar, R. L. Jernigan and E. W. Yu, Nature, 2010, 467, 484–488 CrossRef CAS PubMed.
- J. T. Rubino, P. Riggs-Gelasco and K. J. Franz, J. Biol. Inorg. Chem., 2010, 15, 1033–1049 CrossRef CAS PubMed.
- Y. Shenberger, H. E. Gottlieb and S. Ruthstein, J. Biol. Inorg. Chem., 2015, 20, 719–727 CrossRef CAS PubMed.
| This journal is © The Royal Society of Chemistry 2016 |