Spontaneous osteogenic differentiation of mesenchymal stem cells on electrospun nanofibrous scaffolds†
Ning Zhang,
Qian-Ru Xiao,
Xin-Yao Man,
Hai-Xia Liu,
Lan-Xin Lü and
Ning-Ping Huang*
State Key Laboratory of Bioelectronics, School of Biological Science and Medical Engineering, Southeast University, Nanjing, 210096, P. R. China. E-mail: nphuang@seu.edu.cn
First published on 19th February 2016
Abstract
In bone tissue engineering, stem cell-scaffold constructs play an important role in bone regeneration. Desirable biomimetic scaffolds that can facilitate the committed differentiation of stem cells into osteoblasts at high efficiency in the absence of inducing media are essentially useful for the application. In this study, we have investigated the potency of spontaneous osteogenic differentiation of bone marrow-derived mesenchymal stem cells (MSCs) on PHBV electrospun nanofibrous scaffolds loaded with hydroxyapatite nanoparticles, and studied the underlying mechanisms. By studying cell morphology and the cytoskeleton, as well as osteogenic marker gene expression, we found that MSCs cultured on the above-mentioned nanofibrous scaffolds could efficiently differentiate into osteoblasts in the absence of inducing media. Moreover, we examined the activity of several signaling pathways involved in osteoblast differentiation, including Wnt/β-catenin, BMP-Smad, and MAPK (ERK1/2 and p38) pathways. This study uncovers the regulatory mechanisms of MSC differentiation into osteoblasts stimulated by biomimetic nanofibrous scaffolds, which will help understand bone tissue repair for therapeutic applications and optimize the biomaterial scaffolds.
Introduction
In bone tissue engineering, the nanostructured surfaces of a biomedical implant play an important role in cell behavior and function.1–7 The development of the electrospinning technique helps to fabricate nanoscaled fibrous scaffolds that may mimic the native extracellular matrix (ECM) in structures and thus promote cell adhesion and proliferation.8–11 Great progress made in stem cell biology has opened up new vistas for tissue engineering. Bone marrow-derived mesenchymal stem cells (MSCs) are multipotent stem cells that can differentiate into a variety of cell lineages, including osteoblasts, chondrocytes, and neurons in specific conditions.12–14 Previous studies have proved that MSCs can be induced into osteoblasts by adding some chemical factors into the culture media, or by adjusting the chemical property, topography, or stiffness of the supporting substrates.15–19 Hydroxyapatite (HA), a main inorganic component of bone, has displayed osteoconductive and “inherent” osteoinductive effects.20 The osteoinductive role of HA has been investigated by monitoring its ability to induce pluripotent MSCs to differentiate into osteoblasts.21–23 A recent study reported that the incorporation of HA into the PCL (poly(ε-caprolactone)) nanofibrous scaffolds could regulate the osteogenic differentiation of human MSCs in total absence of osteogenic supplements.24 In our previous study,25,26 we found that PHBV (poly(3-hydroxybutyrate-co-3-hydroxyvalerate)) nanofibrous scaffolds loaded with HA nanoparticles could induce rat bone marrow derived-MSCs to differentiate into osteoblasts. Moreover, 3D PHBV/HA scaffolds made from aligned or random oriented nanofibers were implanted into critical-sized rabbit radius defects and exhibited significant effects on the repair of critical bone defects. However, the mechanisms of spontaneous osteogenic differentiation of MSCs on developed nanofibrous scaffolds haven't been well understood, which may hamper the optimization of biomimetic scaffolds used for bone tissue engineering.Osteoblast differentiation from MSCs is a well-orchestrated process. Osteoblast commitment and differentiation are controlled by complex activities involving signal transduction and transcriptional regulation of gene expression.27 The Wnt/β-catenin signaling pathway which plays an essential role in bone mass and bone cell function is involved in the response of cells to implant surface properties.28–31 It has also been shown that the BMP-Smad signaling pathway mediates the biological effects of the implant surfaces.32–34 Mitogen-activated protein kinases (MAPKs), including extracellular signal-regulated kinases 1 and 2 (ERK1/2), c-Jun N-terminal kinase and p38 families, are key mediators of cellular responses to a variety of extracellular stimuli.35 Nonetheless, signaling pathway activated in osteogenic differentiation of MSCs cultured on HA-containing nanofibrous scaffolds remains unclear.
In this study, random-oriented or aligned PHBV and PHBV/HA nanofibrous scaffolds were fabricated by electrospinning technique. Osteogenic differentiation of MSCs on nanofibrous scaffolds were investigated by detecting the expression of the runt-related transcription factor 2 (Runx2), alkaline phosphatase (ALP), osteocalcin (OCN) and collagen type I (Col I), respectively. Moreover, the roles of the Wnt/β-catenin signaling pathway, BMP-Smad signaling pathway and MAPK signaling pathway in osteogenic differentiation were studied through immunofluorescence staining of key components in these signaling pathways.
Experimental
Preparation and characterization of PHBV-based nanofibrous scaffolds
According to the procedures described in our previous studies,25,26 random-oriented and aligned PHBV and PHBV/HA nanofibers were produced via the electrospinning technology. In brief, to prepare the electrospun solution, a 2 wt% PHBV solution was prepared as follows: PHBV (Sigma-Aldrich, USA) and poly(ethylene oxide) (PEO, Guoren Chemical Co., China), at a mass ratio of 9![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, were mixed in 2,2,2-trifluoroethanol (TFE, Darui Finechem Ltd., China) at room temperature. For PHBV/HA solution, hydroxyapatite (HA, diameter less than 200 nm, Sigma-Aldrich, USA), with mass equal to PEO in 2% PHBV solution, was uniformly dispersed into TFE (the same volume as PHBV solution) in ultrasonic bath. To prepare random-oriented nanofibers (NF), solution was loaded into a 20 mL syringe with 6# metal needle (inner diameter of 0.5 mm) and continually driven by syringe driver at a speed of 5 mL per hour. The aluminum foil connecting the cathode was used for collecting random-oriented nanofibers, whereas a roller coated with aluminum foil at a rotating rate of 2500 rpm was used as a collector for aligned nanofibers (A-NF). A 12 kV high DC voltage was applied between the collector and the needle at a woking distance of 20 cm. To remove residual solvent, all collected nanofibers were dried in vacuum desiccator for 48 h at 60 °C. All nanofibrous scaffolds were sterilized by autoclave at 121 °C for 30 min for cell experiments.
1, were mixed in 2,2,2-trifluoroethanol (TFE, Darui Finechem Ltd., China) at room temperature. For PHBV/HA solution, hydroxyapatite (HA, diameter less than 200 nm, Sigma-Aldrich, USA), with mass equal to PEO in 2% PHBV solution, was uniformly dispersed into TFE (the same volume as PHBV solution) in ultrasonic bath. To prepare random-oriented nanofibers (NF), solution was loaded into a 20 mL syringe with 6# metal needle (inner diameter of 0.5 mm) and continually driven by syringe driver at a speed of 5 mL per hour. The aluminum foil connecting the cathode was used for collecting random-oriented nanofibers, whereas a roller coated with aluminum foil at a rotating rate of 2500 rpm was used as a collector for aligned nanofibers (A-NF). A 12 kV high DC voltage was applied between the collector and the needle at a woking distance of 20 cm. To remove residual solvent, all collected nanofibers were dried in vacuum desiccator for 48 h at 60 °C. All nanofibrous scaffolds were sterilized by autoclave at 121 °C for 30 min for cell experiments.
Scanning electron microscope (SEM, ultra plus Zeiss, Germany) was used to display the morphology of electrospun nanofibers. The average diameter and orientation degree of fibers were determined by measuring diameters and angles of 200 nanofibers in SEM images using the Image J software. HA particles dispersed inside fibers were tested by transmission electron microscope (TEM, Tecnai G2 S-TWIN, The Netherlands).
Isolation and culture of MSCs
All animal experiments were approved by the institutional animal care committee of Southeast University and carried out in accordance with the institutional guidelines for care and use of laboratory animals. MSCs were extracted from rat bone marrow as has been described previously.25,26,36 Briefly, the tibias and femurs from 4 week-old Sprague-Dawley rats were dissected. Both ends of the bones were cut down along the epiphysis, and then marrow was flushed with 10 mL of cell culture medium consisting of α-minimal essential medium (α-MEM, Thermo Scientific HyClone, USA) supplemented with 10% FBS and 1% penicillin/streptomycin antibiotics (Gibco, USA). To obtain MSCs, bone marrow cells were transferred into a culture flask and incubated at 37 °C with 5% CO2. For all experiments, cells at Passage three were used at a density of 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 cells per cm2.
000 cells per cm2.
Scanning electron microscopy observation and cell morphology
The morphology of MSCs seeded on the electrospun nanofibers was inspected by field-emission scanning electron microscopy (SEM, ultra plus Zeiss, Germany). After 7 days of culture, MSCs on different surfaces of random-oriented/aligned PHBV and PHBV/HA nanofibrous meshes were gently rinsed three times with PBS buffer and fixed with 2.5% glutaraldehyde (Sigma-Aldrich, USA) for 2 h at room temperature. After fixation, the samples were rinsed again with PBS and underwent dehydration with gradient ethanol (30, 50, 70, 80, 90, 95, and 100%) for 10 min each step. The samples were then dried and examined by SEM.ALP staining
Alkaline phosphatase detection kit (BCIP/NBT, 5-bromo-4-chloro-3-indolyl phosphate/p-nitroblue tetrazolium chloride, Amresco, USA) was used to determine the ALP activity. Seven days after cell seeding, the samples were fixed with 4% paraformaldehyde (Lingfeng Chemical Reagent Co., Ltd., China) for 30 min and rinsed three times with PBS. BCIP/NBT was then added and they were incubated for 30 min. Finally, samples were rinsed once with water and observed under a bright-field microscope.Confocal laser scanning microscopy (CLSM)
Osteocalcin and the role of the Wnt/β-catenin, BMP-Smad, MAPK signaling pathway were examined by immunofluorescent staining of key proteins monitored through confocal laser scanning microscopy. To stain the activated components on different fibrous meshes, samples were permeabilized by 0.5% formaldehyde (Xilong Chemical Technology Co., Ltd., China) coupled with 0.2% Triton X-100 (SunShine Biotechnology Co., Ltd., China) for 5 min at room temperature, and then followed by fixation in 4% formaldehyde for 20 min. 3% bovine serum albumin (BSA, SunShine Biotechnology Co. Ltd., China) in PBS was used as blocking solution to prevent nonspecific binding of antibody. After three times of rinsing with PBS, samples were immersed in primary antibody [osteocalcin, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 100 in 3% BSA, rabbit polyclonal antibody of rat, Santa Cruz Biotechnology, USA; β-catenin, phospho-Smad1/5 (Ser463/465) antibody (p-Smad1/5), phospho-p44/42 MAPK (Erk1/2)(Thr202/Tyr204) antibody (p-Erk), phospho-p38 MAPK (Thr180/Tyr182) antibody (p-p38), 1
100 in 3% BSA, rabbit polyclonal antibody of rat, Santa Cruz Biotechnology, USA; β-catenin, phospho-Smad1/5 (Ser463/465) antibody (p-Smad1/5), phospho-p44/42 MAPK (Erk1/2)(Thr202/Tyr204) antibody (p-Erk), phospho-p38 MAPK (Thr180/Tyr182) antibody (p-p38), 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 100 in 3% BSA, rabbit polyclonal antibody of rat, Cell Signaling Technology, USA] and incubated at 4 °C for at least 12 h. After washed with PBS, samples were incubated in secondary antibody (1
100 in 3% BSA, rabbit polyclonal antibody of rat, Cell Signaling Technology, USA] and incubated at 4 °C for at least 12 h. After washed with PBS, samples were incubated in secondary antibody (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 200 in 3% BSA, Alexa-Fluor 488 goat antirabbit IgG, Invitrogen, USA) for 1 h at 37 °C protected from light. Samples were washed twice and then incubated with Alexa-Fluor 633 phalloidin (Invitrogen, USA) at a dilution of 1
200 in 3% BSA, Alexa-Fluor 488 goat antirabbit IgG, Invitrogen, USA) for 1 h at 37 °C protected from light. Samples were washed twice and then incubated with Alexa-Fluor 633 phalloidin (Invitrogen, USA) at a dilution of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 100 in the dark for 30 min followed by rinsing twice with PBS. Finally, to stain nucleus, samples were immersed in hoechst 33
100 in the dark for 30 min followed by rinsing twice with PBS. Finally, to stain nucleus, samples were immersed in hoechst 33![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 342 (Sigma, USA) at a concentration of 10 μg mL−1 for 30 min protected from light. Finally, cells were observed with a CLSM (Revolution XD, Andor Technology, Northern Ireland).
342 (Sigma, USA) at a concentration of 10 μg mL−1 for 30 min protected from light. Finally, cells were observed with a CLSM (Revolution XD, Andor Technology, Northern Ireland).
Acquisition of the fluorescently labeled images and quantification of the biomarkers
Fluorescence was analyzed using the following wavelengths: Hoechst 33![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 342 (ex = 346 nm and em = 460 nm), Alexa-488 (ex = 495 nm and em = 519 nm) and Alexa-633 (ex = 632 nm and em = 647 nm). ALP and immunofluorescent staining images were analyzed by Image J (National Institutes of Health, USA).37 To measure the surface coverage of activated proteins, staining images were quantified according to the number of fluorescent pixels over the total amount of pixels.38 The channels required for quantification were converted into gray scale. Make sure to set the same threshold for images and the activated protein coverage was summarized.
342 (ex = 346 nm and em = 460 nm), Alexa-488 (ex = 495 nm and em = 519 nm) and Alexa-633 (ex = 632 nm and em = 647 nm). ALP and immunofluorescent staining images were analyzed by Image J (National Institutes of Health, USA).37 To measure the surface coverage of activated proteins, staining images were quantified according to the number of fluorescent pixels over the total amount of pixels.38 The channels required for quantification were converted into gray scale. Make sure to set the same threshold for images and the activated protein coverage was summarized.
Quantitative real time PCR
After 7 and 14 days of cell culture, the osteogenic gene expressions were investigated. The total RNA of the cells on nanofibers was isolated by Trizol reagent (Invitrogen, USA). The quality of RNA was detected by spectrophotometric (BioTek, USA). 1 μg of total RNA was converted to cDNA according to M-MLV reserve transcriptase instructions (Promega, USA). The real-time PCR reactions were performed using SYBR Premix ExTaq (TaKaRa) on ABI 7500 real time PCR system (Application Biosystems, USA), in order to evaluate the gene expression of ALP, OCN, Col I and Runx2. GAPDH was used as a housekeeping gene. The primers are listed in Table 1.| Gene | Forward primer sequence (5′–3′) | Reverse primer sequence (5′–3′) |
|---|---|---|
| ALP | GGATCCTGACAAAGAATCCCAA | CTCATGCAGCGCCTGCTT |
| OCN | AGCTGAAGCTGCCGTTGG | AGTAAGGTGGTGAATAGACTCCG |
| Col I | GCGAAGGCAACAGTCGATTC | CTTGGTGGTTTTGTATTCGATGAC |
| Runx2 | CTTCGTCAGCGTCCTATCAGTTC | CAGCGTCAACACCATCATTCTG |
| GAPDH | TATGACTCTACCCACGGCAA | TACTCAGCACCAGCATCACC |
Statistical analysis
All data were expressed as mean ± standard deviation from at least three independent experiments. One-way ANOVA combined with Student's t-test was used to compare the data among different groups. The threshold significance level was set at 0.05. Thus, p (probability) values lower than 0.05 were considered statistically significant.Results and discussion
Fabrication of PHBV-based nanofibrous scaffolds
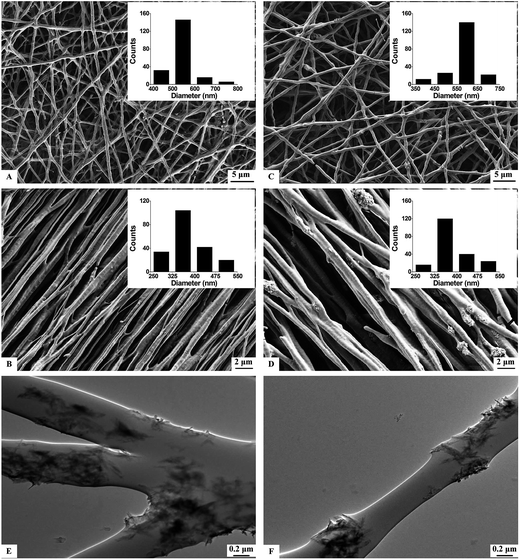
In this study, we used electrospinning technology to create random-oriented/aligned PHBV (NF/A-NF) and PHBV/HA nanofibers (HA-NF/HA-A-NF). Fig. 1A–D show the morphologies of NF, A-NF, HA-NF, and HA-A-NF observed under SEM, with the diameter of 532 ± 75 nm, 372 ± 65 nm, 563 ± 71 nm, and 396 ± 73 nm, respectively. The insets in Fig. 1A–D show the diameter distribution of PHBV-based nanofibers, from which it can be seen that fibers are uniform in diameter. Most A-NF and HA-A-NF fibers are oriented parallel to each other. The angle distribution of aligned fibers indicates that most fibers have the same orientation in both A-NF and HA-A-NF (Fig. S1†). The SEM images also indicate that the introduction of HA does not influence the morphology of nanofibers. Furthermore, TEM images clearly show the distribution of HA particles dispersed in HA-NF (Fig. 1E) and HA-A-NF (Fig. 1F).Morphology of MSCs on PHBV-based nanofibrous scaffolds
After 7 days of culture, the morphology of MSCs on nanofibers was observed by SEM (Fig. 2). The cell numbers attached on the four nanofibrous scaffolds were similar. The magnified SEM images display that on the random-oriented nanofibers (NF) and HA-containing nanofibers (HA-NF), MSCs spread in random directions while on the aligned nanofibers (A-NF) and HA-containing aligned nanofibers (HA-A-NF), MSCs elongated along the nanofibers. Cell filopodia extending along the fibers could be observed on all types of nanofibrous scaffolds. | ||
| Fig. 2 SEM images of MSCs after 7 days of culture on (A and B) NF, (C and D) A-NF, (E and F) HA-NF, and (G and H) HA-A-NF. Red arrows denote the orientation of nanofibers. | ||
Differentiation of MSCs on PHBV-based nanofibrous scaffolds
HA has been recently reported osteoinductive.24 In our study, the osteoinductive property of HA was found by in vitro investigating the osteogenic differentiation of MSCs on various nanofibrous scaffolds without any additional osteogenic factors.ALP expression of cells was tested by BCIP/NBT kit after 7 days of culture on different nanofibers. Optical microscopy images (Fig. 3) shows that ALP expression on HA-containing PHBV nanofibers (both random-oriented and aligned nanofibers) was significantly higher than on PHBV nanofibers without HA. The quantified ALP activity on PHBV/HA nanofibers was twice higher than that on PHBV. Fig. 3A also shows that MSCs elongated along the nanofibers on the aligned A-NF and HA-A-NF surfaces.
The immunostaining and quantified surface coverage results suggest that the OCN expression on HA-NF and HA-A-NF was significantly higher than that on NF and A-NF after 7 days and 14 days of culture (Fig. 4), which was in agreement with the case of ALP expression. In addition, more OCN expression was observed on nanofibrous scaffolds at day 14.
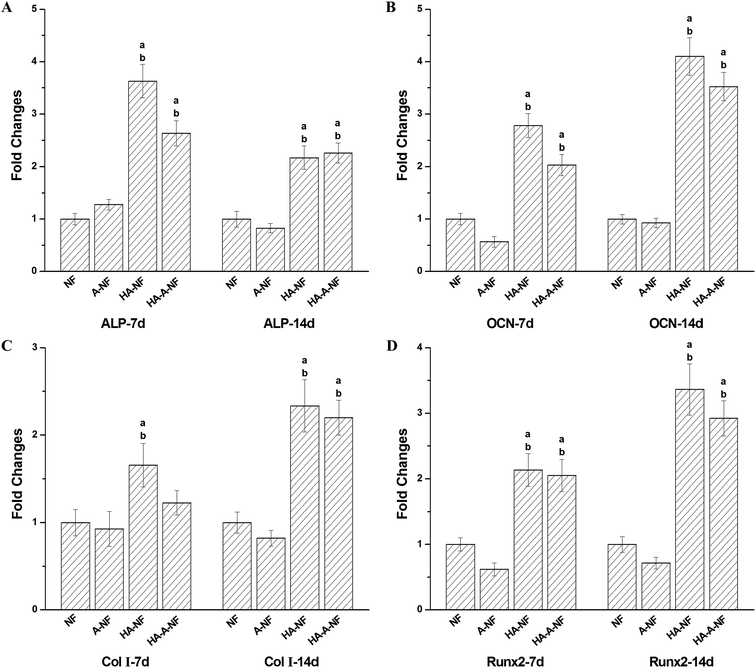
Furthermore, we studied gene expression of ALP, OCN, Col I, and Runx2 by quantitative real time PCR analysis after 7 and 14 days of culture. The analysis data are shown in Fig. 5. Compared to NF and A-NF, the level of ALP, OCN, Col I and Runx2 expression were upregulated on HA-NF and HA-A-NF at day 7 and day 14. The expression level of ALP was lower at day 14 but higher at day 7. Whereas the expression level of OCN, Col I, and Runx2 of cells cultured on HA-containing nanofibers for 14 days were significantly higher than those cultured for 7 days (fold changes > 2, p < 0.05). However, no significant difference of gene expression has been observed between random-oriented and aligned nanofibrous scaffolds.
ALP is considered as a marker for early osteoblast differentiation.39 Higher ALP expression was found on PHBV/HA scaffolds compared to that on PHBV scaffolds regardless of the fiber orientation. This indicates that HA plays an important role in stimulating the osteogenic differentiation of MSCs at early stage. OCN expression appeared late, accompanied with mineralization.27 The positive OCN expression on PHBV/HA scaffolds and on PHBV scaffolds verify that the scaffolds loaded with HA nanoparticles exhibit significant osteoinductivity. Col I is a major protein in bone and a biochemical marker in osteoblast differentiation.40 Runx2 is an essential transcription factor for osteoblast differentiation.27 The fact that upregulation of the expression of these genes demonstrates that the loading of HA nanoparticles can promote the osteoblast differentiation.
Mechanisms of MSCs differentiation into osteoblasts on PHBV/HA nanofibrous scaffolds
PHBV/HA nanofibrous scaffolds could significantly induce the osteogenic differentiation of MSCs. However, the biological mechanisms are still not well understood. Several signaling pathways are known to play important roles in osteoblast differentiation, including Wnt/β-catenin, BMP-Smad, and MAPK signaling pathway. We investigated the role of those three pathways in osteogenic differentiation on PHBV/HA scaffolds through immunofluorescence staining after 3 days and 7 days of cell seeding.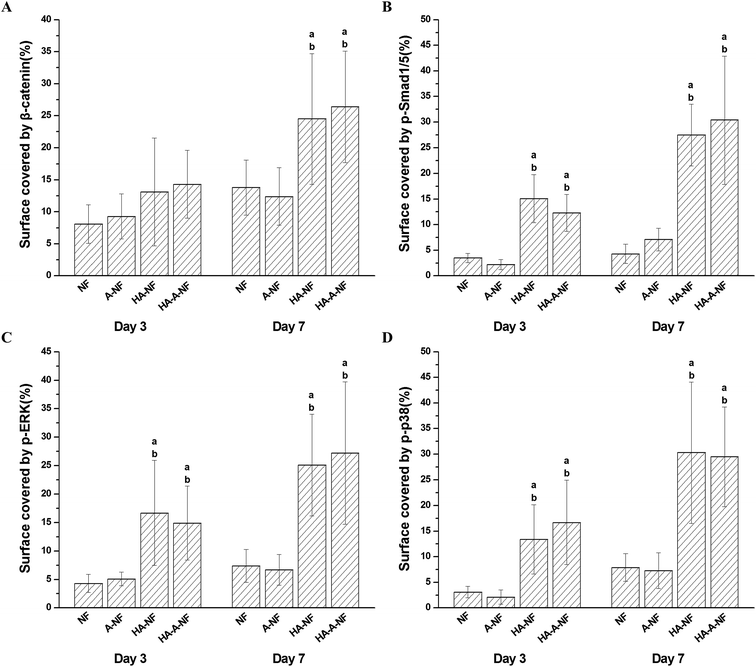 | ||
| Fig. 6 Quantification of the surface coverage by expressed β-catenin (A), p-Smad1/5 (B), p-ERK (C) and p-p38 (D). Statistical significance: (a) p < 0.05 compared to NF, (b) p < 0.05 compared to A-NF. | ||
Wnt/β-catenin pathway is important in regulating cell growth, differentiation, function, and death related to bone biology.41 The Wnt/β-catenin signaling pathway is stimulated by Wnt proteins which bind to the FZD/LRP5/6 complex at the cell surface. Signals are transduced through the proteins Disheveled, Axin, and Frat-1, which leads to the inhibition of glycogen synthase kinase 3 (GSK3). As a result, GSK3 is unable to phosphorylate β-catenin, and thus β-catenin could accumulate in the cytoplasm and translocate into the nucleus to activate target genes.27,42 In this study, a higher level of β-catenin expression in nucleus was found when MSCs were cultured on PHBV/HA scaffolds compared to that on PHBV scaffolds (Fig. S2†) suggesting that HA in electrospun nanofibrous scaffolds activated the Wnt/β-catenin signaling pathway in MSCs.
Bone morphogenetic proteins (BMPs), a member of the transforming growth factor beta (TGF beta) superfamily, are multi-functional growth factors. Bone morphogenetic protein-2 (BMP-2) is a powerful inducer of bone formation. Previous studies have demonstrated that the cytoplasmic signaling molecules, Smad1 and Smad5, play key roles in mediating the osteogenic action of BMP-2.32 Upon stimulation of BMP-2, Smad1/Smad5 is activated and forms a complex with the effector Smad4.33,34 Subsequently, a complex of Smad1/Smad4 or Smad5/Smad4 is transferred to the nucleus and regulate transcription of Runx2 to enhance osteogenic differentiation.32 In our study, with the addition of HA into the nanofibrous scaffolds, more p-Smad1/5 proteins were expressed in nucleus, demonstrating that the BMP-Smad signaling pathway is activated. The expression of p-Smad1/5 protein is complete after 7 days of cell seeding compared to that when cell cultured for 3 days.
Mitogen-activated protein kinases (MAPKs) regulate diverse cellular programs including embryogenesis, proliferation, differentiation and apoptosis based on cues derived from the cell surface, the metabolic state, and microenvironment of the cell.43 In mammalian cells, three MAPK families have been clearly characterized: namely extracellular signal-regulated kinases 1 and 2 (ERK1/2), c-Jun N-terminal kinase and p38 families. The MAPK pathways involving a series of protein kinase cascades play a critical role in regulation of cell proliferation. In our study, we mainly focused on the ERK1/2 and p38. ERK1/2 and p38 have been the best characterized kinases involved in MAPK signaling pathway. The activation of ERK1/2 and p38 is closely related to osteogenic differentiation of stem cells.44 In this study, we demonstrated that activation of ERK1/2 and p38 occurred on HA-NF and HA-A-NF scaffolds is more significant than that on NF and A-NF scaffolds. Our data also support that the phosphorylated ERK1/2 and p38 participate in osteoblast differentiation induced by scaffolds with HA.
Conclusions
In this study, we prepared PHBV and PHBV/HA nanofibrous scaffolds with different orientations. Upon the addition of HA to scaffolds, the expression level of osteogenic markers is significantly higher than that on PHBV scaffolds. The study shows that the introduction of HA nanoparticles accelerates osteogenic differentiation of MSCs better than PHBV without HA. However, no significant difference has been observed between random-oriented and aligned nanofibrous scaffolds. This study indicates that osteogenic differentiation of MSCs induced on HA-containing PHBV nanofibrous scaffolds involves Wnt/β-catenin, BMP-Smad, and MAPK (ERK1/2 and p38) signaling pathways. Further studies are needed to decipher details of the three signaling pathways. Promisingly, the elucidation of interaction between electrospun nanofibrous scaffolds and mesenchymal stem cells provides a guidance for design of desirable biomaterial scaffolds, which is essentially useful for the further application in bone tissue engineering and regeneration.Acknowledgements
This work was financially supported by the National Science Foundation of China (No. 31340049) and National High Technology Research & Development Program of China (2015AA020502).References
- D. M. Fan, G. R. Akkaraju, E. F. Couch, L. T. Canham and J. L. Coffer, Nanoscale, 2011, 3, 354–361 RSC.
- S. Pina, J. M. Oliveira and R. L. Reis, Adv. Mater., 2015, 27, 1143–1169 CrossRef CAS PubMed.
- Y. X. Luo, A. Lode, C. T. Wu, J. Chang and M. Gelinsky, ACS Appl. Mater. Interfaces, 2015, 7, 6541–6549 CAS.
- J. J. Li, N. Kawazoe and G. P. Chen, Biomaterials, 2015, 54, 226–236 CrossRef CAS PubMed.
- L. G. Xia, K. L. Lin, X. Q. Jiang, B. Fang, Y. J. Xu, J. Q. Liu, D. L. Zeng, M. L. Zhang, X. L. Zhang, J. Chang and Z. Y. Zhang, Biomaterials, 2014, 35, 8514–8527 CrossRef CAS PubMed.
- S. W. Crowder, D. Prasai, R. Rath, D. A. Balikov, H. Bae, K. I. Bolotin and H. J. Sung, Nanoscale, 2013, 5, 4171–4176 RSC.
- A. M. Ross, Z. X. Jiang, M. Bastmeyer and J. Lahann, Small, 2012, 8, 336–355 CrossRef CAS PubMed.
- W. J. Lu, J. S. Sun and X. Y. Jiang, J. Mater. Chem. B, 2014, 2, 2369–2380 RSC.
- B. Sun, Y. Z. Long, H. D. Zhang, M. M. Li, J. L. Duvail, X. Y. Jiang and H. L. Yin, Prog. Polym. Sci., 2014, 39, 862–890 CrossRef CAS.
- V. Leszczak, L. W. Place, N. Franz, K. C. Popat and M. J. Kipper, ACS Appl. Mater. Interfaces, 2014, 6, 9328–9337 CAS.
- Y. D. Liu, H. T. Cui, X. L. Zhuang, Y. Wei and X. S. Chen, Acta Biomater., 2014, 10, 5074–5080 CrossRef CAS PubMed.
- C. Spadaccio, A. Rainer, M. Trombetta, G. Vadala, M. Chello, E. Covino, V. Denaro, Y. Toyoda and J. A. Genovese, Ann. Biomed. Eng., 2009, 37, 1376–1389 CrossRef PubMed.
- E. K. Yim, S. W. Pang and K. W. Leong, Exp. Cell Res., 2007, 313, 1820–1829 CrossRef CAS PubMed.
- N. Chevallier, F. Anagnostou, S. Zilber, G. Bodivit, S. Maurin, A. Barrault, P. Bierling, P. Hernigou, P. Layrolle and H. Rouard, Biomaterials, 2010, 31, 270–278 CrossRef CAS PubMed.
- C. P. Zheng, J. S. Wang, Y. N. Liu, Q. Q. Yu, Y. Liu, N. Deng and J. Liu, Adv. Funct. Mater., 2014, 24, 6872–6883 CrossRef CAS.
- M. Rodrigues, H. Blair, L. Stockdale, L. Griffith and A. Wells, Stem Cells, 2013, 31, 104–116 CrossRef CAS PubMed.
- W. Chen, L. Kaili, C. Jiang and S. Jiao, Biomaterials, 2013, 34, 64–77 CrossRef PubMed.
- J. L. Wang, M. Y. Yang, Y. Zhu, L. Wang, A. P. Tomsia and M. Chuanbin, Adv. Mater., 2014, 26, 4961–4966 CrossRef CAS PubMed.
- M. J. Dalby, N. Gadegaard, R. Tare, A. Andar, M. O. Riehle, P. Herzyk, C. D. W. Wilkinson and R. O. C. Oreffo, Nat. Mater., 2007, 6, 997–1003 CrossRef CAS PubMed.
- R. Z. LeGeros, Chem. Rev., 2008, 108, 4742–4753 CrossRef PubMed.
- K. H. Park, K. Na, S. W. Kim, B. K. Sun, D. G. Woo, H. N. Yang and H. M. Chung, Biomaterials, 2007, 28, 2631–2637 CrossRef PubMed.
- E. Tsiridis, A. Bhalla, A. Zubier, N. Gurav, M. Heliotis, S. Deb and L. DiSilvio, Injury, 2006, 37, S25–S32 CrossRef PubMed.
- L. R. McCabe, R. Shu, R. McMullen and M. J. J. Baumann, J. Biomed. Mater. Res., Part A, 2003, 67A, 1196–1204 CrossRef PubMed.
- A. Polini, D. Pisignano, M. Parodi, R. Quarto and S. Scaglione, PLoS One, 2011, 6, e26211 CAS.
- L. X. Lü, Y. Y. Wang, X. Mao, Z. D. Xiao and N. P. Huang, Biomed. Mater., 2012, 7, 015002 CrossRef PubMed.
- L. X. Lü, X. F. Zhang, Y. Y. Wang, L. Ortiz, X. Mao, Z. L. Jiang, Z. D. Xiao and N. P. Huang, ACS Appl. Mater. Interfaces, 2013, 5, 319–330 Search PubMed.
- W. Huang, S. Y. Yang, J. Z. Shao and Y. P. Li, Front. Biosci., Landmark Ed., 2007, 12, 3068–3092 CrossRef CAS.
- F. Liu, S. Kohlmeier and C. Y. Wang, Cell Signal, 2008, 20, 999–1009 CrossRef CAS PubMed.
- J. J. Guan, J. Y. Zhang, S. C. Guo, H. Y. Zhu, Z. Z. Zhu, H. Y. Li, Y. Wang, C. Q. Zhang and J. Chang, Biomaterials, 2015, 55, 1–11 CrossRef CAS PubMed.
- X. Y. Ma, Y. F. Feng, Z. S. Ma, X. Li, J. Wang, L. Wang and W. Lei, Biomaterials, 2014, 35, 7259–7270 CrossRef CAS PubMed.
- C. T. Wu, L. G. Xia, P. P. Han, M. C. Xu, B. Fang, J. C. Wang, J. Chang and Y. Xiao, Carbon, 2015, 93, 116–129 CrossRef CAS.
- R. Nishimura, K. Hata, S. E. Harris, F. Ikeda and T. Yoneda, Bone, 2002, 31, 303–312 CrossRef CAS PubMed.
- C. H. Heldin, K. Miyazono and P. ten Dijke, Nature, 1997, 390, 465–471 CrossRef CAS PubMed.
- J. Massague, Cell, 1996, 85, 947–950 CrossRef CAS PubMed.
- W. Wang, Q. Liu, Y. M. Zhang and L. Z. Zhao, Acta Biomater., 2014, 10, 3705–3715 CrossRef CAS PubMed.
- X. Mao, C. L. Chu, Z. Mao and J. J. Wang, Tissue Cell, 2005, 37, 349–357 CrossRef CAS PubMed.
- C. A. Schneider, W. S. Rasband and K. W. Eliceiri, Nat. Methods, 2012, 9, 671–675 CrossRef CAS PubMed.
- V. Milleret, T. Hefti, H. Hall, V. Vogel and D. Eberli, Acta Biomater., 2012, 8, 4349–4356 CrossRef CAS PubMed.
- M. P. Whyte, Endocr. Rev., 1994, 15, 439–461 CAS.
- S. Viguet-Carrin, P. Garnero and P. D. Delmas, Osteoporosis Int., 2006, 17, 319–336 CrossRef CAS PubMed.
- J. J. Westendorf, R. A. Kahler and T. M. Schroeder, Gene, 2004, 341, 19–39 CrossRef CAS PubMed.
- W. Wang, L. Z. Zhao, Q. L. Ma, Q. T. Wang, P. K. Chu and Y. M. Zhang, Biomaterials, 2012, 33, 7993–8002 CrossRef CAS PubMed.
- M. Raman, W. Chen and M. H. Cobb, Oncogene, 2007, 26, 3100–3112 CrossRef CAS PubMed.
- S. Gallea, F. Lallemand, A. Atfi, G. Rawadi, V. Ramez, S. Spinella-Jaegle, S. Kawai, C. Faucheu, L. Huet, R. Baron and S. Roman-Roman, Bone, 2001, 28, 491–498 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c5ra22578g |
| This journal is © The Royal Society of Chemistry 2016 |