Targeting human telomeric G-quadruplex DNA with curcumin and its synthesized analogues under molecular crowding conditions†
Abstract
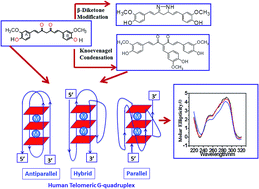
The formation of telomeric G-quadruplexes has been shown to inhibit telomerase activity. Indeed, a number of small molecules capable of π-stacking with G-tetrads have shown the ability to inhibit telomerase activity through the stabilization of G-quadruplexes. Curcumin displays a wide spectrum of medicinal properties ranging from anti-bacterial, anti-viral, anti-protozoal, anti-fungal and anti-inflammatory to anti-cancer activity. We have investigated the interactions of curcumin and its structural analogues with the human telomeric sequence AG3(T2AG3)3 under molecular crowding conditions. Experimental studies indicated the existence of a AG3(T2AG3)3/curcumin complex with binding affinity of 0.72 × 106 M−1 under molecular crowding conditions. The results from UV-visible absorption spectroscopy, a fluorescent TO displacement assay, circular dichroism and molecular docking studies, imply that curcumin and their analogues interact with G-quadruplex DNA via groove binding. While other analogs of curcumin studied here bind to G-quadruplexes in a qualitatively similar manner their affinities are relatively lower in comparison to curcumin. The Knoevenagel condensate, a methoxy-benzylidene derivative of curcumin, also exhibited significant binding to G-quadruplex DNA, although with two times decreased affinity. Our study establishes the potential of curcumin as a promising natural product for G-quadruplex specific ligands.

 Please wait while we load your content...
Please wait while we load your content...