Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Protein-specific localization of a rhodamine-based calcium-sensor in living cells†
Marcel
Best
a,
Isabel
Porth
a,
Sebastian
Hauke
b,
Felix
Braun
c,
Dirk-Peter
Herten
c and
Richard
Wombacher
*a
aRuprecht-Karls-University Heidelberg, Institute for Pharmacy und Molecular Biotechnology, Im Neuenheimer Feld 364, 69120 Heidelberg, Germany. E-mail: wombacher@uni-heidelberg.de; Fax: +49(0)621 544879; Tel: +49 (0)621 544879
bEuropean Molecular Biology Laboratory, Cell Biology and Biophysics Unit, Meyerhofstraße 1, 69117 Heidelberg, Germany
cRuprecht-Karls-University Heidelberg, Institute for Physical Chemistry, Im Neuenheimer Feld 267, 69120 Heidelberg, Germany
First published on 7th April 2016
Abstract
A small synthetic calcium sensor that can be site-specifically coupled to proteins in living cells by utilizing the bio-orthogonal HaloTag labeling strategy is presented. We synthesized an iodo-derivatized BAPTA chelator with a tetramethyl rhodamine fluorophore that allows further modification by Sonogashira cross-coupling. The presented calcium sensitive dye shows a 200-fold increase in fluorescence upon calcium binding. The derivatization with an aliphatic linker bearing a terminal haloalkane-function by Sonogashira cross-coupling allows the localization of the calcium sensor to Halo fusion proteins which we successfully demonstrate in in vitro and in vivo experiments. The herein reported highly sensitive tetramethyl rhodamine based calcium indicator, which can be selectively localized to proteins, is a powerful tool to determine changes in calcium levels inside living cells with spatiotemporal resolution.
Introduction
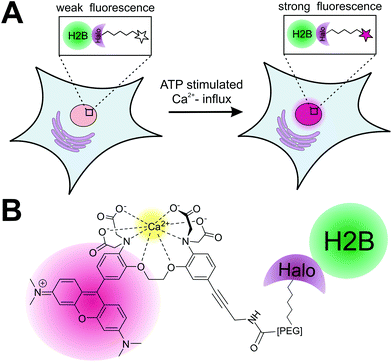
Calcium plays a key role as second messenger in living cells and both the spatial and temporal changes of its endogenous concentration [Ca2+]i have a significant effect on signaling transduction in excitable and non-excitable cells.1 Different approaches to measure calcium fluctuations in live cell experiments have been developed: the genetically encoded calcium indicators (GECIs)2 utilize calcium sensitive fluorescent proteins or fusion proteins with the so-called “cameleons”3 and “camgaroos”4 as well known examples. GECIs have the advantage that they can be genetically targeted to any protein of interest (POI), but they lack fast response to quick changes of [Ca2+]i in living cells.5 Alternatively, small synthetic calcium indicators6 as Indo-1/Fura 1–3,7 Rhod-1 + 2/Fluo1–38 and Fluo-49 have been developed for calcium imaging in live cells and organisms with the advantage of being much smaller in size compared to fluorescent proteins and therefore less disruptive. Furthermore they can be chemically modified to adapt both the chemical and physical properties to the experimental needs. Over the last decades, numerous small synthetic calcium sensors have been developed and used for calcium measurements in living systems.10 However, their application is limited to the measurements of [Ca2+]i in the cytosolic environment as they lack the ability to be specifically localized within the cells. Recently, protein labeling strategies for site-specific localization of small synthetic calcium sensors to a POI utilizing the FlAsH-5 and SNAP-tag protein11 labeling strategy have been reported. Whereas the calcium sensor for FlAsH-tag applications is based on fluorescein combined with the calcium binding APTRA (2-aminophenol-N,N,O-tri-acetat)12 motive, the SNAP-tag based approach utilizes a BODIPY (boron dipyrromethene)13 conjugated to the well-known BAPTA (1,2 bis-(ortho-aminophenoxy)ethane-N,N,N′,N′-tetra-acetate)14 calcium-ion chelator. The fluorophores used in these studies need to be excited in the blue range of the spectrum, whereas calcium indicators for protein localization, which can be excited at longer wavelengths, have not been reported yet. Here we introduce a new sensitive synthetic calcium indicator based on the fluorophore rhodamine that can be specifically localized to a POI in living cells utilizing the Halo-tag protein labeling strategy (Fig. 1).15 The new calcium sensor shows a >200-fold increase in fluorescence upon calcium binding and can be excited in the green spectrum (Exmax = 552 nm; Emmax = 580 nm). In a first proof of principle, we demonstrate the specific localization of the calcium sensitive fluorophore to a POI by labeling of a purified Halo fusion protein. For live cell application of the presented strategy, we performed specific labeling of nucleus localized Halo tagged histone protein (H2B) with the calcium sensor. We show that the H2B localized calcium sensor can be used to monitor nuclear calcium changes in living cells. The herein presented localizable rhodamine derived calcium indicator can be used to measure rapid spatiotemporal changes of [Ca2+]i in living cells.Results and discussion
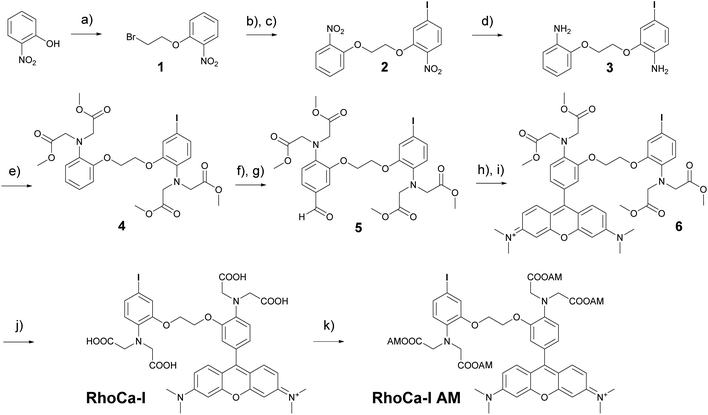
The considerations for the development of the calcium sensor were based on the high photostability and brightness of the fluorophore tetramethyl rhodamine (TAMRA) in aqueous systems16 and the high selectivity of the BAPTA chelator in the presence of magnesium ions.14 The commercially available Rhod-2 calcium indicator8 which is based on these both chemical components has a Kd-value in the nanomolar range and became a standard tool for calcium measurements in living cells.8,17 However, the mitochondrial localization18 of rhodamine derivatives limits its application. Therefore we sought to develop a calcium indicator with the advantageous photophysical properties of a rhodamine, the excellent calcium selectivity and sensitivity of BAPTA and an additional functionality for further chemical modification. The additional functionality will then be used to attach a haloalkane which serves as a substrate for the HaloTag and enables the covalent attachment to Halo fusion proteins. The highly selective protein localization will then suppress the unwanted mitochondrial localization of the rhodamines. For this reason, we first synthesized a BAPTA core with an iodine modification that allows the selective functionalization by Sonogashira cross-coupling in a final step. Scheme 1 outlines the synthesis of RhoCa-I, the iodine modified BAPTA calcium chelator coupled to rhodamine. First ortho-nitrophenol was alkylated with 1,2-dibromoethane under basic conditions to yield 1. Further alkylation with 5-iodo-2-nitrophenol and reduction of the nitro functions resulted in the diamine 3. Alkylation with methyl bromoacetate followed by Vilsmeier–Haack formylation yielded 5 which was then converted to the BAPTA rhodamine 6 by treatment of 5 with 3-(dimethylamino)-phenol under acetic conditions and subsequent oxidation.19 Finally, the methyl esters of 6 were saponified to obtain the calcium sensitive RhoCa-I. To render the newly synthesized calcium indicator cell permeable for its application in living cells, 6 was converted to the respective acetoxymethyl ester (AM-ester) RhoCa-I AM.Calcium titration (in an EGTA-buffered solution with free calcium concentrations ranging from 0 μM to 39 μM, Fig. S1A,†λex = 552 nm) of RhoCa-I revealed a 215-fold fluorescence enhancement upon calcium binding and a Kd-value of 2.50 ± 0.17 μM (Fig. S1B†). The relative quantum yield20 was determined to be 20.9 ± 0.6% at [Ca2+]free = 39 μM and 0.07 ± 0.01% at [Ca2+]free = 0 μM in reference to Rhodamine B.21 As expected, the Kd-value of 2.5 μM is significantly higher than the Kd-value of Rhod-2 (∼570 nM),10 which has a methyl group in para-position to the alkyl amine groups. We assume that that the higher Kd-value is due to the iodine substituent as electron withdrawing groups are known to decrease the calcium affinity of the BAPTA chelator.22
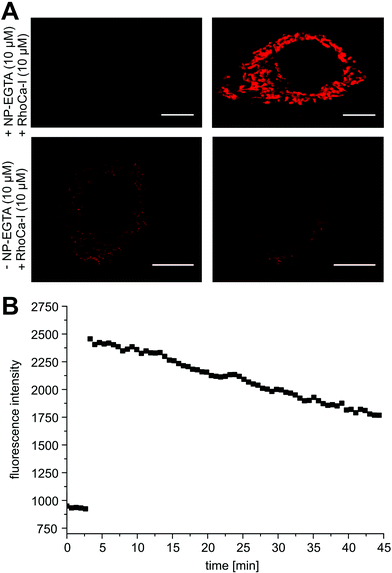
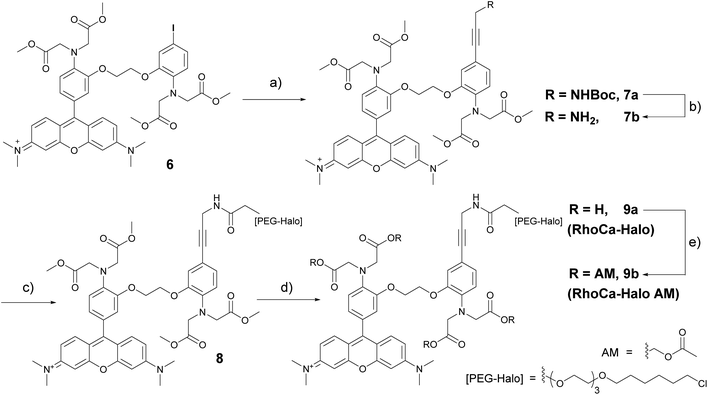
Next, we evaluated the calcium sensitivity of RhoCa-I in living cells. Therefore, HeLa cells were incubated with the AM ester of the photo-cleavable o-nitrophenyl EGTA (NP-EGTA23 AM) which has a high affinity for calcium ions (Kd = 80 nM). One of the chelating moieties can be cleaved off by UV-light radiation, thereby lowering the Ca2+ affinity by a factor of more than 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000. To release calcium ions from cellular internal stores, the HeLa cells were stimulated with 100 μM ATP24 followed by the incubation with 10 μM of the AM ester25 of Rho-Ca-I (RhoCa-I AM). As presented in Fig. 2, the calcium sensor shows a significant increase of fluorescence intensity after calcium release from NP-EGTA after UV-radiation. As mentioned before, rhodamine dyes tend to localize to mitochondria what we also observed for the calcium sensor RhoCa-I (Fig. 2A). To overcome that limitation and to actively choose the subcellular site for localization, we further synthesized the calcium sensor with a haloalkane functionality for protein labeling utilizing the bioorthogonal HaloTag-strategy (RhoCa-Halo, Scheme 2). First we derivatized 6 in a Sonogashira cross coupling reaction with Boc-protected propargylamine. After deprotection of the amine, the haloalkane functionality for protein localization was introduced by peptide bond formation. Finally, the methyl esters were converted to the free acids by saponification to obtain RhoCa-Halo.
000. To release calcium ions from cellular internal stores, the HeLa cells were stimulated with 100 μM ATP24 followed by the incubation with 10 μM of the AM ester25 of Rho-Ca-I (RhoCa-I AM). As presented in Fig. 2, the calcium sensor shows a significant increase of fluorescence intensity after calcium release from NP-EGTA after UV-radiation. As mentioned before, rhodamine dyes tend to localize to mitochondria what we also observed for the calcium sensor RhoCa-I (Fig. 2A). To overcome that limitation and to actively choose the subcellular site for localization, we further synthesized the calcium sensor with a haloalkane functionality for protein labeling utilizing the bioorthogonal HaloTag-strategy (RhoCa-Halo, Scheme 2). First we derivatized 6 in a Sonogashira cross coupling reaction with Boc-protected propargylamine. After deprotection of the amine, the haloalkane functionality for protein localization was introduced by peptide bond formation. Finally, the methyl esters were converted to the free acids by saponification to obtain RhoCa-Halo.
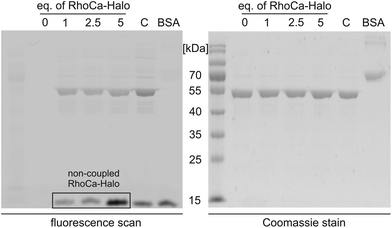
Next, we evaluated the suitability of the newly synthesized RhoCa-Halo for protein labeling by an in vitro experiment. RhoCa-Halo was incubated with different concentrations of a purified HaloTag fused to green fluorescent protein (GFP-Halo) and the covalent attachment of the probe to the model protein GFP-Halo was visualized by SDS-PAGE. Fig. 3 shows both the in-gel fluorescence scan of the SDS-gel and the Coomassie stain. From the fluorescence-scan it becomes clear, that GFP-Halo gets efficiently labeled with RhoCa-Halo. BSA served as negative control and did not show any labeling by the calcium sensor. As expected, the positive control with a non-calcium sensitive Halo-rhodamine, a commercially available substrate for the HaloTag (see ESI† for structure) resulted in labeling of the Halo fusion protein. When fluorescence emission was recorded at λem = 580 nm, we still detected fluorescence from GFP. Therefore, we chose to measure the fluorescence at λem = 670 nm, a range where GFP does not emit fluorescence light when excited, to selectively record fluorescence from RhoCa-Halo. Fig. 3 shows no fluorescence for the sample containing only GFP-Halo (c (RhoCa-Halo = 0 μM)), which shows that the detected fluorescence only accounts for RhoCa-Halo. The detected fluorescence of the calcium sensor is due to calcium traces in the SDS running buffer and the increased calcium affinity at higher pH – values26 (pH = 8.8 in the resolving gel).
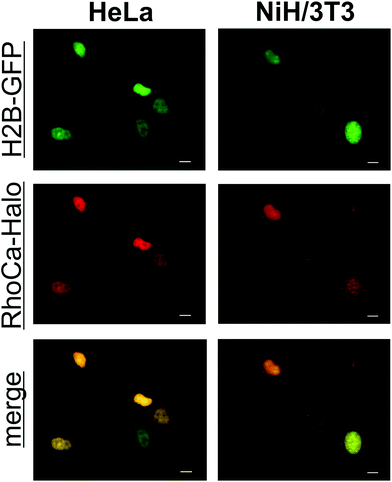
To determine the spectral properties of the calcium sensor, covalently attached to a protein, we incubated GFP-Halo with 5-fold excess of RhoCa-Halo for two hours and subsequently removed the excess of RhoCa-Halo by spin-filtration. The labeled protein was then used for titration with calcium to determine the increase of fluorescence upon calcium binding. An average fluorescence turn-on upon calcium binding of 76-fold with a quantum yield of 14.0 ± 1.6% at [Ca2+]free = 39 μM and 0.16 ± 0.01% at [Ca2+]free = 0 μM was determined (Fig. S2A,†λex = 552 nm). In a separate experiment we determined a 240-fold fluorescence enhancement upon calcium binding for the free RhoCa-Halo (not bound to the protein). The observed lower fluorescence enhancement for the protein bound sensor might be caused by interactions between the calcium sensor and protein. As it is well known that the environment of a fluorophore has strong impact on its photo-physical properties. The Kd-value of the protein bound sensor was found to be 1.51 ± 0.13 μM (Fig. S2B†) which makes it well suitable for investigations of calcium oscillations ([Ca2+]i up to 1 μM)27 and calcium sparks ([Ca2+]i up to 10 μM) in biological systems.28 After having established a method for the site specific labeling of the model protein GFP-Halo with the calcium-sensitive probe RhoCa-Halo, the newly developed calcium sensor found application in live cell experiments. Therefore, NIH/3T3 and HeLa cells were transiently transfected with H2B-GFP and Halo-H2B and incubated with the according AM-Ester (RhoCa-Halo AM). Epifluorescent microscopy indicated successful labeling of the nucleus with the localizable calcium sensor in both cell lines (Fig. 4).
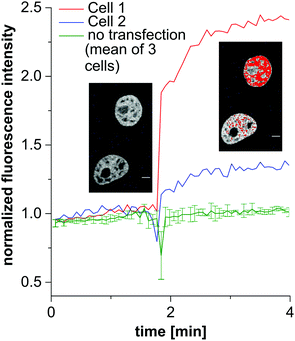
Next, we investigated the behavior of the calcium sensor in live cells after stimulation with ATP, inducing cytosolic influx of calcium ions from endogenous stores. Therefore, HeLa cells were transfected with Halo-H2B and incubated with RhoCa-Halo AM. Non-transfected cells were distinguished from the transfected cells by confocal microscopy. Cytosolic influx of free calcium ions was then stimulated with 100 μM ATP and a noticeable increase of fluorescence was observed in the nucleus of the transfected and labeled cells directly upon addition of ATP (Fig. 5). The non-transfected cells did not show any detectable increase of fluorescence in the nucleus. This clearly demonstrated the potential of the fluorescent sensor RhoCa-Halo as a localizable calcium sensor to monitor spatiotemporal changes of free calcium ions in live cell experiments.
Conclusion
In conclusion, we developed a new localizable rhodamine based calcium sensor which can be applied in live cell imaging. RhoCa-Halo is a localizable highly sensitive calcium sensor which exhibits an increase of fluorescence upon calcium-binding, thereby enabling monitoring spatiotemporal changes of calcium concentrations in living cells. We demonstrated its application in in vitro and live cell experiments. Moreover, RhoCa-I serves as a general building block that can be readily derivatized by Sonogashira cross-coupling to introduce alternative functionalities to the rhodamine calcium sensor. RhoCa-Halo is the first localizable small molecule based calcium sensor with excitation in the green spectrum and represents a valuable addition to the toolbox of calcium imaging. The sensor can be combined with other sensors which are spectrally separated. Furthermore, the application of the HaloTag labeling strategy allows multi-color calcium imaging of different organelles in living cells when combined with the orthogonal SNAP-tag based localizable calcium sensor,11 an Indo-1 derivative with excitation in the near-UV (Exmax = 350 nm; Emmax = 475 nm). The specific localization of a fluorescent calcium sensor to any POI has great potential to study the spatiotemporal regulation of calcium as a second messenger. Furthermore we would like to point out that the high photo-stability and specific photo-physical properties of the rhodamine moiety makes the newly developed RhoCa-Halo probe a potential candidate for application in super-resolution microscopy.29Acknowledgements
This work was supported by the Deutsche Forschungs-Gemeinschaft DFG (SPP1623, WO 1888/1-2). We thank the Nikon Imaging Center Heidelberg for access to fluorescence microscopes. We gratefully acknowledge Carsten Schultz (EMBL) for access to confocal microscopes and for providing NP-EGTA AM. We thank the Department of Infectious Diseases (Medical Biology and Hygiene, group of Konrad Bode) for access to the in-gel fluorescence scanner. We acknowledge the support from the Inorganic Chemical Institute of the University of Heidelberg (Prof. Dr Markus Enders and Beate Termin) for providing access to the NMR facility for spectroscopic measurements. We thank Mathis Baalmann, Achim Wieczorek and Michael Heimes for scientific support and Heiko Rudy for technical support.Notes and references
- D. E. Clapham, Cell, 2007, 131, 1047–1058 CrossRef CAS PubMed.
- (a) J. Zhang, R. E. Campbell, A. Y. Ting and R. Y. Tsien, Nat. Rev. Mol. Cell Biol., 2002, 3, 906–918 CrossRef CAS PubMed; (b) M. Mank and O. Griesbeck, Chem. Rev., 2008, 108, 1550–1564 CrossRef CAS PubMed; (c) T. Rose, P. M. Goltstein, R. Portugues and O. Griesbeck, Front. Mol. Neurosci., 2014, 7, 88 Search PubMed.
- A. Miyawaki, J. Llopis, R. Heim, J. M. McCaffery, J. A. Adams, M. Ikura and R. Y. Tsien, Nature, 1997, 388, 882–887 CrossRef CAS PubMed.
- O. Griesbeck, G. S. Baird, R. E. Campbell, D. A. Zacharias and R. Y. Tsien, J. Biol. Chem., 2001, 276, 29188–29194 CrossRef CAS PubMed.
- O. Tour, S. R. Adams, R. A. Kerr, R. M. Meijer, T. J. Sejnowski, R. W. Tsien and R. Y. Tsien, Nat. Chem. Biol., 2007, 3, 423–431 CrossRef CAS PubMed.
- M. Oheim, M. van 't Hoff, A. Feltz, A. Zamaleeva, J. M. Mallet and M. Collot, Biochim. Biophys. Acta, 2014, 1843, 2284–2306 CrossRef CAS PubMed.
- G. Grynkiewicz, M. Poenie and R. Y. Tsien, J. Biol. Chem., 1985, 260, 3440–3450 CAS.
- A. Minta, J. P. Kao and R. Y. Tsien, J. Biol. Chem., 1989, 264, 8171–8178 CAS.
- K. R. Gee, K. A. Brown, W. N. Chen, J. Bishop-Stewart, D. Gray and I. Johnson, Cell Calcium, 2000, 27, 97–106 CrossRef CAS PubMed.
- R. M. Paredes, J. C. Etzler, L. T. Watts, W. Zheng and J. D. Lechleiter, Methods, 2008, 46, 143–151 CrossRef CAS PubMed.
- (a) M. Bannwarth, I. R. Correa, M. Sztretye, S. Pouvreau, C. Fellay, A. Aebischer, L. Royer, E. Rois and K. Johnsson, ACS Chem. Biol., 2009, 4, 179–190 CrossRef CAS PubMed; (b) M. Kamiya and K. Johnsson, Anal. Chem., 2010, 82, 6472–6479 CrossRef CAS PubMed.
- B. Raju, E. Murphy, L. A. Levy, R. D. Hall and R. E. London, Am. J. Physiol., 1989, 256, C540–C548 CAS.
- I. J. Arroyo, R. R. Hu, G. Merino, B. Z. Tang and E. Pena-Cabrera, J. Org. Chem., 2009, 74, 5719–5722 CrossRef CAS PubMed.
- R. Y. Tsien, Biochemistry, 1980, 19, 2396–2404 CrossRef CAS PubMed.
- G. V. Los, L. P. Encell, M. G. McDougall, D. D. Hartzell, N. Karassina, C. Zimprich, M. G. Wood, R. Learish, R. F. Ohana, M. Urh, D. Simpson, J. Mendez, K. Zimmerman, P. Otto, G. Vidugiris, J. Zhu, A. Darzins, D. H. Klaubert, R. F. Bulleit and K. V. Wood, ACS Chem. Biol., 2008, 3, 373–382 CrossRef CAS PubMed.
- (a) C. Eggeling, J. Widengren, R. Rigler and C. A. M. Seidel, Anal. Chem., 1998, 70, 2651–2659 CrossRef CAS PubMed; (b) B. Hein, K. I. Willig, C. A. Wurm, V. Westphal, S. Jakobs and S. W. Hell, Biophys. J., 2010, 98, 158–163 CrossRef CAS PubMed.
- J. Qiu, Y. W. Tan, A. M. Hagenston, M. A. Martel, N. Kneisel, P. A. Skehel, D. J. A. Wyllie, H. Bading and G. E. Hardingham, Nat. Commun., 2013, 4 Search PubMed.
- (a) G. Hajnoczky, L. D. Robbgaspers, M. B. Seitz and A. P. Thomas, Cell, 1995, 82, 415–424 CrossRef CAS PubMed; (b) D. R. Trollinger, W. E. Cascio and J. J. Lemasters, Biochem. Biophys. Res. Commun., 1997, 236, 738–742 CrossRef CAS PubMed.
- B. Agnew, J. Beechem, K. Gee, R. Haugland, T. Steinberg and W. Patton, PCT/US2004/036968, 2004 Search PubMed.
- J. N. Demas and G. A. Crosby, J. Phys. Chem., 1971, 75, 991–1024 CrossRef.
- R. F. Kubin and A. N. Fletcher, J. Lumin., 1982, 27, 455–462 CrossRef.
- R. Pethig, M. Kuhn, R. Payne, E. Adler, T. H. Chen and L. F. Jaffe, Cell Calcium, 1989, 10, 491–498 CrossRef CAS PubMed.
- G. C. Ellis-Davies, Nat. Methods, 2007, 4, 619–628 CrossRef CAS PubMed.
- (a) A. Luckhoff and R. Busse, J. Cell. Physiol., 1986, 126, 414–420 CrossRef CAS PubMed; (b) F. A. Gonzalez, E. Rozengurt and L. A. Heppel, Proc. Natl. Acad. Sci. U. S. A., 1989, 86, 4530–4534 CrossRef CAS PubMed; (c) R. Charest, P. F. Blackmore and J. H. Exton, J. Biol. Chem., 1985, 260, 5789–5794 Search PubMed.
- D. G. Lambert, Calcium signaling protocols, Springer Science & Business Media, 2006 Search PubMed.
- U. Quast, A. M. Labhardt and V. M. Doyle, Biochem. Biophys. Res. Commun., 1984, 123, 604–611 CrossRef CAS PubMed.
- A. T. Harootunian, J. P. Kao, S. Paranjape and R. Y. Tsien, Science, 1991, 251, 75–78 CAS.
- M. B. Cannell and C. Soeller, J. Gen. Physiol., 1999, 113, 373–376 CrossRef CAS PubMed.
- (a) G. T. Dempsey, J. C. Vaughan, K. H. Chen, M. Bates and X. Zhuang, Nat. Methods, 2011, 8, 1027–1036 CrossRef CAS PubMed; (b) M. Heilemann, S. van de Linde, M. Schuttpelz, R. Kasper, B. Seefeldt, A. Mukherjee, P. Tinnefeld and M. Sauer, Angew. Chem., Int. Ed., 2008, 47, 6172–6176 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c6ob00365f |
| This journal is © The Royal Society of Chemistry 2016 |