Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Formation of target-specific binding sites in enzymes: solid-phase molecular imprinting of HRP†
J.
Czulak
*a,
A.
Guerreiro
*b,
K.
Metran
b,
F.
Canfarotta
c,
A.
Goddard
b,
R. H.
Cowan
d,
A. W.
Trochimczuk
a and
S.
Piletsky
b
aFaculty of Chemistry, Department of Polymer and Carbonaceous Materials, Wroclaw University of Technology, 27 Wyspianskiego Str., 50-373 Wroclaw, Poland. E-mail: joanna.czulak@pwr.edu.pl
bDepartment of Chemistry, College of Science and Engineering, University of Leicester, LE1 7RH, UK. E-mail: ag398@le.ac.uk
cMIP Diagnostics Ltd, Fielding Johnson Building, University of Leicester, Leicester, LE1 7RH, UK
dDepartment of Molecular and Cell Biology, College of Medicine, Biological Sciences and Psychology, University of Leicester, LE1 9HN, UK
First published on 28th April 2016
Abstract
Here we introduce a new concept for synthesising molecularly imprinted nanoparticles by using proteins as macro-functional monomers. For a proof-of-concept, a model enzyme (HRP) was cross-linked using glutaraldehyde in the presence of glass beads (solid-phase) bearing immobilized templates such as vancomycin and ampicillin. The cross-linking process links together proteins and protein chains, which in the presence of templates leads to the formation of permanent target-specific recognition sites without adverse effects on the enzymatic activity. Unlike complex protein engineering approaches commonly employed to generate affinity proteins, the method proposed can be used to produce protein-based ligands in a short time period using native protein molecules. These affinity materials are potentially useful tools especially for assays since they combine the catalytic properties of enzymes (for signaling) and molecular recognition properties of antibodies. We demonstrate this concept in an ELISA-format assay where HRP imprinted with vancomycin and ampicillin replaced traditional enzyme–antibody conjugates for selective detection of templates at micromolar concentrations. This approach can potentially provide a fast alternative to raising antibodies for targets that do not require high assay sensitivities; it can also find uses as a biochemical research tool, as a possible replacement for immunoperoxidase-conjugates.
Introduction
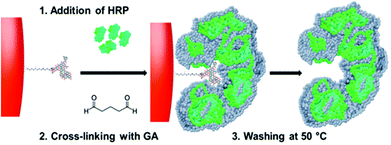
The routine laboratory use of immunoassays in clinical diagnostics, and agricultural, environmental as well as food and forensic industries largely relies on the specific interactions between antibodies and antigens to generate a quantifiable signal for the recognition and quantification of analytes.1,2 The arrival of monoclonal antibodies through the development of the hybridoma technique afforded assays with single epitope specificity and enhanced sensitivity.3 The competitive assay, of which the radioimmunoassay (RIA) can be considered an archetype, relies on the competition for antibody binding sites, between the free analyte and a radiolabelled form of the analyte. Non-competitive assays typically involve the formation of a ‘sandwich’ comprising of an antibody–antigen–antibody complex.4 One of the most popular methods of immunoassays is the enzyme-linked immunosorbent assay (ELISA), here the free analyte is added to the immobilised capture antibodies, and subsequently a secondary enzyme-labelled antibody (conjugate) is added. The enzyme is used as a reporter, catalysing a reaction which produces coloured products which are proportional to the analyte concentration.5,6 Although antibodies are the current benchmark biomolecules when it comes to molecular recognition, they do possess limitations. Antibodies are generally expensive and comparatively difficult to obtain, requiring often either live animals or hybridoma cell lines for their production. Due to their complex structure, large size with multiple domains, antibodies are usually susceptible to thermal degradation. Furthermore, special handling or/and costly production is required for making/storing efficient antibody–enzyme conjugates; this process often leads to adverse changes in the molecular recognition properties of the conjugate. As an alternative, molecular imprinting using cross-linked acrylic polymers can be used to produce synthetic receptors; these have been proven suitable as replacement for antibodies in the pseudo-immunoassay development.7–9 Molecularly imprinted polymers (MIPs) were incorporated into assays and used to determine the presence of theophylline, diazepam or vancomycin in serum with specificity and cross-reactivity profiles similar or superior to those of antibodies.7,9 Owing to their synthetic nature, stability, ease of preparation and shelf-life are significantly better than that of protein-based ligands. Drawbacks include lack of commercial availability and the necessity to use additional enzyme-target conjugates for detection/signalling. One way to circumvent the latter is to use a stable enzyme analogue such as iron oxide. This was demonstrated recently with a totally abiotic ELISA-type assay10 based on MIP nanoparticles containing a catalytic core of Fe3O4 with peroxidase-mimicking properties.11–13 In this assay, core–shell nanoparticles containing MIP at the surface are used for their combined recognition and signalling function. An alternative way to create molecular constructs for diagnostic applications that combine recognition and catalytic functionalities is to “shape” additional target-specific binding sites on a suitable enzyme (reporter) molecule. Modification of proteins to create binders from non-immunoglobulins is usually accomplished by introducing variable amino acid sequences into existing protein scaffolds such as the B-domain of protein A (affibodies) though a wide range of scaffolds.14 These can be engineered by a variety of approaches such as loop/surface randomization, random insertion, error-prone PCR or rational design.15 Though these are useful methods for the production of affinity materials, all are complex approaches, requiring intricate production and screening methods. Adapting conventional molecular imprinting techniques by using native enzymes/proteins as monomers can potentially be used as a simpler method to generate affinity protein ligands. Imprinting using biomolecules (bioimprinting) involves typically precipitation/lyophilisation of an enzyme solution in the presence of one of its substrates (template). The precipitation process increases the conformational rigidity of the enzyme, “shaping” the active site to preferentially accept the substrate used as a template (leading to increased enzymatic specificity) in a non-aqueous environment.16 This can be reversed if the protein is exposed to moisture. In order to produce stable/water compatible imprinted enzymes, the protein precipitates can be further chemically cross-linked, thus stabilizing the intended active site structure even under aqueous conditions.17,18 However, this process can only be used to fine-tune enzymatic specificity towards a substrate or close analogues bearing common functional groups, and it is not suitable to generate ligands against unrelated targets.Herein we describe a new concept for the generation of specific binding sites in enzyme molecules by cross-linking protein chains in the presence of immobilized templates. We used horse radish peroxidase (HRP) as a model enzyme (as used for signalling purposes in assays) which was imprinted with two analytes – vancomycin and ampicillin-creating what can be considered as an analogue of an HRP–antibody conjugate. Molecular imprinting was achieved using a modified bioimprinting technique performed in the presence of immobilized templates on a solid-phase support. HRP was then added to the solid-phase containing the template, where it adsorbs to its surface, and then cross-linked with glutaraldehyde (GA). This links together intra and inter-protein chains, including those involved in interactions with the template, creating permanent target-specific recognition sites. Unlike traditional bioimprinting, here the target molecule (template) is not one of the enzyme substrates. This cross-linked enzyme system exhibits hybrid features, i.e. catalytic and recognition properties, and can produce a colour change upon reaction with a chromogenic HRP substrate. The proposed solid-phase approach (Fig. 1) with immobilized templates is convenient for separating imprinted from non-imprinted molecules, as it includes an affinity based separation step where poor binders are removed. Furthermore, it eliminates the possibility of template leaching, and increases the homogeneity of the population of imprinted binding sites (with similar binding characteristics).19–21
The proposed method holds great potential for the generation of hybrid bio-nanosystems useful in assay development or as bio-chemical research tools.
Results and discussion
The HRP imprinting was performed on a solid-phase with an immobilized template (Fig. 1) following a modified imprinting procedure published earlier.7 The possible mechanism of formation of specific binding sites within the enzyme molecules can be attributed to an initial adsorption onto the solid-phase surface as common for proteins, likely based on electrostatic and hydrophobic interactions with the immobilized template and the solid-phase surface. This initial (and expected) adsorption was confirmed by performing the 3,3′,5,5′-tetramethylbenzidine (TMB) assay on the vancomycin-derivatised solid-phase after it was incubated with HRP for one hour, then washed with 1/3 volume of the wash buffer used to collect imprinted HRP. As the TMB substrate was added to the washed solid-phase, formation of blue colour was observed immediately (data not shown). These interactions between the protein and surface can affect the spatial arrangement of functional groups on the protein, which are then stabilized by cross-linking with glutaraldehyde. This process is analogous to previous work done with linear polymers with no particular selectivity to a target template, but after being chemically cross-linked in the presence of such a template, the resulting material clearly displays an imprinting effect. Examples include cinchonidine receptors obtained by cross-linking linear poly(methacrylic acid),22 methylene blue receptors with cross-linked amylose23 and tryptophan receptors using polypeptides cast in a polymeric membrane.24 Similar imprinting effects can be obtained by freeze drying proteins in the presence of a target molecule, and this can be performed even with templates with no specific pre-affinity for the protein, though obviously such memory effects are erased under aqueous conditions.25 In the present case, cross-linking can affect protein chains in the same enzyme molecule or lead to cross-linking of several proteins. As reported in Fig. 2, the population of imprinted enzyme nanoparticles varies from ca. 20 to 80 nm, and are made up of tens of proteins linked together. The results obtained with nanoparticle tracking analysis (NTA) show the main size distribution to be within 36 to 104 nm, see the ESI.† The appearance of a peak at ca. 320 nm was attributed to nanoparticle aggregation, and this could be reversed by sonicating the sample. With the exception of this peak, the solution of cross-linked HRP was stable without indication of further chemical cross-linking.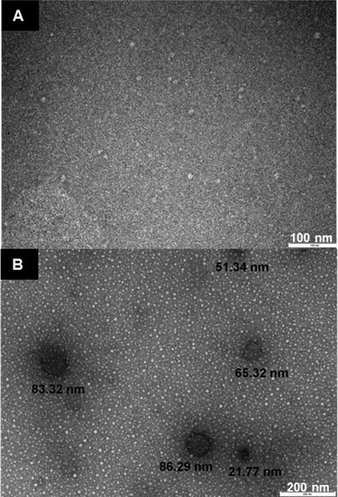 | ||
| Fig. 2 TEM images of HRP: (A) non-cross-linked HRP, which does not show the presence of nanostructures (individual protein molecules are too small for imaging); (B) cross-linked HRP nanoparticles. | ||
After the cross-linking step, a cold wash is performed on the solid-phase containing the template and bound HRP to remove protein molecules which have no affinity for the template. At 50 °C, the non-covalent interactions between the high-affinity nanoparticles and the templates are disrupted and a pure fraction of high-affinity cross-linked HRP can be eluted; in the present case 0.04 mg is produced when using 20 g of the solid-phase containing the immobilized template. Though this work is essentially a proof-of concept study, it is anticipated that the yield and particle size can possibly be improved by using a solid-phase with a larger surface area (such as smaller beads), by using cross-linking agents with different molecule lengths or by assessing different enzymes/enzyme fractions. Imprinted particle size could possibly be tuned by varying the amount of available protein, or by using simply protein adsorbed at the surface of the solid-phase, by varying its hydrophobicity. It is worth noticing that if the same procedure is applied to HRP in the absence of glutaraldehyde, as expected, practically all the enzyme is washed out from the solid-phase containing the template during the cold washes. Thus cross-linking is absolutely necessary for creating and preserving the affinity sites in the protein molecules. Conversely, if HRP is cross-linked in the absence of a template, no molecular recognitions are observed, just like for native HRP.
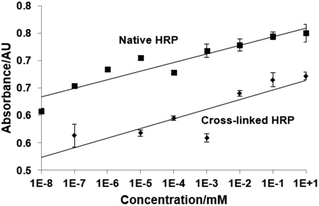
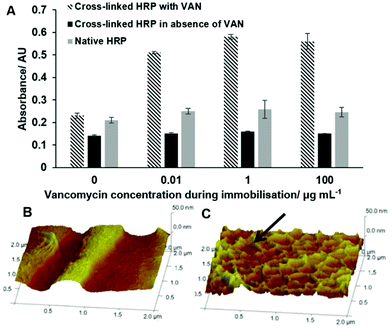
The influence of cross-linking on the stability of the enzyme can be measured by thermal analysis of native and cross-linked HRP using circular dichroism (CD) spectroscopy. The samples were heated from 20 °C to 94 °C and chiral dichroism was recorded at 200–260 nm. The thermal denaturation profile shows an increase of the melting temperature (Tm) for cross-linked HRP (81 ± 0.4 °C) compared with the native one (Tm = 75.8 ± 0.1 °C), see the ESI† for the CD spectra of native HRP and HRP processed through the solid-phase, but in the absence of GA. As indicated by the positive Van't Hoff enthalpy (ΔHVH) values, the protein unfolding is an endothermic process, since the heat is absorbed in order to disrupt the interactions maintaining the protein's secondary conformation. In the case of native HRP, this value is very large (244.0 ± 4.8 kJ mol−1) indicating that the equilibrium is shifted towards the “unfolded” end. In contrast, the cross-linked form exhibits a 69% decrease in ΔHVH (168.4 ± 6.1 kJ mol−1) compared to the native protein, suggesting that although the equilibrium still favours the unfolded protein, it is closer to the equilibrium point compared to the native. Thus, the cross-linking not only retains the inherent stability of the native HRP but as expected, it also improves the latter quite significantly. It has been previously shown that the bioimprinting process can affect the catalytic properties of enzymes.17 In our experiments we found that vancomycin affects the reaction of HRP with its chromogenic substrate (TMB), however this effect is non-specific and not linked to imprinting procedure (Fig. 3). It can be seen that the presence of vancomycin leads to an increase in the optical density during the HRP/TMB assay. Teicoplanin (which was to be used to test assay selectivity) had similar, even larger effect on the assay (data not shown due to difficult quantification, see below). Ampicillin on the other hand had no effect on colour development. The effect of vancomycin and teicoplanin, however, is not related to an actual activation of the enzyme but rather to their impact on the colour development during TMB oxidation. Thus, teicoplanin added to the microplate well forms a visible white precipitate in the presence of TMB which makes spectrophotometric quantification highly variable and for this reason it has been excluded from further work. The formation of specific binding sites in HRP by imprinting had been proven in competitive assays with immobilized target molecules. Polystyrene microplates were coated with BSA by physical adsorption producing a hydrophilic surface exposing carboxylic groups. Templates were covalently immobilized on BSA-coated microplates using standard EDC/NHS coupling protocols. Three concentrations of vancomycin were used for immobilization: 0.01, 1 and 100 μg mL−1. Subsequently, the wells were blocked with 50 mM ethanolamine (EA). Unfortunately it was not possible to measure the exact amount of the immobilized template due to the lack of an appropriate analytical technique. It should however be noted that various concentrations were used purely in order to determine the ideal amount of vancomycin to be used during immobilisation which would later result in the highest colorimetric response from the bound HRP. The procedure was repeated for both cross-linked and native HRP. The results show that there is a substantial difference in binding of cross-linked HRP (imprinted) when compared with cross-linked (but non-imprinted) and native enzymes to immobilized vancomycin (Fig. 4).
In contrast to native and cross-linked but non-imprinted enzymes, the binding of imprinted HRP to the microplate well clearly raises above background levels as soon as vancomycin is immobilised at the well surface. This clearly indicates the presence of specific binding sites for vancomycin within the cross-linked and imprinted HRP.
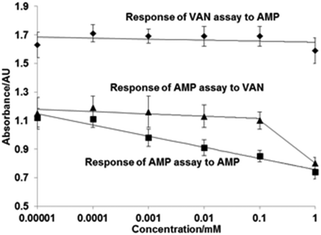
Atomic force microscopy (AFM) was used to characterize the surface topography of the microplate wells after capturing the cross-linked HRP. A non-modified polystyrene plate was characterized by a smooth surface which is corroborated by the very low roughness parameter (Ra) of 1.9 nm (Fig. 4). BSA coating and vancomycin immobilization increased the Ra to 4.6 nm. After capturing the cross-linked HRP, the AFM images displayed a different surface topology (Fig. 4C). A denser distribution of differently shaped peaks was detected. In particular, the Ra increased by 60% with a value of 5.95 nm. In light of these results, it might be assumed that most of the larger cross-linked HRP nanoparticles (with diameters of up to 130 nm) are not bound to the microplate surface (possibly due to steric reasons), and binding of smaller constructs is favoured. A competitive assay was run in microplates with immobilized vancomycin as described in the Experimental section. Fig. 5 shows the calibration plot for the enzyme-linked competitive assay for vancomycin. For concentrations below 1 μM there is no observable response, whilst for values above 10 mM saturation was observed. The assay therefore showed linearity from 1 μM to 10 mM of free vancomycin. No effect was observed upon addition of vancomycin when the cross-linked HRP was replaced with the native enzyme. Neither free ampicillin had any effects on vancomycin assay (Fig. 6). This proves the formation of specific binding sites in the protein by cross-linking in the presence of the template molecule. To assess the generic nature of the imprinting approach presented here, HRP was cross-linked in the presence of ampicillin and the assay was repeated with microplates bearing immobilized ampicillin (Fig. 6).
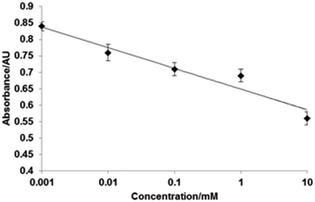 | ||
| Fig. 5 Calibration plot determined for the enzyme-linked competitive assay for vancomycin performed with HRP cross-linked in the presence of vancomycin. Experiments were performed in triplicate. | ||
This assay based on HRP imprinted against ampicillin showed linearity from 0.01 μM to 1 mM, though there was some effect of the vancomycin on the ampicillin assay at concentrations above 0.1 mM. As reported for the previous template, no effect of ampicillin was observed when the cross-linked HRP was re-placed with the native enzyme. These assays were performed purely for demonstration of the selective molecular recognition properties introduced into the cross-linked HRP by the immobilized template; the aim of the present work being the introduction of a new bioimprinting protocol. However, the performance of cross-linked HPR in connection with assay development can possibly be improved by using different cross-linking chemistries or by using linkers with different lengths, in addition to alternative enzymes.
The stability of the bioimprinted HRP was tested in parallel to the native enzyme. No evident change in the assay performance was detected after 3 weeks. However, the cross-linked enzyme lost catalytic affinity in parity with native molecules when stored in buffer. For this reason, it might be advisable to store the cross-linked enzyme lyophilized.
Experimental
Materials
Glass beads with 90 μm average diameter (GB), Spheri-glass A-Glass 2429, were obtained from Potters, UK. N-[3-(Trimethoxysilyl)propyl]ethylenediamine (TPEA), glutaraldehyde (GA, 50% w/w in H2O), 1-ethyl-3-(3-dimethylaminopropyl)carbodiimide (EDC), cysteamine, N-hydroxysuccinimide (NHS), HPLC grade water, bovine serum albumin (BSA), vancomycin, ampicillin, teicoplanin, horseradish peroxidase (HRP) and ethanolamine (EA) were obtained from Sigma-Aldrich, UK. Phosphate Buffered Saline (PBS) was prepared from PBS buffer tablets (Sigma-Aldrich, UK) and consisted of phosphate buffer (0.01 M), potassium chloride (0.0027 M) and sodium chloride (0.137 M), pH 7.4. 3,3′,5,5′-Tetramethylbenzidine (TMB) was used in the form of a liquid substrate system for ELISA (Sigma-Aldrich, UK). Amicon centrifuge filtration devices with a 30 kDa molecular weight cut-off (MWCO) were obtained from Millipore, UK. Nunclon 96 well polystyrene microplates were purchased from Thermo Scientific, UK.Template immobilization on the solid-phase (GB)
All GB incubation steps were performed with 0.4 mL solution per gram of GB. The solid-phase (GB, 180 g) was boiled in 2 M NaOH for 15 minutes, then washed with deionized water until neutral pH, followed by 200 mL acetone, and then dried at 60 °C. Subsequently, GB were incubated in a solution of TPEA 2% (v/v) in anhydrous toluene overnight, then thoroughly washed by suction filtration with acetone, methanol and dried at 80 °C. Afterwards, the beads were incubated for 2 h in a 7% v/v GA solution in PBS, pH 7.2 and then washed with deionized water. The immobilization of the templates (vancomycin or ampicillin) was performed by incubation of GA-derivatized GB in a template solution (1 mg mL−1 in PBS) at 4 °C overnight. Afterwards, the GB with immobilized template were washed with water and ACN, dried under vacuum and stored at 4 °C until further use.Solid-phase bioimprinting
Twenty grams of GB with either immobilized template were placed in a glass vial and mixed with 10 mL of HRP solution in PBS (4 mg mL−1), followed by 100 μL of GA solution. The mixture was incubated under shaking for 2 hours at RT. Then 30 μL of ethanolamine was added to GB/HRP (final concentration 50 mM) and left to react for another 1 hour at room temperature. Then the whole contents of the vial were poured into a fritted SPE cartridge connected to a vacuum manifold and washed using 290 mL of PBS solution at RT to remove low affinity particles. The high affinity particles of the cross-linked HRP were collected using 200 mL of the PBS solution at 50 °C. A control sample of the non-cross-linked HRP was prepared in the same manner without using GA, and a sample of the cross-linked HRP prepared in the absence of a template was prepared in solution (no solid-phase). The latter was then purified from excess EA and GA by filtration on a 30 kDa MWCO centrifuge cartridge. All samples were kept at 4 °C before analysis. The concentration of the cross-linked HRP was determined by means of a colorimetric reaction with TMB. The samples were diluted (×100![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000) and 50 μL of each sample were placed in the PS microplate wells. Then 50 μL of TMB was added and, after 2 minutes, the reaction was stopped by addition of 50 μL of 0.5 M H2SO4. The concentration was measured using a microplate reader at 450 nm by comparing data obtained from a calibration curve made against weighted lyophilised HRP samples. Each measurement was performed in triplicate.
000) and 50 μL of each sample were placed in the PS microplate wells. Then 50 μL of TMB was added and, after 2 minutes, the reaction was stopped by addition of 50 μL of 0.5 M H2SO4. The concentration was measured using a microplate reader at 450 nm by comparing data obtained from a calibration curve made against weighted lyophilised HRP samples. Each measurement was performed in triplicate.
Template immobilisation onto the surface of microplates
Microplates were incubated with 100 μL of BSA solution in PBS 0.1% (w/w) overnight at 4 °C and then washed 3 times with 300 μL of PBS solution. Afterwards 50 μL of EDC/NHS solution (4 mg mL−1 and 6 mg mL−1 respectively) were added and kept for 15 minutes at RT. The microplates were washed 3 times using 300 μL of water, then 50 μL of either template solution was added and incubated for 2 hours at RT. The remaining NHS esters were blocked with 50 μL ethanolamine (50 mM in PBS) for 1 hour at RT. Three different concentrations of template solutions were initially used to determine the optimum conditions (0.1; 1 and 100 μg mL−1, see the next section). The optimum was found at 1 μg mL−1 and used in further experiments.Determination of the optimal template concentration for immobilization on microplate wells
A volume of 50 μL of the cross-linked and non-cross-linked HRP was added to a microplate coated with either BSA or one of the immobilized templates (0.1, 1 and 100 μg mL−1) and incubated for 2 hours at RT. The non-bound HRP was washed out using 300 μL of PBS solution (3 times). Then 50 μL of TMB was added and incubated for 10 minutes and then the reaction was stopped using 0.5 M H2SO4. The plate was read using a microplate reader at 450 nm. The amount of bound HRP was determined by comparing the data obtained from a calibration curve made against a weighted lyophilized HRP sample.Competitive binding assay
For binding analysis, the template was immobilized on the microplates at 1 μg mL−1. Then 25 μL of the cross-linked and non-cross-linked HRP (0.4 mg L−1) were added to the wells (in PBS buffer), mixed with 25 μL of free template solution (10 nM–10 mM in PBS buffer) and incubated for 2 hours at room temperature. Then the solution was removed and the microplate was washed 3 times with 300 μL of PBS. The amount of bound HRP was determined using a colorimetric reaction with 50 μl TMB (10 min incubation time) and measured using a microplate reader at 450 nm.Effect of antibiotics on HRP/TMB assay
Cross-linked and non-cross-linked HRP (200 μL, 10 μg L−1) were mixed with 200 μL of vancomycin or ampicillin solutions (0–10 mM) (in PBS) and incubated for 2 h at RT. The solutions (50 μL) were then added to a polystyrene microplate, followed by TMB (50 μL). After 5 min incubation, the reaction was quenched with 50 μL of H2SO4 (0.5 M) and the absorbance was measured at 450 nm. All measurements were made in quadruplicate.AFM visualization of the HRP-modified microplates
The wells with immobilized vancomycin were washed with PBS (3 × 300 μL), and incubated with 50 μL HRP (0.2 mg L−1) for 2 h at room temperature. The solution was removed and the wells were washed with PBS (3 × 300 μL). The flat bottom-surface of a well was cut out and fixed onto a glass slide. A Veeco Dimension 3100 Scanning Probe Microscope (Veeco Instruments Inc. USA), operating NanoScope 6.12 Software was used for AFM imaging, performed at RT. Tapping mode was selected, using a Tap300 antimony-doped silicon probe tip (Bruker, UK) with a scan size of 2.0 × 2.0 μm and scan rate of 0.2 Hz. The total number of sample lines was fixed at 256, and the amplitude set point was 0.8 V, whilst the integral and proportional gain was optimized at 0.04 and 0.06 respectively.Size determination by nanoparticle tracking analysis
A cross-linked HRP sample in deionized water was filtered through a syringe filter with a pore size of 0.8 μm and introduced into the sample chamber of a NanoSight LM10 (Malvern Instruments, UK), and left to equilibrate to the chamber temperature (22 °C) for 30 s. The camera was calibrated to 166 nm per pixel at 30 frames per s, 1800 frames collected. Data was processed with the software package Nanoparticle Tracking analysis 2.3 (Malvern Instruments, UK).TEM visualisation of HRP samples
A sample of 3 μL of cross-linked HRP was deposited on a carbon-coated grid and allowed to settle for approximately 2 min. Excess of liquid was blotted with tissue paper and then a drop of uranyl acetate stain (1% v/v aq.) was deposited over and blotted dry with tissue paper. The excess of staining agent was washed out with demineralized water. Visualization was conducted with a JEOL 1400 transmission electron microscope (JEOL, USA) with an accelerating voltage of 80 kV.Stability studies of cross-linked HRP by circular dichroism spectroscopy (CD)
Native or cross-linked HRP (250 μL, 0.04 mg mL−1) were added to a quartz cuvette with a 1 mm path length. The spectra were obtained using a Chirascan Plus CD spectrometer (Applied Photophysics) and recorded with a 1 nm bandwidth over a wavelength range of 195–260 nm. The thermal melt was conducted in temperature stepped ramp mode over a range of 20–94 °C using 1 °C steps with 45 s equilibration time between spectra measurements. Sample temperatures were recorded using a platinum resistance thermocouple placed inside the CD cuvette. In order to improve the signal-to-noise ratio, two repeats were conducted at each data point. Phase diagram analysis was used to select appropriate unfolding models, which were fitted to the data using Global3 (Applied Photophysics).26Conclusions
This study demonstrates that target-specific binding sites can be imprinted into HRP by cross-linking with glutaraldehyde in the presence of a solid-phase bearing immobilised templates. The bioimprinted nanoparticles (“enzybodies”) display a combination of catalytic properties (conferred by the enzyme) and specific molecular recognition properties (conferred by the imprinting process). The proposed approach is virtually generic in nature and could be extended to analytes other than the ones employed in this study, as well as taking advantage of other cross-linking chemistries. This system may be particularly useful when antibodies are difficult to obtain or exhibit poor recognition properties towards target molecules.Acknowledgements
Project supported by Wroclaw Centre of Biotechnology, programme The Leading National Research Centre (KNOW) for years 2014–2018.References
- S. L. Bullock and K. W. Walls, J. Infect. Dis., 1977, 136, S279–S285 CrossRef PubMed.
- K. W. Walls, S. L. Bullock and D. K. English, J. Clin. Microbiol., 1977, 5, 273–277 CAS.
- C. Milstein, Sci. Am., 1980, 243, 66–74 CrossRef CAS PubMed.
- B. Kudlak, N. Szczepanska, K. Owczarek, Z. Mazerska and J. Namiesnik, Crit. Rev. Anal. Chem., 2015, 45, 191–200 CrossRef CAS PubMed.
- S. D. Gan and K. R. Patel, J. Invest. Dermatol., 2013, 133, E10–E12 CrossRef PubMed.
- R. M. Lequin, Clin. Chem., 2005, 51, 2415–2418 CAS.
- I. Chianella, A. Guerreiro, E. Moczko, J. S. Caygill, E. V. Piletska, I. M. P. De Vargas Sansalvador, M. J. Whitcombe and S. A. Piletsky, Anal. Chem., 2013, 85, 8462–8468 CrossRef CAS PubMed.
- Z. X. Xu, H. J. Gao, L. M. Zhang, X. Q. Chen and X. G. Qiao, J. Food Sci., 2011, 76, R69–R75 CrossRef CAS PubMed.
- G. Vlatakis, L. I. Andersson, R. Muller and K. Mosbach, Nature, 1993, 361, 645–647 CrossRef CAS PubMed.
- R. V. Shutov, A. Guerreiro, E. Moczko, I. P. de Vargas-Sansalvador, I. Chianella, M. J. Whitcombe and S. A. Piletsky, Small, 2014, 10, 1086–1089 CrossRef CAS PubMed.
- N. Wang, L. Zhu, M. Wang, D. Wang and H. Tang, Ultrason. Sonochem., 2010, 17, 78–83 CrossRef CAS PubMed.
- Y. Shi, P. Su, Y. Wang and Y. Yang, Talanta, 2014, 130, 259–264 CrossRef CAS PubMed.
- L. Gao, J. Zhuang, L. Nie, J. Zhang, Y. Zhang, N. Gu, T. Wang, J. Feng, D. Yang, S. Perrett and X. Yan, Nat. Nanotechnol., 2007, 2, 577–583 CrossRef CAS PubMed.
- H. K. Binz, P. Amstutz and A. Pluckthun, Nat. Biotechnol., 2005, 23, 1257–1268 CrossRef CAS PubMed.
- J. Löfblom, J. Feldwisch, V. Tolmachev, J. Carlsson, S. Ståhl and F. Y. Frejd, FEBS Lett., 2010, 584, 2670–2680 CrossRef PubMed.
- I. Mingarro, C. Abad and L. Braco, Proc. Natl. Acad. Sci. U. S. A., 1995, 92, 3308–3312 CrossRef CAS.
- F. Peißker and L. Fischer, Bioorg. Med. Chem., 1999, 7, 2231–2237 CrossRef.
- N. A. E. Kronenburg, J. A. M. de Bont and L. Fischer, J. Mol. Catal. B: Enzym., 2001, 16, 121–129 CrossRef CAS.
- F. Canfarotta, M. J. Whitcombe and S. A. Piletsky, Biotechnol. Adv., 2013, 31, 1585–1599 CrossRef CAS PubMed.
- R. Lorenzo, A. Carro, C. Alvarez-Lorenzo and A. Concheiro, Int. J. Mol. Sci., 2011, 12, 4327 CrossRef CAS PubMed.
- S. Ambrosini, S. Beyazit, K. Haupt and B. Tse Sum Bui, Chem. Commun., 2013, 49, 6746–6748 RSC.
- J. Matsui, N. Minamimura, K. Nishimoto, K. Tamaki and N. Sugimoto, J. Chromatogr., B: Biomed. Appl., 2004, 804, 223–229 CrossRef CAS PubMed.
- S. Shinkai, M. Yamada, T. Sone and O. Manabe, Tetrahedron Lett., 1983, 24, 3501–3504 CrossRef CAS.
- M. Yoshikawa, J.-I. Izumi, T. Kitao and S. Sakamoto, Macromolecules, 1996, 29, 8197–8203 CrossRef CAS.
- K. Dabulis and A. M. Klibanov, Biotechnol. Bioeng., 1992, 39, 176–185 CrossRef CAS PubMed.
- I. M. Kuznetsova, K. K. Turoverov and V. N. Uversky, J. Proteome Res., 2004, 3, 485–494 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available: Additional circular dichroism data and nanoparticle tracking analysis trace. See DOI: 10.1039/c6nr02009g |
| This journal is © The Royal Society of Chemistry 2016 |