Open Access Article
Open Access ArticleCausation or only correlation? Application of causal inference graphs for evaluating causality in nano-QSAR models†
Natalia
Sizochenko
ab,
Agnieszka
Gajewicz
a,
Jerzy
Leszczynski
b and
Tomasz
Puzyn
*a
aLaboratory of Environmental Chemometrics, Faculty of Chemistry, University of Gdansk, Wita Stwosza 63, 80-308, Gdansk, Poland. E-mail: t.puzyn@qsar.eu.org
bInterdisciplinary Center for Nanotoxicity, Department of Chemistry, Jackson State University, 1400 J. R. Lynch Street, P. O. Box 17910, Jackson, MS 39217, USA
First published on 29th February 2016
Abstract
In this paper, we suggest that causal inference methods could be efficiently used in Quantitative Structure–Activity Relationships (QSAR) modeling as additional validation criteria within quality evaluation of the model. Verification of the relationships between descriptors and toxicity or other activity in the QSAR model has a vital role in understanding the mechanisms of action. The well-known phrase “correlation does not imply causation” reflects insight statistically correlated with the endpoint descriptor may not cause the emergence of this endpoint. Hence, paradigmatic shifts must be undertaken when moving from traditional statistical correlation analysis to causal analysis of multivariate data. Methods of causal discovery have been applied for broader physical insight into mechanisms of action and interpretation of the developed nano-QSAR models. Previously developed nano-QSAR models for toxicity of 17 nano-sized metal oxides towards E. coli bacteria have been validated by means of the causality criteria. Using the descriptors confirmed by the causal technique, we have developed new models consistent with the straightforward causal-reasoning account. It was proven that causal inference methods are able to provide a more robust mechanistic interpretation of the developed nano-QSAR models.
Introduction
Quantitative Structure–Activity/Property Relationships (QSAR/QSPR) analyses are widely used by chemoinformaticians for investigating the biological activity of various compounds. QSAR/QSPR based on the assumption that the molecular structures of a series of compounds contain key information about the factors responsible for their physical, chemical or biological properties.1 In QSAR/QSPR analysis one takes theoretical or experimental information related to the molecular structures of the studied compounds (structural descriptors) and correlates them with values of the investigated activity/property.Nowadays one of the biggest challenges in QSAR/QSPR studies is related to assessing the reliability of mechanistic interpretation of the identified relationships.2 There are an extremely large number of descriptors available and a huge array of statistical methods may serve to select the most appropriate descriptors and to correlate them with the studied property. Hence, there are a substantial number of possible ways of building QSAR/QSPR equations that yield approximately equal measures of statistical quality (i.e. correlation coefficients).
Johnson3 noticed that a large number of publications devoted to QSAR/QSPR modeling contain a logical fallacy related to the fact that causality was incorrectly assigned to the variables which were only correlated. It is widely known that any statistical relationship does not automatically imply causation, because just having a high correlation coefficient between two variables is not a sufficient condition for pointing out the existence of causation. Saying that in mathematical language, the correlation is a necessary part, but it is not sufficient to confirm the existence of the causal relationship. Therefore, the usual way of interpreting QSAR/QSPR models that are based on the values of correlation coefficients does not necessarily confirm the existence of a cause–effect relationship. Unfortunately, specific algorithms that infer causal interactions between particular variables have recently been developed and applied only to genomics data.4 Possible applications of those methods in QSAR have not been extensively studied yet.
Nowadays an emerging field for employing QSAR/QSPR methods is the risk assessment of newly designed nanoparticles (nano-[Q]SAR).5–11 As such, in our previous contribution we have applied causal inference methods for interpretation of the classification nano-SAR model that predicted cytotoxicity towards BEAS-2B and RAW 264.7 cell lines for a series of metal oxide nanoparticles.7 However, causal inference methods have never been used in quantitative SAR studies before. Thus, an expansion of these methods onto nano-QSAR studies might be of the high benefit and the high importance.8
In current contribution, we investigated causality of the descriptors from the recently developed nano-QSAR models for predicting toxicity of metal oxide nanoparticles towards E. coli bacteria.5,6,9–11 Our intention was to demonstrate the usefulness of causal inference methods in nano-QSAR modeling.
Materials and methods
Causal inference methods
Causality refers to the relationship between two sets of events, where one set of events (the effects) is a direct consequence of another set of events (the causes). The goal of causal modeling is to provide coarse descriptions of mechanisms, at a level sufficient to predict the result of changes.8,12Causal inference methods are based on several fundamental mathematical concepts. They include conditional probability and its joint distribution, as well as directed and undirected graphs.8,13 Conditional probability measures the probability of an event given that another event has occurred. Statistically, conditional probability is an update of the probability of an event based on new information. A probability distribution denotes a probability each measurable event. Given two jointly distributed random variables, X and Y, the conditional probability distribution of Y given X represents the probability distribution of Y when X is known to be a particular value. For example, the probability of obtaining a random number from a dataset of size 5 is equal to 1/5, and would be denoted by P(X|Y).
The main aim of the causal inference method is to separate the cause from the effect if the given two variables X and Y have a causal relationship. Cause–effect relationships are represented by directed graphs.8,15 A directed graph must equal a product of terms, one term for each variable, with each term giving the conditional probability of that variable on its parent variables in the graph. For instance, in a system with three variables A, B, and C, there are six possible types of graph:8
1. Unconnected graph: A, B, C with the independent structure A ⊥ B, A ⊥ C, B ⊥ C;
2. Single arrow chain: A → C, B or C → A, B with the independent structures A ⊥ B and C ⊥ B;
3. Chain: A → B → C or A ← B ← C with the independent structures A ⊥ C and C ⊥ A;
4. Fork: A ← B → C with the independent structure A ⊥ C|B;
5. Collider: A → B ← C with the independent structures A ⊥ B and C ⊥ B.
6. Fully connected graph: A → B → C and A → C without independencies.
Described above are fundamental ideas of probability theory. For readers who are interested in the details, several fundamental references can be recommended.8,14,16
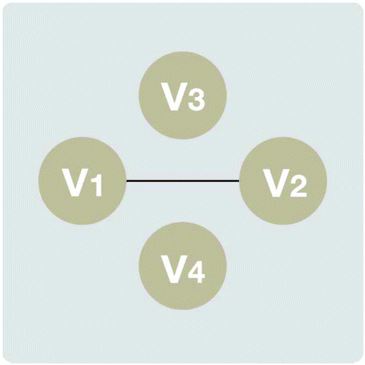
In the current paper we utilized the Peter Spirtes and Clark Glymour (PC)15 algorithm to determine causal relationships using causal inference graphs (CIGs). This algorithm is also based on conditional independence tests. PC starts with the assumption of a complete undirected graph. Then, a series of conditional independence tests is done and edges are deleted. The result is a skeleton, in which every edge is still undirected. The orientation of edges is found by repeatedly applying the above rules. Thus, a form of digraphs consists of a set of vertices (V), and a set of directed edges between the vertices, e(v1, v2) where v1 and v2 are parts of whole set V. As a representation for causal relationships, the vertices correspond to the variables (or attributes) that are in a dataset. Each directed edge from v1 to v2 corresponds to a causal influence from v1 to v2.16 For example, as one can see in Fig. 1, there is a causal relationship between vertices v1 and v2, while the remaining part of the set, V (vertices v3 and v4), does not demonstrate causal relationships.
Source code for the described algorithm is available on the website of the Max-Planck-Institute for Intelligent Systems Tübingen.17 During the last two decades many various algorithms that infer causal interactions from observational data have been developed. For those readers who are interested in the details an excellent review can be recommended.14
Existing nano-QSAR models of toxicity of metal oxide nanoparticles towards bacteria E. coli
In our previous paper5 we developed a nano-QSAR model that linked the experimental data on the toxicity of 17 nano-sized metal oxide nanoparticles to their structure (Model I). The toxicity has been expressed as the negative decimal logarithm of MIC50 – the minimum inhibitory concentration where 50% of the population exhibit a response (note: in the original Paper5 it is referred as EC50 – minimum concentration which affects the reduction of bacteria viability of 50%). The model employs only one descriptor – the enthalpy of formation of a gaseous cation having the same oxidation state as that of the metal oxide structure (ΔHMe+) derived from quantum-mechanical calculations (semi-empirical PM6 method):| pMIC50 = 2.59 − 0.50ΔHMe+ | (1) |
Toropov et al.9 developed another nano-QSAR model based on the same experimental toxicity data (Model II). In contrast, they utilized the SMILES-based optimal descriptor DCW containing information on the two SMILES attributes: double bonds and oxygen atoms (encoded as ‘=’, and ‘O’):
| pMIC50 = 3.4056 + 0.4000DCW | (2) |
Similarly, Kar et al.10 proposed two nano-QSAR models (Models III and IV), that employ periodic table-based descriptors: the charge of the metal cation corresponding to a given oxide (Z) and metal electronegativity (χ)
| pMIC50 = 4.781 − 1.380Z | (3) |
| pMIC50 = 4.401 − 1.324Z + 0.176χ | (4) |
It is worth highlighting that all of the above nano-QSAR models are linear. Statistical parameters of the mentioned models are presented in Table 1.
| Model I5 | Model II9 | Model III10 | Model IV10 | Model V11 | |
|---|---|---|---|---|---|
| R 2 – coefficient of determination; RMSE – root-mean-square error. | |||||
| R 2 (training set) | 0.85 | 0.74–0.83 | 0.73–0.90 | 0.81–0.90 | 0.93 |
| RMSE (training set) | 0.20 | 0.17–0.23 | 0.16–0.27 | 0.16–0.22 | 0.13 |
| R 2 (test set) | 0.83 | 0.83–0.96 | 0.65–0.91 | 0.73–0.96 | 0.78 |
| RMSE (test set) | 0.19 | 0.14–0.33 | 0.15–0.29 | 0.15–0.26 | 0.32 |
Finally, in one of our recent contributions,11 we have applied a more comprehensive, non-linear classification Random forest approach to build a model describing the toxicity of metal oxides (Model V). This model uses several types of descriptors, such as fragmental descriptors calculated using the Simplex Representation of Molecular Structure (SiRMS)18 approach, size-dependent descriptors based on the Liquid Drop Model (LDM)19,20 and metal–ligand binding characteristics (MLB).21 Statistical parameters characterizing Model V are presented in Table 1 as well. Descriptor values from all listed contributions are presented in Table 2.
| Metal oxide nanoparticle | pMIC50 | Model I5 | Model II9 | Models III and IV10 | Model V11 | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ΔHMe+ | DCW | χ | Z | S 1 | S 2 | S 3 | r w | SV | CPP | ρ | ||
| Al2O3 | 2.49 | 1187.83 | −1.817 | 1.61 | 3 | 1 | 13.54 | 0 | 0.183 | 0.033 | 0.168 | 3960 |
| Bi2O3 | 2.82 | 1137.40 | −1.817 | 2.02 | 3 | 1 | 14.36 | 0 | 0.232 | 0.021 | 0.087 | 8900 |
| CoO | 3.51 | 601.80 | −0.127 | 1.88 | 2 | 0 | 5.32 | 0 | 0.144 | 0.012 | 0.062 | 6000 |
| Cr2O3 | 2.51 | 1268.70 | −1.817 | 1.66 | 3 | 1 | 13.64 | 0 | 0.191 | 0.025 | 0.146 | 5210 |
| CuO | 3.20 | 706.25 | −0.127 | 1.90 | 2 | 0 | 5.34 | 0 | 0.143 | 0.035 | 0.055 | 6450 |
| Fe2O3 | 2.29 | 1408.29 | −1.817 | 1.83 | 3 | 1 | 13.98 | 0 | 0.194 | 0.048 | 0.164 | 5250 |
| In2O3 | 2.81 | 1271.13 | −1.817 | 1.78 | 3 | 1 | 13.88 | 0 | 0.210 | 0.056 | 0.113 | 7180 |
| La2O3 | 2.87 | 1017.22 | −1.817 | 1.10 | 3 | 1 | 12.52 | 0 | 0.229 | 0.040 | 0.087 | 6510 |
| NiO | 3.45 | 596.70 | −0.127 | 1.91 | 2 | 0 | 5.35 | 0 | 0.134 | 0.054 | 0.058 | 7450 |
| Sb2O3 | 2.64 | 1233.06 | −1.817 | 2.05 | 3 | 1 | 14.42 | 0 | 0.238 | 0.013 | 0.118 | 5190 |
| SiO2 | 2.20 | 1686.38 | −3.252 | 1.90 | 4 | 1 | 8.78 | 1 | 0.176 | 0.094 | 0.400 | 2650 |
| SnO2 | 2.01 | 1717.32 | −3.252 | 1.96 | 4 | 1 | 8.84 | 1 | 0.173 | 0.030 | 0.232 | 7010 |
| TiO2 | 1.74 | 1575.73 | −3.252 | 1.54 | 4 | 1 | 8.42 | 1 | 0.174 | 0.093 | 0.302 | 3600 |
| V2O3 | 3.14 | 1097.73 | −1.817 | 1.63 | 3 | 1 | 13.58 | 0 | 0.194 | 0.048 | 0.141 | 4870 |
| Y2O3 | 2.87 | 837.15 | −1.817 | 1.22 | 3 | 1 | 12.76 | 0 | 0.223 | 0.047 | 0.100 | 4840 |
| ZnO | 3.45 | 662.44 | −0.127 | 1.65 | 2 | 0 | 5.09 | 0 | 0.151 | 0.017 | 0.054 | 5700 |
| ZrO2 | 2.15 | 1357.66 | −3.252 | 1.33 | 4 | 1 | 8.21 | 1 | 0.173 | 0.029 | 0.222 | 5680 |
Evaluating causality in the previously published QSAR models
All models (Model I–V) are comparable to each other in terms of the statistical quality. However, by using inference causal methods, one can verify whether all of the models discover really causal relationships.Therefore, we propose the following procedure to evaluate causality of the observed structure–activity relationships:
1. Development of CIGs to find the causal relationships between independent variables (here: descriptors) and a dependent variable (here: toxicity towards bacteria E. coli);
2. Selection of first elements (main causes) in the CIG chain;
3. Evaluation: measuring the Pearson's correlation coefficient (in the case of monotonic relationships) or the interclass correlation coefficient (in the case of non-monotonic relationships) for chain first elements and co-dependent variables;22
4. Selection of the best elements of CIGs to develop the best QSAR equation.
Results and discussion
In order to evaluate causality in the recently published nano-QSAR models we have developed CIGs and estimated correlation measurers for the most significant cases (Fig. 2).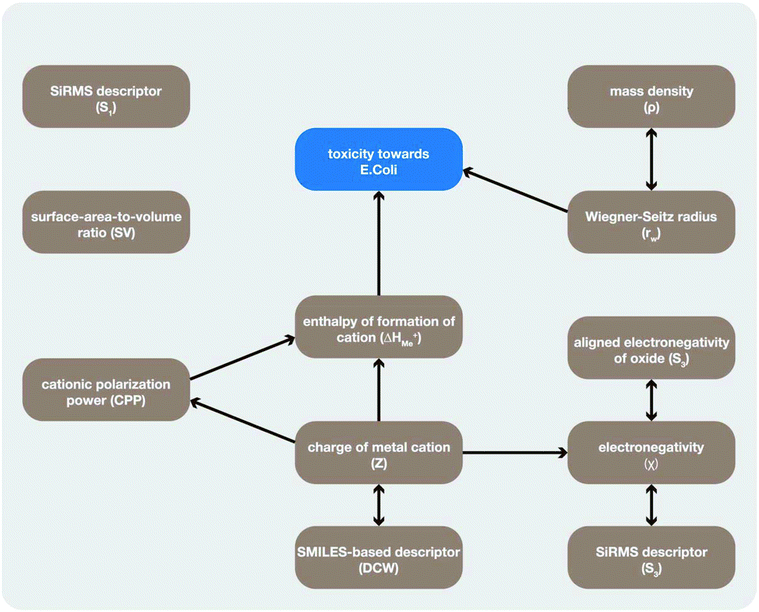 | ||
| Fig. 2 A causal graph illustrating the relationships between the descriptors used in the studied models and the toxicity of nanoparticles towards bacteria E. coli. | ||
Descriptors showing cause–effect relationships with toxicity (Fig. 2) are the descriptors forming the first edges of the CIG, namely: ΔHMe+ (Model I) and rw (Model V). We suggest that these descriptors are the best choices to develop a model with both correlational and causal relationships.
The enthalpy of formation of metal cations (enthalpy, ΔHMe+) is the most relevant parameter describing toxicity of metal oxide nanoparticles towards bacteria E. coli. The enthalpy ΔHMe+ linearly correlates with pMIC50 with R2 = 0.85 (Pearson's correlation coefficient r = −0.92). Model I employed only one descriptor. Therefore Model I is the perfect case in terms of causality and correlation.
The enthalpy of formation of metal cations has a straight single connection to the toxicity. Moreover, there is the collider graph structure with the charge (Z, Models III and IV) and the cation polarization power (CPP, Model V). The enthalpy (ΔHMe+) is closely related to the lattice energy, which increases with increasing charge (Z) of the metal ion.5 The charge (Z) correlates with the enthalpy (ΔHMe+) with R2 = 0.85 (Pearson's correlation coefficient r = −0.92). Single arrows connect cation polarization power (CPP) with charge (Z) and then with the enthalpy (ΔHMe+). The fact that CPP (eqn (5)) represents the combination of the charge of the ion (Z) and the Pauling radius (r) might explain this observation:21
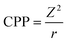 | (5) |
At the same time, the cation polarization power (CPP) is directly related to the enthalpy and the best way to describe the relationship between CPP and ΔHMe2+ (r = −0.83) is the exponential function (eqn (6)):
| CPP = f(ΔHMe+) | (6) |
As we previously noticed,11 cation polarization power (CPP) reflects electrostatic interactions, important to the process of inducing toxicity. CPP might reflect a metal cation release. This is in agreement with the assumption that the enthalpy of formation of a gaseous cation describes the release of metal cations from the nanoparticle surface.5 It means that the descriptors ΔHMe+, Z and CPP are closely related to each other and they indirectly describe the same processes. Since the calculations of Z and CPP descriptors are simpler and faster when compared with the computation of enthalpy, it might be more efficient to use them in modeling instead of enthalpy. KNIME node for these calculations is available on the website of the NanoBRIDGES project.23
All mutually-related descriptors are related to each other (mechanically or statistically). For example, the Wigner–Seitz radius (rw) and mass density (ρ) are not directly correlated to each other in statistical terms (the Pearson's correlation coefficient is 0.07). However, the relationship between ρ and rw has straightforward mechanistic behavior and can be deducted from the basic LDM equation (eqn (7)).19,20
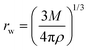 | (7) |
Wigner–Seitz radius also do not demonstrate simple linear relationships with the target toxicity (the Pearson's correlation coefficient is r = −0.25, Fig. 2). Using numerical analysis, polynomial regression for the Wigner–Seitz radius (R2 = 0.69) was identified.24,25 Polynomials are non-monotonic non-linear functions. Pearson's correlation coefficient is not applicable to such types of relationship.22,26 Therefore, there are some limitations to the mechanistic interpretation of polynomial equations. To estimate correlation measures for the Wigner–Seitz radius, the intraclass correlation coefficient was measured.27 It operates on data structured as groups, rather than data structured as paired observations. Intraclass correlation evaluates the level of agreement between raters in measurements, where the measurements might be intervals.28 We observed individual intraclass correlations equal to −0.921.
Next, we have utilized the Wigner–Seitz radius as a similarity measure for studied nanoparticles. Similarity in the chosen descriptor space (1D) was measured using the k-means algorithm.29 Using rw we divide nanoparticles into four clusters: separate clusters for MeO and MeO2 and two clusters for Me2O3 (see ESI†).
This observation is in agreement with the mentioned toxicity trend: toxicity decreases in the following order of the formal charge of the metal cations:3 Me2+ > Me3+ > Me4+. It means that rw represents not only a fraction of the free molecules on the nanocluster's surface, but also the charge of the metal ions. It brings us back to the importance of ionic properties, as was demonstrated by other descriptors in our previous contributions.9–11 As it was mentioned in earlier works, one of the main mechanisms of metal oxide nanoparticles’ toxicity towards E. coli bacteria is related to the release of the metal ions from the nanoparticle's surface.7–11 Therefore, we assume that the release of ions may be a dominant cause.30,31
There are several other mutually related variables. At first, the metal electronegativity (χ, Model IV) is mutually related to SiRMS-based aligned electronegativity of the metal oxide (S2, Model V) and the SiRMS-based descriptor of electronegativity (S3, Model V). This observation is not surprising in the light of the previously proposed mechanism of the toxicity of MeOx to the E. coli bacteria. In other words, different descriptors from particular models are in causal agreement and depict the same basic mechanisms associated with the process of metal cations’ release from the particle surface.
The charge of the metal cation (Z) and the electronegativity (χ) also have a causal relationship. It follows one of the basic concepts of physics: the more electronegative element has a greater share of electrons. As such, the partial negative charge reflects the higher electron density. In the same way, the less electronegative element has a partially positive charge reflecting the lack of electron density.32
The SMILES-based descriptor DCW (Model II) is mutually related to the charge of the metal cation (Z, Models III and IV). However, it is not just a causal relationship. DCW and charge (Z) are highly correlated (r = 1.0). In other words, the SMILES-based DCW descriptor represents the charge of the metal cation in 100%.
Finally, we found that several descriptors, such as surface area-to-volume ratio (SV) and SiRMS-based descriptor of the van-der-Waals interactions (S1) from the Model V are unconnected (independent) and do not demonstrate causal relationships with both the target toxicity and other descriptors (Fig. 2). It means they have little or even no impact on the relevant toxicity of the metal oxide nanoparticles towards bacteria E. coli.
Based on the above discussion, we can conclude that the descriptor ΔHMe+ (Model I) is the best choice for nano-QSAR modeling. Enthalpy was the only main cause, with clear linear behavior and a significant Pearson's correlation coefficient. All other descriptors are somehow related to each other or to target toxicity. Thus, Occam's razor33 must be applied before formulating the scope of new studies. The Occam's razor principle is a heuristic technique based on the idea that among competing hypotheses, the one with the fewest assumptions should be selected. In other words, it would be helpful to formulate the initial causal hypothesis to see relationships between new descriptors with known theories and models.
Conclusions
In this paper, we have illustrated that causal inference methods could be used efficiently in QSAR modeling (particularly in nano-QSAR) as additional criteria for QSAR quality evaluation. Previously developed nano-QSAR models of metal oxides’ toxicity towards bacteria E. coli have been validated by means of the causality criteria. We have verified the character of the relationships between the descriptors and target toxicity. We found that not every descriptor that is statistically correlated with the studied toxicity endpoint, in fact, explains the toxicity.The relationships between the descriptors were discovered based on the developed causal structure. The causal inference technique is useful in situations when descriptors that are more sophisticated are not applicable (e.g., quantum chemistry software and/or appropriate computational resources are not available).
We have analyzed the descriptors selected by causal criteria in terms of a straightforward causal-reasoning account. Selected descriptors reflect specific properties of the nanoparticle's surface: the release of ions and the fraction of available molecules on the nanocluster's surface. Selected descriptors demonstrate high quality in terms of statistics, causality and interpretation.
Causal inference methods are able to clarify the existing associations and complex causal relationships between the descriptors and between the descriptors and the endpoint. Moreover, the causal inference methods are able to provide more robust mechanistic interpretation of the developed nano-QSAR models. The insights described above allow development of better schema for nano-QSAR modeling using the causal discovery approach.
Acknowledgements
This research has received funding from the European Union Seventh Framework Program (FP7/2007-2013) under grant agreement #309837 (NanoPUZZLES project). The authors are thankful for the financial support of the European Commission through the Marie Curie IRSES program, NanoBRIDGES project (FP7-PEOPLE-2011-IRSES, grant agreement #295128). In the USA the authors are thankful for the support from the NSF CREST Interdisciplinary Nanotoxicity Center NSF-CREST – Grant # HRD-0833178 and NSF-EPSCoR Award #:362492-190200-01\NSFEPS-0903787.Notes and references
- V. Consonni and R. Todeschini, in Recent Advances in QSAR Studies, ed. T. Puzyn, J. Leszczynski and M. T. Cronin, Springer, Netherlands, 2010, vol. 8, ch. 3, p. 29 Search PubMed.
- Guidance document on the validation of (Quantitative) Structure-Activity Relationship [(Q)SAR] models, Paris, 2007, vol. 69, ch. 5 Search PubMed.
- S. R. Johnson, J. Chem. Inf. Model., 2007, 48, 25 CrossRef PubMed.
- Z. Zhao, Y. Huang, B. Zhang, Y. Shyr and H. Xu, BMC Genomics, 2012, 13(Suppl 8), S1 CrossRef PubMed.
- T. Puzyn, B. Rasulev, A. Gajewicz, X. Hu, T. P. Dasari, A. Michalkova, H.-M. Hwang, A. Toropov, D. Leszczynska and J. Leszczynski, Nat. Nanotechnol., 2011, 6, 175 CrossRef CAS PubMed.
- A. P. Toropova, A. A. Toropov, R. Rallo, D. Leszczynska and J. Leszczynski, Ecotoxicol. Environ. Saf., 2015, 112, 39 CrossRef CAS PubMed.
- N. Sizochenko, B. Rasulev, A. Gajewicz, E. Mokshyna, V. Kuz'min, J. Leszczynski and T. Puzyn, RSC Adv., 2015, 5, 77739 RSC.
- G. F. Cooper and C. N. Glymour, Computation, causation, and discovery, AAAI Press, MIT Press, Menlo Park, California, 2002 Search PubMed.
- A. A. Toropov, A. P. Toropova, E. Benfenati, G. Gini, T. Puzyn, D. Leszczynska and J. Leszczynski, Chemosphere, 2012, 89, 1098 CrossRef CAS PubMed.
- S. Kar, A. Gajewicz, T. Puzyn, K. Roy and J. Leszczynski, Ecotoxicol. Environ. Saf., 2014, 107c, 162 CrossRef PubMed.
- N. Sizochenko, B. Rasulev, A. Gajewicz, V. Kuz'min, T. Puzyn and J. Leszczynski, Nanoscale, 2014, 6, 13986 RSC.
- J. Koster, Stat. Med., 2003, 22, 2236 CrossRef.
- D. Janzing, J. Mooij, K. Zhang, J. Lemeire, J. Zscheischler, P. Daniu and B. Schölkopf, Artif. Intell., 2012, 182–183, 1 CrossRef.
- J. Pearl, Causality: models, reasoning, and inference, Cambridge University Press, 2000 Search PubMed.
- P. Spirtes, C. Glymour and R. Scheines, in Causation, Prediction, and Search, Springer, New York, 1993, vol. 81, ch. 3, p. 41 Search PubMed.
- P. Spirtes, C. Glymour and R. Scheines, in Causation, Prediction, and Search, Springer, New York, 1993, vol. 81, ch. 5, p. 103 Search PubMed.
- Causal inference at the Max-Planck-Institute for Intelligent Systems Tübingen, http://webdav.tuebingen.mpg.de/causality/.
- V. E. Kuz'min and et al., Challenges and Advances in Computational Chemistry and Physics, 2010, vol. 8, p. 127 Search PubMed.
- B. M. Smirnov, Phys. -Usp, 2011, 54, 691 CrossRef CAS.
- N. Sizochenko, K. Jagiello, J. Leszczynski and T. Puzyn, J. Phys. Chem. C, 2015, 119, 25542 CAS.
- C. Tatara, M. Newman, J. T. McCloskey and P. L. Williams, Aquat. Toxicol., 1998, 42, 255 CrossRef CAS.
- A. Fujita, J. R. Sato, M. A. Demasi, M. C. Sogayar, C. E. Ferreira and S. Miyano, J. Bioinf. Comput. Biol., 2009, 7, 663 CrossRef CAS.
- NanoBRIDGES (http://nanobridges.eu/).
- V. Rastija and M. Medić-Šarić, Eur. J. Med. Chem., 2009, 44, 400 CrossRef CAS PubMed.
- W. D. Baten and J. S. Frame, Am. Math. Monthly, 1959, 66, 283 CrossRef.
- S. Siqueira Santos, D. Y. Takahashi, A. Nakata and A. Fujita, Briefings Bioinform., 2013, 15, 906 CrossRef PubMed.
- A. Donner, J. J. Koval and J. John, Biometrics, 1980, 36, 19 CrossRef CAS PubMed.
- L. G. Portney and M. P. Watkins, Foundations of clinical research. applications and practice, Appleton & Lange, Connecticut, 1993 Search PubMed.
- I. L. Ruiz, G. C. Garcia and M. A. Gomez-Nieto, Curr. Comput. -Aided Drug Des., 2013, 9, 254 CrossRef CAS PubMed.
- F. Russo and J. Williamson, Int. Stud. Phil. Sci., 2007, 21, 157 CrossRef.
- C. V. Phillips and K. J. Goodman, Emerg. Themes Epidemiol., 2006, 3, 5 CrossRef PubMed.
- M. J. Clugston and R. Flemming, Advanced chemistry, Oxford University Press, Oxford, 2000 Search PubMed.
- A. N. Soklanov, Found. Phys. Lett., 2002, 15, 107 CrossRef.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c5nr08279j |
| This journal is © The Royal Society of Chemistry 2016 |