Surface protein imprinted magnetic nanoparticles for specific recognition of bovine hemoglobin
Weiming
Li
,
Miaomiao
Chen
,
Huayu
Xiong
,
Wei
Wen
,
Hanping
He
,
Xiuhua
Zhang
* and
Shengfu
Wang
Hubei Collaborative Innovation Center for Advanced Organic Chemical Materials, Ministry of Education Key Laboratory for the Synthesis and Application of Organic Functional Molecules & College of Chemistry and Chemical Engineering, Hubei University, Wuhan 430062, P. R. China. E-mail: zhangxh@hubu.edu.cn
First published on 5th November 2015
Abstract
Molecular imprinting for the detection of protein has gained great interest in recent years. For this purpose, we prepared magnetic molecularly imprinted polymers (MIPs) for the recognition of bovine hemoglobin (BHb) through the surface imprinting technique with two-stage core–sell sol–gel polymerization on the surface of silica modified Fe3O4 nanospheres. 3-Aminopropyltriethoxylsilane and octyltrimethoxysilane were chosen as monomers to construct the MIP layer. The morphology and structure property of prepared nanoparticles were characterized by TEM, X-ray diffraction, Fourier transform infrared spectrometry, and the vibrating sample magnetometer. The obtained magnetic MIPs with high saturation magnetization (60 emu g−1) made it easy to separate the target protein from solution by an external magnetic field. The adsorption and recognition performance of this magnetic MIPs was discussed through adsorption kinetics, adsorption isotherms, special selectivity, reusability and reproducibility tests. It turned out that the magnetic MIPs showed a relatively high adsorption capacity of 124.86 mg g−1 and excellent selectivity towards BHb with a separation factor of 1.99. Moreover, the adsorption capacity of magnetic MIPs was not significantly reduced after three continuous adsorption and elution processes, which indicated their good reusability for at least three repeated cycles.
Introduction
Molecular imprinting is an effective technique to produce specific recognition sites that are chemically and sterically complementary in shape, size and functionality to the target molecules in polymer networks. Owing to the advantages, such as good physical, chemical, and thermal stability, high selectivity, and strong affinity, molecular imprinted technology have been applied in wide fields, including bioseparation,1 solid phase extraction,2 biosensors,3,4 chromatography,5,6 and drug delivery.7,8So far, molecular imprinted technology has been successfully applied for the analysis, detection, and separation of a wide range of small molecules.9–12 However, the imprinting of macromolecules like proteins has been far less successful. The problems faced for imprinting of the large molecular size, complexity, flexible conformation, solubility, and inactivation of proteins are usually the reasons why the imprinting of proteins remains challengeable. To meet this challenge, people have developed some strategies to resolve these problems, such as bulk imprinting,13–15 surface imprinting,16–19 boronate affinity molecular imprinting,20,21 and epitope imprinting.22,23 Surface imprinting whose recognition sites were formed close to or at the material surface is a high prior strategy to be employed for protein imprinting. As known to us, proteins are easily entrapped and difficult to be removed from the polymer network completely. Thus the thin film prepared by the surface imprinting technique can speed the response kinetics, and enable target protein access to the surface binding sites.
Nanomaterials, having an extremely large surface area, are superior for surface modification. Therefore, silica nanospheres,24 carbon nanotubes,25 and Fe3O4 nanoparticles26,27 have been widely applied for protein imprinting in recent years. Among the various nanosized substrates, Fe3O4 nanoparticles have gained wide attraction due to their easy preparation and low toxicity. Moreover, their easy magnetically susceptible properties can simplify the separation process to a large extent. When Fe3O4 nanoparticles are encapsulated inside the molecularly imprinted polymers (MIPs), the resulting materials are magnetically susceptible and can be easily separated by external magnetic fields after they had finished their adsorption and recognition. They showed obvious simplicity and biocompatibility. Therefore, combining Fe3O4 nanoparticles with the surface imprinting technique has become a subject of intensive research.
In this study, 3-aminopropyltriethoxylsilane (APTES) and octyltrimethoxysilane (OTMS) have been chosen as monomers for the investigation of bovine hemoglobin (BHb) imprinting. For this purpose, magnetic MIPs have been prepared through the surface imprinting technique and two-stage core–sell sol–gel polymerization. The excellent adsorption and recognition performance of this magnetic MIPs demonstrate that the magnetic MIPs are likely for protein detection.
Experimental
Materials and methods
| Q = (c0 − ct)V/m | (1) |
To evaluate the selectivity properties of magnetic MIPs and NIPs, we imported the concept of the imprinting factor (IF) and the separation factor (SF), which could be calculated by the following formula:
| IF = QMIP/QNIP | (2) |
| SF = IFBHb/SFBSA | (3) |
Results and discussion
Synthesis of magnetic MIPs
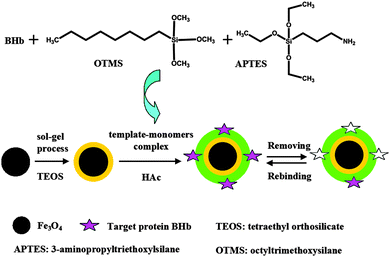
The preparation of magnetic MIPs is illustrated in Fig. 1. The silica shells were deposited on the surface of Fe3O4 by a sol–gel process by using TEOS. TEOS can cause hydrolyzation under alkaline conditions, a process of nucleophilic substitution reaction. The OH− of the alkali attacks the center Si atom of TEOS, to form a large amount of Si(OH)4, which then coats the surface of Fe3O4 nanoparticles in the form of SiO2.29 Fe3O4 nanoparticles having silica shells provided features of good biocompatibility, non-toxic, chemically stable, and high dispersion in all solvents.28 The magnetic dipolar attraction between Fe3O4 nanoparticles could be prevented by the silica shells, which explained the excellent stability of silica modified Fe3O4. Then the template protein (BHb)-monomer (APTES and OTMS) complex was formed through copolymerization. After that, MIP shells were anchored on the surface of silica modified Fe3O4 by a surface imprinted process. In this procedure, HAc was used as catalyst.28 Finally, the BHb was extracted with 0.1 M NaOH to form the imprinted cavities complementary to BHb in shape, size, and functional group orientation.Characterization of the magnetic nanoparticles
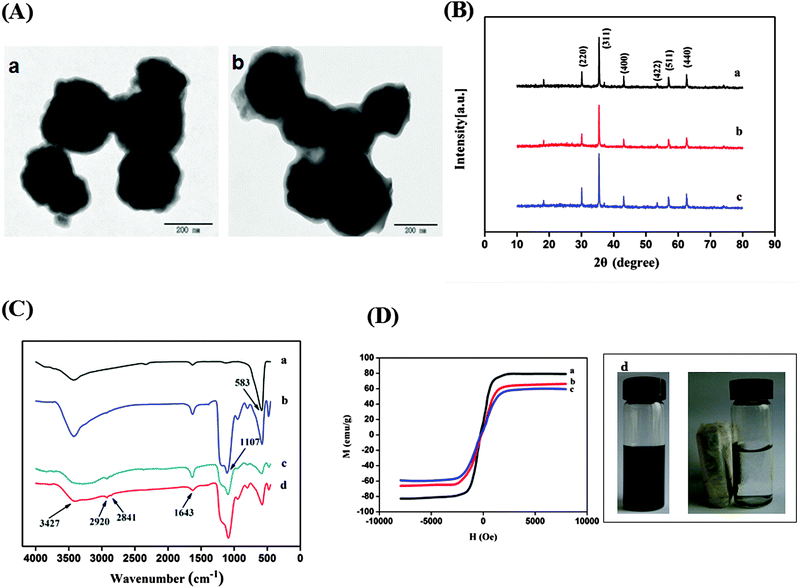
TEM images of silica modified Fe3O4 and magnetic MIPs are shown in Fig. 2A. It was clear that SiO2 shells and MIP layers were coated on the surface of Fe3O4 successively and the core–shell structure was successfully prepared.XRD patterns for the synthesized magnetic nanoparticles are shown in Fig. 2B. In the 2θ range of 10°–80°, six characteristic peaks for Fe3O4 (2θ = 30.38°, 35.38°, 43.14°, 53.48°, 57.08°, and 62.66°) were all observed for the three samples, and the peak position at the corresponding 2θ value were indexed to (220), (311), (400), (422), (511), and (440), respectively, which matched well with the database of magnetite in the JCPDS-International Center for Diffraction Data (JCPDS card: 019-0629) file. The XRD patterns showed that the synthesized process of magnetic MIPs and NIPs does not change the XRD phase of Fe3O4.
FTIR spectroscopy was used to study the chemical structure of Fe3O4, silica modified Fe3O4, and magnetic NIPs and MIPs (Fig. 2C). A sharp and strong Fe–O stretching peak (∼583 cm−1) was observed for all nanoparticles, indicating that the main structure is not changed by modification. The peak at 1107 cm−1 in the spectrum of silica modified Fe3O4 nanoparticles was attributed to the Si–O–Si stretching vibration, and the absorbance band was very strong, which conformed that the silica coating is successfully grafted on the surface of Fe3O4 nanoparticles. In Fig. 2C(c) and C(d), the characteristic peak of magnetic NIPs and MIPs at 2841 cm−1 and 2920 cm−1 were the characteristic peaks of C–H in APTES and OTMS, and the peak at 1643 cm−1 and 3427 cm−1 were the bending vibration and stretching vibration of N–H in APTES.30,31 These results confirmed that the coatings of the BHbmonomer complex layers were successfully formed on the surface of the silica modified Fe3O4.
To study the magnetic properties of nanoparticles, the VSM was employed at room temperature. The magnetic hysteresis curves of the dried Fe3O4, silica modified Fe3O4, and magnetic MIPs are illustrated in Fig. 2D. The saturation magnetization values of silica modified Fe3O4 and magnetic MIPs were 66 emu g−1 (Fig. 2D(b)) and 60 emu g−1 (Fig. 2D(c)), respectively, which were obviously lower than Fe3O4 (79 emu g−1) (Fig. 2D(a)). The reason was that the silica coating and the BHb-monomer complex layers were successively formed on the surface of Fe3O4, which have a shielding effect on the magnetic properties of the nanoparticles. The saturation magnetization of the prepared magnetic MIPs was high enough to make them susceptible to magnetic fields and could be easily and quickly separated from the suspension. As shown in Fig. 2D(d), a dark homogeneous dispersion existed in the absence of an external magnetic field, the black magnetic MIPs were soon attracted to the wall of the vial with the solution becoming clear and transparent when an external magnetic field was applied. The results demonstrated that the high magnetism of the magnetic MIPs can be used for the quick magnetic separation of proteins.
The effect of pH on the adsorption of magnetic MIPs and magnetic NIPs
The zeta potentials and binding capacities of magnetic MIPs and NIPs in the presence of 300 mg L−1 BHb at five different pH values were measured, and the results are shown in Table 1. It was observed that the zeta potential of magnetic MIPs and NIPs was all negative and the adsorption capacities of magnetic MIPs and NIPs to bind BHb depend upon pH. It was obvious that the optimal binding capacities for magnetic MIPs were at pH 7.0. The reason was that magnetic MIPs and BHb carry opposite charges at pH 7.0, the electrostatic interactions between negative polymers and positive BHb (pI 6.8) enhance the adsorption. We could also find that the imprinting factors presented an irregular variation tendency at different pH values, and the imprinting factor was highest at pH 7.0. Therefore, considering the adsorption capacity and the imprinting factor, the incubation buffer of pH 7.0 was adopted in the following adsorption experiments.| pH | Zeta potential (mV) | Q for magnetic MIPs (mg g−1) | Q for magnetic NIPs (mg g−1) |
|---|---|---|---|
| 5.0 | −20.1 | 52.56 | 52.28 |
| 6.0 | −26.1 | 83.70 | 67.85 |
| 7.0 | −32.1 | 124.86 | 62.85 |
| 8.0 | −36.6 | 67.30 | 58.59 |
| 9.0 | −39.5 | 10.85 | 7.23 |
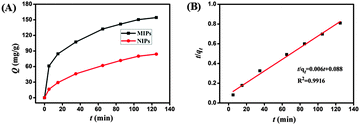
Adsorption kinetics
Adsorption kinetics experiment was carried out by changing the adsorption time from 0 to 125 min, and the initial concentration of BHb was kept constantly at 300 mg L−1. The curve-fitted using a sigmoid curve of the dynamic adsorption is presented in Fig. 3A. This curve revealed a rapid dynamic adsorption of BHb to the magnetic MIPs. In the first 65 min, absorption amounts increased rapidly with the increase of adsorption time, and the adsorption capacity of magnetic MIPs increased slightly in the afterward time. The adsorption trend of magnetic NIPs was similar to magnetic MIPs, but the adsorption capacity of magnetic NIPs was relatively small. There maybe two reasons for this phenomenon, one was that imprinted cavities formed on magnetic MIPs are complementary to the template molecule in both shape and functionality. While magnetic NIPs had no imprinted cavities, the adsorption process mainly was nonspecific adsorption which includes van der Waals' force, and the other was the hydrogen bond that may also exist between proteins and the imprinted cavities.To investigate the adsorption mechanism, the second-order kinetic model was used to analyse the kinetic data. The model can be expressed as follows:
| t/qt = 1/(k2qe2) + t/qe = 1/v0 + t/qe | (4) |
Adsorption isotherms
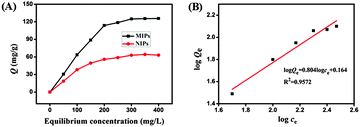
The binding isotherms of BHb onto the magnetic MIPs and magnetic NIPs were determined in the concentration range of 50–400 mg L−1 (initial concentration). Fig. 4A shows the adsorption isotherm of magnetic nanomaterials, from which we could see that binding capacity for magnetic MIPs increased distinctly along with increasing BHb concentrations from 50 to 300 mg L−1. When the initial concentration of BHb was 300 mg L−1, the adsorption capacity of magnetic MIPs reached a value of 124.86 mg g−1, while the adsorption of magnetic NIPs was 62.85 mg g−1. And the magnetic MIPs had higher binding capacity for BHb than that of the magnetic NIPs at the same initial concentration, which was due to the imprinting effect. The imprinted factor was 1.99, indicating that the magnetic MIPs are superior to their non-imprinted counterparts in capturing BHb.The high adsorption property of magnetic MIPs to template proteins, which is higher than some other studies,17,26,29 indicating that the imprinted sites are on the surface of magnetic MIPs. The high adsorption property is essential for applying our imprinted nanoparticles to the detection of BHb.
The adsorption equilibrium of BHb on magnetic MIPs and NIPs was found to obey the Freundlich isotherm:33
| Qe = Qfce1/n | (5) |
lg![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Qe = 1/n lg Qe = 1/n lg![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) ce + lg ce + lg![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Qf Qf | (6) |
| K0 = Qfn | (7) |
The Freundlich isotherm is suitable for multilayer adsorption of a heterogeneous system and not restricted to the formation of the monolayer. The value of R2 is 0.9572 (Fig. 4B), indicating that the Freundlich model is suitable for describing the adsorption process and the adsorption of BHb on magnetic MIPs is heterogeneous.17
Compared with other reported studies, the results are shown in Table 2, from which we could see that our method displayed some advantages in aspects of imprinting effects and showed an encouraging result.
| Matrix | Template molecule | Monomers | Q MIP (mg g−1) | IF | Ref. |
|---|---|---|---|---|---|
| Lyz: lysozyme, γ-APS: γ-aminopropyltriethoxysilane, NIPAM: N-isopropylacrylamide, MAA: methacrylic acid, AAm: acrylamide, DMAEMA: 2-(dimethylamino)ethyl methacrylate, MBA: N,N′-methylenebisacrylamide. | |||||
| Fe3O4 | BHb | γ-APS/TEOS | 10.52 | 4.61 | 26 |
| Fe3O4 | BHb | Dopamine | 4.65 | 1.51 | 29 |
| Fe3O4 | BSA | APTES/OTMS | 42.52 | 9.53 | 28 |
| Fe3O4 | BSA | NIPAM/MAA | 71.85 | 1.70 | 34 |
| SiO2 | Lyz | AAm/MAA/DMAEMA/MBA | 17.7 | 1.46 | 35 |
| Fe3O4 | BHb | APTES/OTMS | 124.86 | 1.99 | This work |
Special selectivity of magnetic MIPs and NIPs
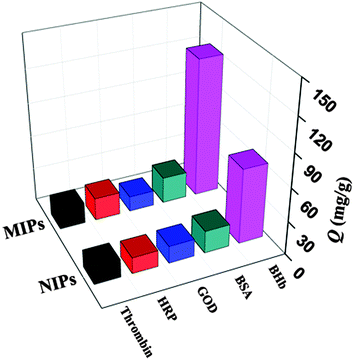
The special selectivity test of magnetic MIPs and NIPs was carried out by using BSA (Mw 68 kDa, pI 4.6), GOD (Mw 13.82 kDa, pI 4.9), HRP (Mw 40 kDa, pI 7.2), and thrombin (Mw 36 kDa, pI 5.0) as the comparative proteins. The results of special selectivity tests of magnetic MIPs and NIPs for the BHb and other comparative proteins are shown in Fig. 5 and Table 3.| Proteins | Q MIP (mg g−1) | Q NIP (mg g−1) | IF | SF |
|---|---|---|---|---|
| Adsorption condition: V = 10 mL, mMIP = mNIP = 20 mg, cBHb = cBSA = cGOD = cHRP = cThrombin = 300 mg L−1, time 65 min, room temperature, Tris-HCl (pH = 7.0, 10 mM). | ||||
| BHb | 124.86 | 62.85 | 1.99 | — |
| BSA | 23.70 | 20.15 | 1.18 | 1.69 |
| GOD | 13.69 | 15.48 | 0.88 | 2.26 |
| HRP | 20.31 | 14.61 | 1.39 | 1.44 |
| Thrombin | 18.61 | 15.83 | 1.18 | 1.69 |
As displayed in Fig. 5 and Table 3, it was clear that the binding capacity of magnetic MIPs for BHb was much higher than those of comparative proteins. It was probable that the size, shape and position of recognition function groups of BHb were complementary to the binding sites in magnetic MIPs, while the structure of other proteins was not complementary to the binding sites. Large protein BSA (Mw 68 kDa, pI 4.6) was difficult to be captured, because the space resistance made them less accessible to magnetic nanoparticles.27 For electrically neutral protein HRP (Mw 40 kDa, pI 7.2), the low Q value resulted from the absence of the electrostatic interaction in the capture process. On the basis of the above observation, multiple interactions between MIPs and the target protein played an important role in the selective capture process.
The imprinted factor was 1.99, which was higher than some other imprinted polymers for BHb.29,35 Although the imprinted factor was not high, the magnetic MIPs showed specific adsorption to template molecules which can be demonstrated from the fast adsorption kinetics, adsorption isotherm, and special selectivity tests.
Reusability and reproducibility of the magnetic MIPs
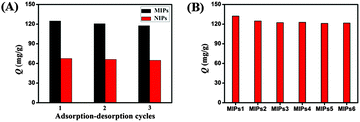
The reusability and reproducibility are the most important properties for the application of imprinted polymers. The cycles of adsorption and elution processes by using the same batch of magnetic MIPs and NIPs are shown in Fig. 6A. The results showed that there was a slight decrease in the adsorption capacity after three continuous adsorption and elution processes for magnetic MIPs, while the binding capacity of magnetic NIPs remained almost unchanged. The reason for this phenomenon may be that some recognition sites in the network of MIPs could be blocked after regeneration or destroyed after rewashing, therefore they were no longer fit for the template. However, the adsorption of BHb to NIPs was nonspecific, so the effect of regeneration or washing was negligible.28 The results demonstrated that the magnetic MIPs had a high stability and remained almost with the same ability to absorb the target protein for at least three regeneration cycles, which is a clear advantage over single-use materials.For magnetic MIPs, especially with protein as the template, the synthesis reproducibility is an important issue to consider.36 In our experiment, the reproducibility of six batches of magnetic MIPs prepared on different days was investigated. From Fig. 6B, it could be seen that the reproducibility of magnetic MIPs was satisfactory, that was because the template immobilization and polymerization under the respectively optimized conditions were beneficial for keeping stable reaction solution. Besides, the prepolymerization between BHb and the monomers probably could improve the conformation stability of the template protein.16
Conclusion
In summary, the magnetic MIPs were synthesized by combining the surface imprinting technique with two-stage core–sell sol–gel polymerization. The synthesis of magnetic MIPs was simple, convenient and low-cost. The results of adsorption experiments indicated that the imprinted nanoparticles, where the imprinted sites are close to or at the surface, had a relatively high adsorption capacity, fast adsorption kinetics, and excellent specific selectivity towards template proteins. Moreover, this type of magnetic MIP showed high stability and are reusable for good repeatability. So the magnetic MIPs showed promising applications in the separation of biomacromolecules.Acknowledgements
This work was financially supported by the National Natural Science Foundation of China (No. 21075029, 21375033), the Natural Science Fund for Creative Research Groups of Hubei Province of China (No. 2011CDA111, 2014CFA015), the Key Project of the Natural Science Foundation of Hubei Province (No. 2015CFA124) and the Program for Excellent Youth Scholars of Innovative Research Team by Hubei Provincial Department of Education (No. T201101).Notes and references
- Z. H. Zhang, X. Yang, X. Chen, M. L. Zhang, L. J. Luo, M. J. Peng and S. Z. Yao, Anal. Bioanal. Chem., 2011, 401, 2855 CrossRef CAS PubMed.
- X. J. Liu, Z. Y. Chen, R. Zhao, D. H. Shangguan, G. Q. Liu and Y. Chen, Talanta, 2007, 71, 1205 CrossRef CAS PubMed.
- K. Reimhult, K. Yoshimatsu, K. Risveden, S. Chen, L. Ye and A. Krozer, Biosens. Bioelectron., 2008, 23, 1908 CrossRef CAS PubMed.
- C. C. Hong, C. C. Lin, C. L. Hong and P. H. Chang, Biomed. Microdevices, 2012, 14, 435 CrossRef CAS PubMed.
- H. Q. Zhang, L. Ye and K. Mosbach, J. Mol. Recognit., 2006, 19, 248 CrossRef CAS PubMed.
- J. Haginaka, Anal. Bioanal. Chem., 2004, 379, 332 CrossRef CAS PubMed.
- B. Sellergren and C. J. Allender, Adv. Drug Delivery Rev., 2005, 57, 1733 CrossRef CAS PubMed.
- M. E. Byrne and V. Salian, Int. J. Pharm., 2008, 364, 188 CrossRef CAS PubMed.
- D. X. Nie, D. W. Jiang, D. Zhang, Y. Liang, Y. Xue, T. S. Zhou, L. T. Jin and G. Y. Shi, Sens. Actuators, B, 2011, 156, 43 CrossRef CAS.
- X. L. San, L. J. Zhang, H. X. Zhang, H. Qian, Y. Z. Zhang, L. L. Tang and Z. J. Li, J. Agric. Food Chem., 2014, 62, 4552 CrossRef PubMed.
- J. Matsui, K. Akamatsu, N. Hara, D. Miyoshi, H. Nawafune, K. Tamaki and N. Sugimoto, Anal. Chem., 2005, 77, 4282 CrossRef CAS PubMed.
- G. H. Yao, R. P. Liang, C. F. Huang, Y. Wang and J. D. Qiu, Anal. Chem., 2013, 85, 11944 CrossRef CAS PubMed.
- M. J. Whitcombe, I. Chianella, L. Larcombe, S. A. Piletsky, J. Noble, R. Porter and A. Horgan, Chem. Soc. Rev., 2011, 40, 1547 RSC.
- Y. Ma, G. Q. Pan, Y. Zhang, X. Z. Guo and H. Q. Zhang, Angew. Chem., Int. Ed., 2013, 52, 1511 CrossRef CAS PubMed.
- S. Shinde, A. Bunschoten, J. A. W. Kruijtzer, R. M. J. Liskamp and B. Sellergren, Angew. Chem., Int. Ed., 2012, 51, 8326 CrossRef CAS PubMed.
- J. X. Liu, Q. L. Deng, D. Y. Tao, K. G. Yang, L. H. Zhang, Z. Liang and Y. K. Zhang, Sci. Rep., 2014, 4, 5487 CAS.
- Y. X. Li, Q. Bin, Z. Y. Lin, Y. T. Chen, H. H. Yang, Z. W. Cai and G. N. Chen, Chem. Commun., 2015, 51, 202 RSC.
- Q. R. Li, K. G. Yang, Y. Liang, B. Jiang, J. X. Liu, L. H. Zhang, Z. Liang and Y. K. Zhang, ACS Appl. Mater. Interfaces, 2014, 6, 21954 CAS.
- R. Q. Lv, T. W. Tan and F. Svec, Biotechnol. Adv., 2013, 31, 1172 CrossRef PubMed.
- L. Li, Y. Lu, Z. J. Bie, H. Y. Chen and Z. Liu, Angew. Chem., Int. Ed., 2013, 52, 7451 CrossRef CAS PubMed.
- R. Hajian, V. Z. Shahabadi, A. Ghaedi, H. Izadi and M. Zafari, Chem. Sci., 2014, 5, 1135 RSC.
- D. F. Tai, C. Y. Lin, T. Z. Wu and L. K. Chen, Anal. Chem., 2005, 77, 5140 CrossRef CAS PubMed.
- L. Xu, Y. F. Hu, F. Shen, Q. S. Li and X. Q. Ren, J. Chromatogr. A, 2013, 1293, 85 CrossRef CAS PubMed.
- L. Li, L. L. Yang, Z. L. Xing, X. J. Lu and X. W. Kan, Analyst, 2013, 138, 6962 RSC.
- R. Liu, M. Sha, S. S. Jiang, J. Luo and X. Y. Liu, Talanta, 2014, 120, 76 CrossRef CAS PubMed.
- X. W. Kan, Q. Zhao, D. L. Shao, Z. R. Geng, Z. L. Wang and J. J. Zhu, J. Phys. Chem. B, 2010, 114, 3999 CrossRef CAS PubMed.
- N. Li, L. Qi, Y. Shen, J. Qiao and Y. Chen, ACS Appl. Mater. Interfaces, 2014, 6, 17289 CAS.
- R. X. Gao, X. R. Mu, J. J. Zhang and Y. H. Tang, J. Mater. Chem. B, 2014, 2, 783 RSC.
- X. P. Jia, M. L. Xu, Y. Z. Wang, D. Ran, S. Yang and M. Zhang, Analyst, 2013, 138, 651 RSC.
- Q. W. Peng, J. Gan, S. F. Wang, L. B. Kong, G. R. Chen, Y. X. Yang and G. J. Huang, Ind. Eng. Chem. Res., 2013, 52, 7713 CrossRef CAS.
- S. Mohapatra, N. Pramanik, S. Mukherjee, S. K. Ghosh and P. Pramanik, J. Mater. Sci., 2007, 42, 7566 CrossRef CAS.
- C. Y. Piao and L. G. Chen, J. Chromatogr. A, 2012, 1268, 185 CrossRef CAS PubMed.
- K. L. Gurunatha and E. Dujardin, J. Phys. Chem. C, 2013, 117, 3489 CAS.
- X. J. Li, B. L. Zhang, W. Li, X. F. Lei, X. L. Fan, L. Tian, H. P. Zhang and Q. Y. Zhang, Biosens. Bioelectron., 2014, 51, 261 CrossRef CAS PubMed.
- H. Y. He, G. Q. Fu, Y. Wang, Z. H. Chai, Y. Z. Jiang and Z. L. Chen, Biosens. Bioelectron., 2010, 26, 760 CrossRef CAS PubMed.
- X. T. Shen, T. C. Zhou and L. Ye, Chem. Commun., 2012, 48, 8198 RSC.
| This journal is © The Royal Society of Chemistry and the Centre National de la Recherche Scientifique 2016 |