High resolution metabolomics technology reveals widespread pathway changes of alcoholic liver disease†
Aihua
Zhang‡
ab,
Guangli
Yan‡
ab,
Xiaohang
Zhou
ab,
Yangyang
Wang
ab,
Ying
Han
ab,
Yu
Guan
ab,
Hui
Sun
*ac and
Xijun
Wang
*ab
aNational TCM Key Laboratory of Serum Pharmacochemistry, Laboratory of Metabolomics, Research Center of Chinmedomics, Heilongjiang University of Chinese Medicine, Heping Road 24, Harbin 150040, China. E-mail: wangxijunomics@126.com; xijunwangls@126.com; Fax: +86-451-82110818; Tel: +86-451-82193039
bResearch Center of Chinmedomics, Heilongjiang University of Chinese Medicine, Heping Road 24, Harbin 150040, China
cDepartment of Pharmaceutical Analysis, School of Pharmacy, Heilongjiang University of Chinese Medicine, Heping Road, Harbin, China
First published on 23rd November 2015
Abstract
Alcoholic liver disease (ALD) is a significant cause of death and morbidity. However little is known regarding the widespread pathway changes of ALD disorder. This study utilized metabolomic profiling to examine the pathogenic mechanisms of ALD based on a rat model. A total of 21 metabolites with significant changes were identified, involving several key metabolic pathways such as pentose and glucuronate interconversions, starch and sucrose metabolism, cysteine and methionine metabolism. Furthermore, the differential proteins corresponding to alterations in metabolism across the metabolic network were identified using iTRAQ-based quantitative proteomics analysis. The proteins appear to be involved in protein binding, metabolism, immune response, and signal conduction. Interestingly, integrated omics profiling firstly reveals that p53 and Fc epsilon RI signaling pathways were closely related to ALD. Our study indicates that most of these proteins were found to play a pivotal role in the regulation of multiple metabolism pathways. Collectively, the current study provides insights into the molecular mechanisms of ALD from widespread pathway changes.
Introduction
Alcohol has been a part of human culture since the beginning of recorded history. Problems associated with excess alcohol consumption include social issues, increased accidents, chronic health problems and mortality.1 Alcohol related morbidity in developed countries is second only to tobacco use and is responsible for 2.5 million deaths globally each year, and costs 1% of the GDP of middle to high income countries.2 Alcohol misuse is a major public health problem worldwide and accounts for elevated social and economic costs. Alcoholic liver disease (ALD) is a common complication of alcohol misuse. The detection of ALD at an early stage could provide opportunities for more optimal management. The biomarkers including the erythrocyte mean cell volume, c-glutamyltransferase and carbohydrate-deficient transferring provide an objective measure of alcohol consumption, and they can assist in the detection of at-risk drinking.3 Clearly, there is a need for more effective and definitive treatment options in order to improve prognosis and outcome of patients with severe ALD. Fortunately, integrated omics technology has been used to explore the particular metabolites, potentially diagnostic and prognostic biomarkers for a deep understanding of the essence of diseases.4–6At the end of the 20th century, genomics wrote out the ‘script of life’; proteomics decoded the script; and metabolomics came into bloom.7 These ‘omics’ quickly became the thrust of life sciences, pushing the discipline to a new high. At present, numerous studies have discovered potential markers of disease using proteomics.8 Isobaric tags for relative and absolute quantitation (iTRAQ), as a quantitative method, is a common tool in proteomics and has been extensively used for biomarker discovery in various disease contexts.9 Metabolomics is the endpoint of genotype functions and the biochemical phenotype in the body, and is linked closely to the alteration of functions in the body, and incorporates a ‘top-down’ strategy to reflect the terminal symptoms of a whole system.10 Recent advances have suggested that metabolite profiles will improve the understanding of the disease mechanisms.11–17 Global metabolomic profiling with alcoholic fibrosis has proceeded very slowly, and changes in the serum proteomes of ALD are rarely reported. There is an urgent need for the discovery of novel molecular signatures to understand the underlying biological basis for ALD. However, to date there have been few studies aimed at gaining deeper insights into ALD through an integrated metabolomics approach. Therefore, unlike other studies, this study was to devise a systematically integrated omics approach to focus and identify widespread pathway changes of ALD in rats.
Materials and methods
Reagents
HPLC grade acetonitrile was obtained from Merck (Darmstadt, Germany); methanol (HPLC grade) was purchased from Fisher Scientific Corporation (Loughborough, UK); water was produced using a Milli-Q Ultra-pure water system (Millipore, Billerica, USA); formic acid was obtained from Honeywell Company (Morristown, New Jersey, USA); leucine enkephalin was purchased from Sigma-Aldrich (St. Louis, MO, USA). The iTRAQ reagent multi-plex kit, containing the iTRAQ reagents, was bought commercially (Applied Biosystems, Foster City, CA, USA). The assay kits for alanine aminotransferase (ALT), aspartate amino transferase (AST), alkaline phosphatase (ALP), triglycerides (TG), total bilirubin (T-BIL), total protein (TP), albumin (ALB), total cholesterol (CHOL), hyaluronic acid (HA), laminin (LN), procollagen Iii (PIIINP), and collagen IV (CIV) were purchased from the Nanjing Jiancheng Biotech Company (Nanjing, China).Animals
Male Wistar rats were maintained in a specific pathogen-free environment. The animals were allowed to acclimatize in metabolic cages for 1 week prior to treatment. The animals were randomly assigned to 4 groups of 8 rats each as follows: control, 7W, 11W and 12W groups. The rats in the model group were orally administrated at a dose of 0.8 ml/100 g mixture (6 g kg−1 alcohol liquor) and high-fat diet (high fat emulsion (10 ml kg−1)) for 12 consecutive weeks. The alcohol-fed model rats were fed ad libitum. Rats were sacrificed by an intraperitoneal injection of 1% pentobarbital sodium (0.15 ml/100 g body weight) at four time points: 4, 7, 11 and 12 weeks after initiation of injection. Livers were collected and washed three times with saline water. Each liver was cut into two pieces. The small piece was immediately fixed in buffered formalin for pathological staining. We collected plasma samples in heparinized tubes, kept them on ice for 1 h and centrifuged them at 5000 rpm for 20 min at 4 °C, flash frozen in liquid nitrogen and stored at −80 °C until the liver function tests and proteomics analyses were performed. Urine was collected daily (at 8:00 am) from the metabolic cages at ambient temperature throughout the entire procedure and centrifuged at 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm at 4 °C for 5 min to remove any solid debris; the supernatants were then stored frozen at −80 °C for subsequent metabolomic analysis. The study was approved by the Ethical Committee of Heilongjiang University of Chinese Medicine and was conducted according to the principles expressed in the Declaration of Helsinki.
000 rpm at 4 °C for 5 min to remove any solid debris; the supernatants were then stored frozen at −80 °C for subsequent metabolomic analysis. The study was approved by the Ethical Committee of Heilongjiang University of Chinese Medicine and was conducted according to the principles expressed in the Declaration of Helsinki.
Liver histology and biochemical assay
We quantified the levels of ALT, AST, ALP, TG, T-BIL, TP, ALB, CHOL, HA, LN, PCIIINP, CIV activities using assay kits according to the manufacturer's instructions. Liver samples from each rat were fixed in 10% neutral buffered formaldehyde solution, embedded in paraffin, stained with hematoxylin-eosin (HE) and Masson trichrome collagen stain and then examined under an optical microscope.Metabolomics analysis
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm for 10 minutes at 4 °C, and then filtered through a 0.22 μm syringe filter, 3 μL of the supernatant were injected into the UPLC/MS.
000 rpm for 10 minutes at 4 °C, and then filtered through a 0.22 μm syringe filter, 3 μL of the supernatant were injected into the UPLC/MS.
iTRAQ-based quantitative proteomic analysis
Results
Histopathology and biochemical analysis
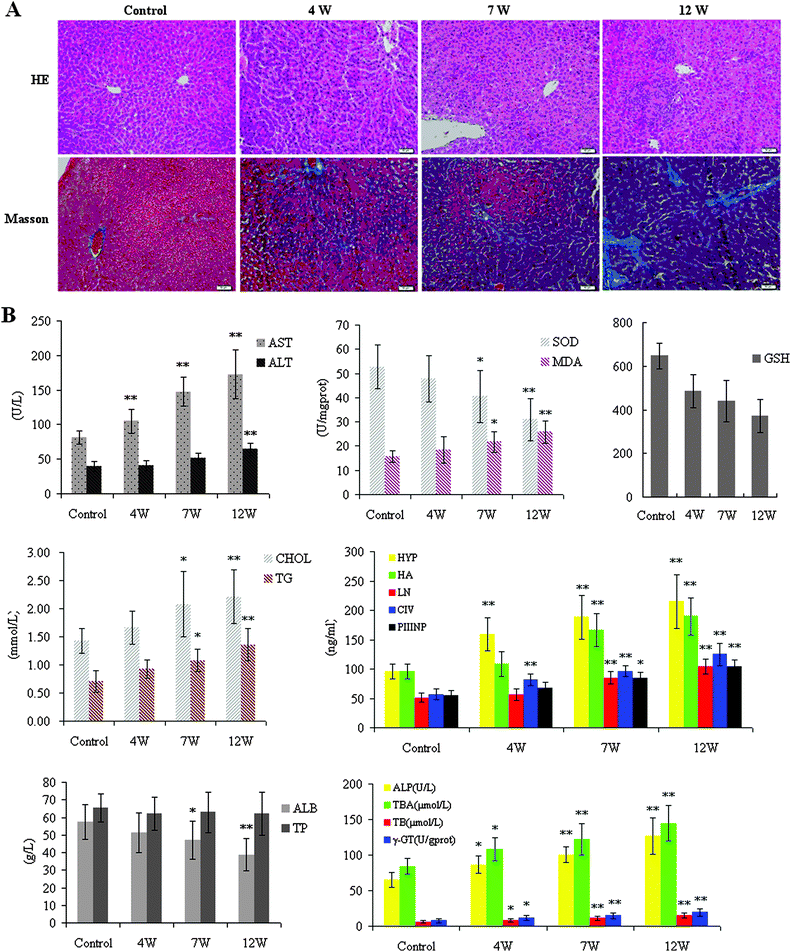
For histopathological analysis, liver samples were quickly obtained and fixed in a 10% neutral buffered formaldehyde solution, embedded in paraffin and sectioned. Paraffin sections were stained with H&E for routine examination, followed by Masson staining for collagen. H&E staining and Masson's staining showed the presence of hepatic fibrosis, marked fatty degeneration, and collagen accumulation in the model group (Fig. 1A). The HE staining showed that ALD was more severe in the 12W-treated group than in the 7W-treated group. Representative figures of HE-stained sections of liver tissue from the control group were evaluated as a normal histology. The liver lobules of the control group showed no pathological changes and the liver cells were normal. Compared with controls, liver cell steatosis was apparent in liver samples at 4W. Foci infiltration around fibrosis tissue can be seen. Large numbers of black and brown particles were deposited and accompanied by fibrosis tissue at 7W, and a similar pattern was evident at 12W. The collagen was shown to be green-colored in the Masson-stained section at 12W.The extent of liver injury was assessed by measuring the enzymes of the liver in the mouse serum. As shown in the liver index, it was significantly different among the experimental groups. The liver index was decreased in rat groups compared to controls (Fig. 1B). AST and ALT concentrations in serum were used as biochemical markers to evaluate hepatic injury. Serum activities of ALT and AST were found to be significantly increased in the 12W-treated group when compared with the control group (p < 0.01). The value of MDA in the serum was decreased (p < 0.01) in the 12W-treated group, but the values of the activities of SOD in the liver tissue were increased (p < 0.01) compared to the control group, and there was no significant change in either group at 4W. Both serum CHOL, TG, and the liver tissue Hyp, HA, LN, PCIIINP, CIV were markedly increased at 12W. The TP content in the 12W-treated group was markedly decreased (p < 0.01), while the level of ALP, T-BA, TB, and r-GT was increased, compared with that of controls. A significant increase levels of the liver enzymes in the serum was occurred at 12W, suggesting that the pathological changes of ALD. Consequently, combined with the histopathological manifestation results, we successfully established ALD in animal models at 12W.
Urine metabolomic profiles
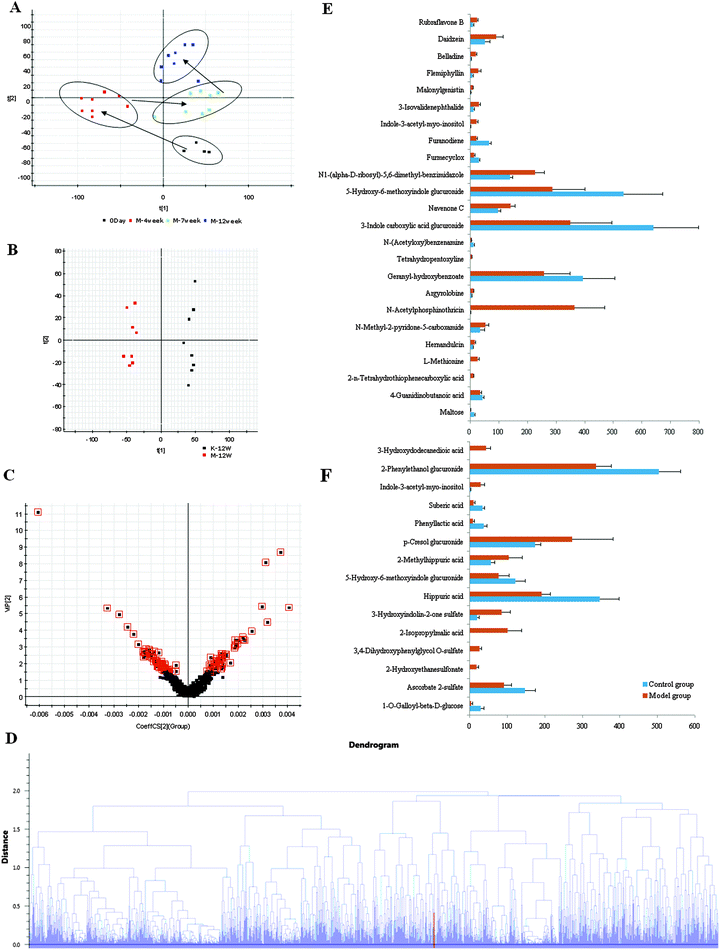
Metabolomics involves studying the processes of all metabolites in diseased samples, thereby revealing disease-related metabolic pathways. The total ion chromatograms exhibited the ideal separation result under the optimized gradient elution procedure. The urine metabolomic profile of each sample consisted of approximately 8000 peaks. Low molecular mass metabolites could be separated well in the short time of 10 min. In order to better visualize the subtle similarities and differences among these complex data sets, multiple pattern recognition methods were employed to phenotype the urine metabolome of rats.| No. | Retention (time/min) | Ion form | Determined (mass/Da) | Calculated (mass/Da) | Error (mDa) | Formula | Identity | VIP-value | t-Test /p-value | Trend |
|---|---|---|---|---|---|---|---|---|---|---|
| a Detected metabolites in both positive and negative modes. | ||||||||||
| 1 | 1.05 | [M + H]+ | 150.0581 | 150.0589 | −0.8 | C5H11NO2S | L-Methionine | 2.9 | 4.65 × 10−8 | ↑ |
| 2 | 1.78 | [M + H]+ | 153.0647 | 153.0664 | −1.7 | C7H8N2O2 | N-Methyl-2-pyridone-5-carboxamide | 1.9 | 3.88 × 10−2 | ↑ |
| 3 | 1.81 | [M + H]+ | 246.0547 | 246.0507 | 4.0 | C7H14NO5P | N-Acetylphosphinothricin | 11.1 | 1.19 × 10−7 | ↑ |
| 4 | 3.59 | [M + H]+ | 152.0698 | 152.0712 | −1.4 | C8H9NO2 | N-(Acetyloxy)benzenamine | 1.5 | 1.03 × 10−3 | ↓ |
| 5 | 4.03 | [M + H]+ | 338.0883 | 338.0876 | 0.7 | C15H15NO8 | 3-Indole carboxylic acid glucuronide | 8.7 | 1.87 × 10−3 | ↓ |
| 6 | 4.26 | [M + H]+ | 340.1020 | 340.1022 | −0.2 | C15H17NO8 | 5-Hydroxy-6-methoxyindole glucuronide | 8.1 | 1.46 × 10−3 | ↓ |
| 7 | 4.61 | [M + H]+ | 279.1328 | 279.1345 | −1.7 | C14H18N2O4 | N1-(alpha-D-ribosyl)-5,6-dimethyl-benzimidazole | 5.3 | 6.08 × 10−6 | ↑ |
| 8 | 5.91 | [M + H]+ | 338.1247 | 338.1240 | 0.7 | C16H19NO7 | Indole-3-acetyl-myo-inositol | 2.7 | 1.08 × 10−7 | ↑ |
| 9 | 0.70 | [M − H]− | 377.0752 | 377.0720 | 3.2 | C13H16O10 | 1-O-Galloyl-beta-D-glucose | 2.1 | 6.19 × 10−6 | ↓ |
| 10 | 0.87 | [M − H]− | 124.9908 | 124.9909 | −0.1 | C2H6O4S | 2-Hydroxyethanesulfonate | 1.8 | 4.51 × 10−7 | ↑ |
| 11 | 0.89 | [M − H]− | 295.0138 | 295.0124 | 1.4 | C8H10O7S | 3,4-Dihydroxyphenylglycol O-sulfate | 2.2 | 2.63 × 10−8 | ↑ |
| 12 | 1.26 | [M − H]− | 221.0656 | 221.0661 | −0.5 | C7H12O5 | 2-Isopropylmalic acid | 4.2 | 4.78 × 10−6 | ↑ |
| 13 | 1.87 | [M − H]− | 227.9975 | 227.9967 | 0.8 | C8H6NO5S | 3-Hydroxyindolin-2-one sulfate | 3.4 | 3.04 × 10−6 | ↑ |
| 14a | 3.61 | [M − H]− | 178.0515 | 178.0504 | 1.1 | C9H9NO3 | Hippuric acid | 5.2 | 2.71 × 10−6 | ↓ |
| 15 | 4.26 | [M − H]− | 338.0865 | 338.0876 | −1.1 | C15H17NO8 | 5-Hydroxy-6-methoxyindole glucuronide | 2.4 | 5.408 × 10−3 | ↓ |
| 16 | 4.28 | [M − H]− | 192.0649 | 192.0661 | −1.2 | C10H11NO3 | 2-Methylhippuric acid | 2.6 | 3.11 × 10−3 | ↑ |
| 17 | 5.10 | [M − H]− | 283.0810 | 283.0818 | −0.8 | C13H16O7 | p-Cresol glucuronide | 5.5 | 2.45 × 10−6 | ↑ |
| 18 | 5.51 | [M − H]− | 173.0815 | 173.0814 | 0.1 | C8H14O4 | Suberic acid | 2.0 | 4.17 × 10−6 | ↓ |
| 19 | 5.91 | [M − H]− | 336.1071 | 336.1083 | −1.2 | C16H19NO7 | Indole-3-acetyl-myo-inositol | 2.2 | 3.2 × 10−6 | ↑ |
| 20 | 6.77 | [M − H]− | 297.0970 | 297.0974 | −0.4 | C14H18O7 | 2-Phenylethanol glucuronide | 5.4 | 1.08 × 10−5 | ↓ |
| 21 | 8.01 | [M − H]− | 291.1290 | 291.1287 | 0.3 | C12H22O5 | 3-Hydroxydodecanedioic acid | 2.8 | 7.99 × 10−8 | ↑ |
iTRAQ-proteomics analysis
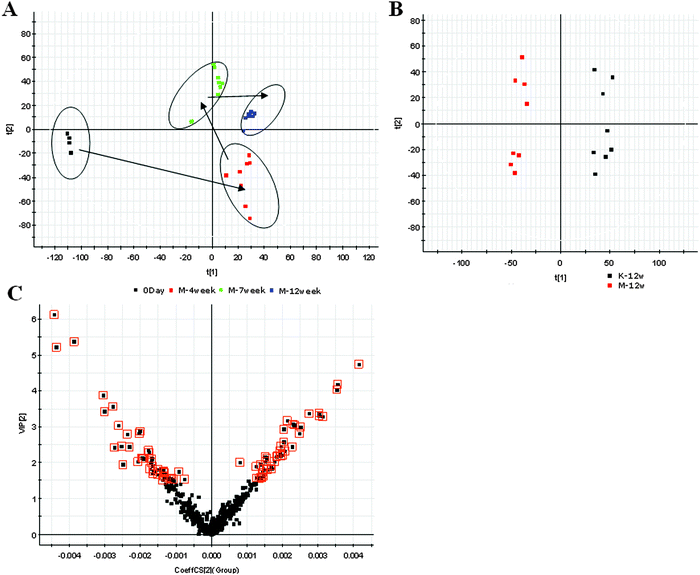
ICA aggregates the set of proteins studied into one point per sample in the two-dimensional coordinate system spanned by the two most relevant independent components (Fig. S2, ESI†). These trajectories reveal that the proteome profiles markedly change immediately after ALD. ICA can really reflect a clear separation between the 12W and the control group.| No. | Pathway | Pathway ID | Proteins |
|---|---|---|---|
| 1 | p53 signaling pathway | ko04115 | P15473, P08025 |
| 2 | PPAR signaling pathway | ko03320 | P06759, P04639, P07871, P04638 |
| 3 | Fatty acid metabolism | ko00071 | P07871, P06757 |
| 4 | TGF-beta signaling pathway | ko04350 | P49000, Q9WUK5 |
| 5 | mTOR signaling pathway | ko04150 | Q63484, P08025 |
| 6 | Cytokine–cytokine receptor interaction | ko04060 | P49000, Q9WUK5 |
| 7 | Purine metabolism | ko00230 | Q9Z244, Q64640 |
| 8 | Natural killer cell mediated cytotoxicity | ko04650 | P15978, P20762 |
| 9 | Neurotrophin signaling pathway | ko04722 | P68511, P35213, Q63484 |
| 10 | Fc epsilon RI signaling pathway | ko04664 | P20762, Q63484 |
Discussion
Alcohol consumption is one of the world's major risk factors for disease development. Ongoing studies show that alcohol consumption is associated with fatty liver disease in mammals. Recent advances in metabolomic technologies have enabled us to elucidate the effect of alcohol consumption on metabolism. Jaremek and co-workers have investigated the relation of alcohol intake and serum metabolite concentrations and identified potential biomarkers that could predict high levels of intake.18 It suggests that alcohol affects mostly the sphingolipid, glycerophospholipid and ether lipid metabolism. Clugston and co-workers analyzed altered hepatic lipid metabolism in mice fed with alcohol using a targeted lipidomic approach.19 They found that chronic alcohol consumption is associated with increased hepatic free fatty acid levels and decreased fatty acyl-CoA levels associated with decreased mitochondrial fatty acid oxidation and decreased fatty acyl-CoA synthesis, increased hepatic ceramide levels associated with higher levels of the precursor molecules sphingosine and sphinganine; and increased hepatic levels of the endocannabinoid anandamide associated with decreased expression of its catabolic enzyme fatty acid amide hydrolase. Kalhan et al. examined the plasma metabolomic profile in nonalcoholic fatty liver disease.20 Their results suggested markedly higher levels of glycocholate, taurocholate and glycochenodeoxycholate in subjects. A study showed that the expression of two proteins associated with alcoholic liver disease, peroxiredoxin 6 and aldehyde dehydrogenase 2, was down-regulated in ethanol fed rats.21 As far as we are aware, no metabolomics–proteomics profiling analyses of ALD rats with alcohol consumption have yet been conducted.In the present study, special attention was paid to the time points of the 1W and 12W which are closely associated with ALD, in the experimental animal model. The results showed that histopathology and biochemical results had significant differences between the controls and 12W. Through biochemical analysis combined with the histopathological results, we successfully established ALD in animal models at 12W. Additionally, alcoholic-induced ALD rat may be a useful model for the determination of ALD. Metabolic changes are associated with a number of complex diseases. A panel of biomarkers to characterize disease could be useful for ALD diagnostics. In this paper, UPLC-MS combined with the pattern recognition analysis approach was used to simplify and quicken the identification of the metabolites of ALD. UPLC-MS based metabolomics could be an advanced tool to help us find metabolites due to its capacity of processing large datasets, and classifying of sample groups, as well as its indiscriminative nature of metabolites. By using our metabolomics platform, PCA revealed a statistically significant separation between the ALD and control samples. An OPLS model was built to find biomarker candidates of ALD and 21 statistically important variables with VIP > 1.5 were defined, many are in various stages of progress at the ALD. Differential metabolites from the urine metabolome indicate the disrupted pentose and glucuronate interconversions, starch and sucrose metabolism, cysteine and methionine metabolism pathways, etc., would be helpful for understanding the occurrence and development of ALD.
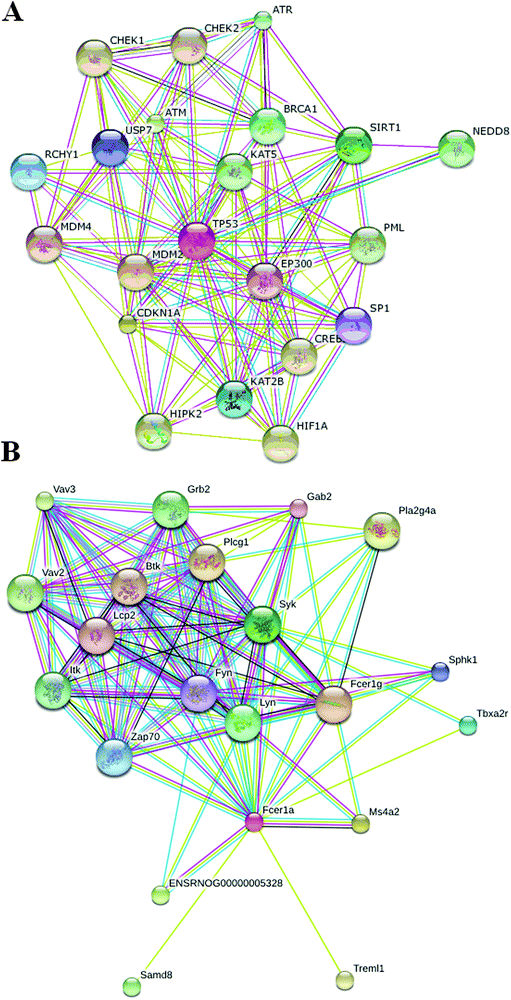
Proteomics, which measures mature proteins, could be used to closely observe biological functions in the body.22 Proteomics research is a potentially useful and effective tool for studying pathogenesis, establishing prognosis and determining treatment outcomes in a variety of diseases.23 Though proteomics is widely performed, the changes in the serum proteomes of ALD are rarely reported due to the complicated mechanism of ALD, therefore this study was also to describe the changes in protein levels in ALD using iTRAQ proteomics. The authors hypothesized that this approach had the potential to ultimately be beneficial for the identification of proteins that were important in the progression of ALD. We constructed the mouse model of the early stage of ALD using alcohol. Based on proteomics analysis, we investigated the proteomic changes and pathways leading to ALD by alcohol. For data analysis, we optimized the normalization of iTRAQ signals and quantified the expression of proteins identified. iTRAQ identified 85 different proteins that had ≥1.2-fold differences in the expression level between the ALD and the controls. Finally, the most significant pathway elucidated by bioinformatics methods was subsequently validated. Bioinformatics analysis of the DEPs illustrated the enrichment of metabolism related processes, such as the macromolecule metabolic process, the primary metabolic process and the cellular metabolic process etc. Studies have shown that the p53 signaling pathway, the PPAR signaling pathway, fatty acid metabolism, the TGF-beta signaling pathway, the mTOR signaling pathway, the cytokine–cytokine receptor interaction, purine metabolism, natural killer cell mediated cytotoxicity, the neurotrophin signaling pathway, and the Fc epsilon RI signaling pathway are critical in the pathogenesis of ALD. Interestingly, the p53 signaling pathway and the Fc epsilon RI signaling pathway were firstly reported in ALD. This study helps to achieve a better understanding of the complexity of metabolic regulations of ALD, which may shed light on metabolism to ALD. GO analysis indicated that these proteins are involved in transport, biological regulation, cellular processes, immune response, and metabolic processes. The DEPs may be involved in the host response to ALD at the molecular level and may be potential diagnostic biomarkers for ALD. It also provides insights into the molecular basis for the use of PPAR signaling agonists, which has been advocated for the treatment of ALD.
The findings yield a valuable tool that can engender new insights into the pathophysiology of ALD and advance the early diagnosis and monitor the progression of ALD, and showed that omics has the potential as a promising screening tool for exploring the essence of ALD disease. In addition, future research will be directed to the biological interpretation: which pathways were involved in the biochemical changes associated with the onset, development and progression of ALD, and whether these changes are the same during onset and progression, or if different changes of biochemistry occur at the different stages of ALD. Thus, the findings presented contribute to a better understanding of the molecular signature of ALD and may provide the biological background to pharmacological interventions in the future.
Conclusions
ALD represents a significant cause of morbidity and mortality worldwide and constitute a problem of global public health importance. Therefore, it needs more reliable metabolism information for effective treatment. We successfully reproduced ALD in animal models from liver histology combined with biochemical results. Omics is a powerful approach for the comprehensive assessment of endogenous metabolites or proteins and attempts were made to systematically identify them from biological samples. In this study, the proteomics and metabolomics improve discrimination between ALD and control. The metabolomics approach coupled with multivariate statistical methods showed 21 altered metabolic biomarkers indicating that ALD could cause more severe disturbances in pentose and glucuronate interconversions, starch and sucrose metabolism, cysteine and methionine metabolism, etc. Using iTRAQ based proteomic methods, we identified 85 differentially expression proteins related to ALD, which might provide clues to clarify novel mechanisms underlying alcohol-induced ALD. Of these, 33 were down-regulated and 52 were up-regulated in ALD. Interestingly, integrated proteomic and metabolomic profiling reveal that the p53 signaling pathway and the Fc epsilon RI signaling pathway were firstly found in ALD. These data suggest that the interruption of these pathways may provide a means to the development of molecularly targeted therapies for ALD.Conflicts of interest
The authors declare no competing financial interests.Acknowledgements
This work was supported by grants from the Key Program of Natural Science Foundation of State (Grant No. 81430093, 81173500, 81373930, 81302905, 81202639), National Key Subject of Drug Innovation (Grant No. 2015ZX09101043-005, 2015ZX09101043-011), Natural Science Foundation of Heilongjiang Province of China (H2015038), Youth Innovative Talent Program of Heilongjiang Province of China (UNPYSCT-2015118), Specialized Research Fund for the Doctoral Program of Higher Education (Grant No. 20122327120006), Fund Project of Heilongjiang Provincial Department of Education (Grant No. 12521498).References
- O. George, C. Sanders, J. Freiling, E. Grigoryan, S. Vu, C. D. Allen, E. Crawford, C. D. Mandyam and G. F. Koob, Recruitment of medial prefrontal cortex neurons during alcohol withdrawal predicts cognitive impairment and excessive alcohol drinking, Proc. Natl. Acad. Sci. U. S. A., 2012, 109, 18156–18161 CrossRef CAS PubMed.
- N. A. Nasrallah, J. J. Clark, A. L. Collins, C. A. Akers, P. E. Phillips and I. L. Bernstein, Risk preference following adolescent alcohol use is associated with corrupted encoding of costs but not rewards by mesolimbic dopamine, Proc. Natl. Acad. Sci. U. S. A., 2011, 108, 5466–5471 CrossRef CAS PubMed.
- T. Neumann, A. Helander, H. Dahl, T. Holzmann, B. Neuner, E. Weiss-Gerlach, C. Müller and C. Spies, Value of ethyl glucuronide in plasma as a biomarker for recent alcohol consumption in the emergency room, Alcohol Alcohol, 2008, 43, 431–435 CrossRef CAS PubMed.
- A. Sreekumar, L. M. Poisson, T. M. Rajendiran, A. P. Khan, Q. Cao, J. Yu, B. Laxman, R. Mehra, R. J. Lonigro, Y. Li, M. K. Nyati, A. Ahsan, S. Kalyana-Sundaram, B. Han, X. Cao, J. Byun, G. S. Omenn, D. Ghosh, S. Pennathur, D. C. Alexander, A. Berger, J. R. Shuster, J. T. Wei, S. Varambally, C. Beecher and A. M. Chinnaiyan, Metabolomic profiles delineate potential role for sarcosine in prostate cancer progression, Nature, 2009, 457, 910–914 CrossRef CAS PubMed.
- N. L. Kuehnbaum, J. B. Gillen, M. J. Gibala and P. Britz-McKibbin, Personalized metabolomics for predicting glucose tolerance changes in sedentary women after high-intensity interval training, Sci. Rep., 2014, 4, 6166 CrossRef CAS PubMed.
- X. Wang, A. Zhang and H. Sun, Power of metabolomics in diagnosis and biomarker discovery of hepatocellular carcinoma, Hepatology, 2013, 57, 2072–2077 CrossRef CAS PubMed.
- E. Sabidó, O. Quehenberger, Q. Shen, C. Y. Chang, I. Shah, A. M. Armando, A. Andreyev, O. Vitek, E. A. Dennis and R. Aebersold, Targeted proteomics of the eicosanoid biosynthetic pathway completes an integrated genomics-proteomics-metabolomics picture of cellular metabolism, Mol. Cell. Proteomics, 2012, 11, M111.014746 Search PubMed.
- J. F. Kellie, R. E. Higgs, J. W. Ryder, A. Major, T. G. Beach, C. H. Adler, K. Merchant and M. D. Knierman, Quantitative measurement of intact alpha-synuclein proteoforms from post-mortem control and Parkinson's disease brain tissue by intact protein mass spectrometry, Sci. Rep., 2014, 4, 5797 CAS.
- L. Sun, M. M. Bertke, M. M. Champion, G. Zhu, P. W. Huber and N. J. Dovichi, Quantitative proteomics of Xenopus laevis embryos: expression kinetics of nearly 4000 proteins during early development, Sci. Rep., 2014, 4, 4365 Search PubMed.
- T. J. Wang, M. G. Larson, R. S. Vasan, S. Cheng, E. P. Rhee, E. McCabe, G. D. Lewis, C. S. Fox, P. F. Jacques, C. Fernandez, C. J. O'Donnell, S. A. Carr, V. K. Mootha, J. C. Florez, A. Souza, O. Melander, C. B. Clish and R. E. Gerszten, Metabolite profiles and the risk of developing diabetes, Nat. Med., 2011, 17, 448–453 CrossRef CAS PubMed.
- G. J. Patti, O. Yanes, L. P. Shriver, J. P. Courade, R. Tautenhahn, M. Manchester and G. Siuzdak, Metabolomics implicates altered sphingolipids in chronic pain of neuropathic origin, Nat. Chem. Biol., 2012, 8, 232–234 CrossRef CAS PubMed.
- Y. Chen, J. Xu, R. Zhang, G. Shen, Y. Song, J. Sun, J. He, Q. Zhan and Z. Abliz, Assessment of data pre-processing methods for LC-MS/MS-based metabolomics of uterine cervix cancer, Analyst, 2013, 138(9), 2669–2677 RSC.
- T. Pacchiarotta, R. J. Derks, E. Nevedomskaya, W. van der Starre, J. van Dissel, A. Deelder and O. A. Mayboroda, Exploratory analysis of urinary tract infection using a GC-APCI-MS platform, Analyst, 2015, 140(8), 2834–2841 RSC.
- X. Wang, A. Zhang, Y. Han, P. Wang, H. Sun, G. Song, T. Dong, Y. Yuan, X. Yuan, M. Zhang, N. Xie, H. Zhang, H. Dong and W. Dong, Urine metabolomics analysis for biomarker discovery and detection of jaundice syndrome in patients with liver disease, Mol. Cell. Proteomics, 2012, 11, 370–380 Search PubMed.
- L. Nadal-Desbarats, N. Aïdoud, P. Emond, H. Blasco, I. Filipiak, P. Sarda, F. Bonnet-Brilhault, S. Mavel and C. R. Andres, Combined 1H-NMR and 1H-13C HSQC-NMR to improve urinary screening in autism spectrum disorders, Analyst, 2014, 139(13), 3460–3468 RSC.
- H. Sun, A. Zhang, G. Yan, C. Piao, W. Li, C. Sun, X. Wu, X. Li, Y. Chen and X. Wang, Metabolomic analysis of key regulatory metabolites in hepatitis C virus-infected tree shrews, Mol. Cell. Proteomics, 2013, 12, 710–719 CAS.
- A. Zhang, H. Sun and X. Wang, Urinary metabolic profiling of rat models revealed protective function of scoparone against alcohol induced hepatotoxicity, Sci. Rep., 2014, 4, 6768 CrossRef PubMed.
- M. Jaremek, Z. Yu, M. Mangino, K. Mittelstrass, C. Prehn, P. Singmann, T. Xu, N. Dahmen, K. M. Weinberger, K. Suhre, A. Peters, A. Döring, H. Hauner, J. Adamski, T. Illig, T. D. Spector and R. Wang-Sattler, Alcohol-induced metabolomic differences in humans, Transl. Psychiatry, 2013, 3, e276 CrossRef CAS PubMed.
- R. D. Clugston, H. Jiang, M. X. Lee, R. Piantedosi, J. J. Yuen, R. Ramakrishnan, M. J. Lewis, M. E. Gottesman, L. S. Huang, I. J. Goldberg, P. D. Berk and W. S. Blaner, Altered hepatic lipid metabolism in C57BL/6 mice fed alcohol: a targeted lipidomic and gene expression study, J. Lipid Res., 2011, 52, 2021–2031 CrossRef CAS PubMed.
- S. C. Kalhan, L. Guo, J. Edmison, S. Dasarathy, A. J. McCullough, R. W. Hanson and M. Milburn, Plasma metabolomic profile in nonalcoholic fatty liver disease, Metabolism, 2011, 60, 404–413 CrossRef CAS PubMed.
- B. W. Newton, W. K. Russell, D. H. Russell, S. K. Ramaiah and A. Jayaraman, Liver proteome analysis in a rodent model of alcoholic steatosis, J. Proteome Res., 2009, 8, 1663–1671 CrossRef CAS PubMed.
- H. W. Rhee, P. Zou, N. D. Udeshi, J. D. Martell, V. K. Mootha, S. A. Carr and A. Y. Ting, Proteomic mapping of mitochondria in living cells via spatially restricted enzymatic tagging, Science, 2013, 339, 1328–1331 CrossRef CAS PubMed.
- R. Wagner, J. Li, E. Kenar, O. Kohlbacher, F. Machicao, H. U. Häring, A. Fritsche, G. Xu and R. Lehmann, Clinical and non-targeted metabolomic profiling of homozygous carriers of Transcription Factor 7-like 2 variant rs7903146, Sci. Rep., 2014, 4, 5296 CAS.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c5mb00603a |
| ‡ These authors contributed equally to this work. |
| This journal is © The Royal Society of Chemistry 2016 |