Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Multiplexed droplet Interface bilayer formation†
Nathan E.
Barlow
ab,
Guido
Bolognesi
 ab,
Anthony J.
Flemming
bc,
Nicholas J.
Brooks
ab,
Laura M. C.
Barter
ab and
Oscar
Ces
*ab
ab,
Anthony J.
Flemming
bc,
Nicholas J.
Brooks
ab,
Laura M. C.
Barter
ab and
Oscar
Ces
*ab
aDepartment of Chemistry, Imperial College London, Exhibition Road, South Kensington, London, SW7 2AZ, UK. E-mail: o.ces@imperial.ac.uk
bInstitute of Chemical Biology, Imperial College London, Exhibition Road, South Kensington, London, SW7 2AZ, UK. E-mail: l.barter@imperial.ac.uk
cSyngenta, Jealott's Hill International Research Centre, Bracknell, Berkshire, RG42 6EY, UK. E-mail: anthony.flemming@syngenta.com
First published on 10th November 2016
Abstract
We present a simple method for the multiplexed formation of droplet interface bilayers (DIBs) using a mechanically operated linear acrylic chamber array. To demonstrate the functionality of the chip design, a lipid membrane permeability assay is performed. We show that multiple, symmetric DIBs can be created and separated using this robust low-cost approach.
A droplet interface bilayer (DIB), first developed by Bayley1 and Takeuchi,2 is a fluid bilayer structure that can be employed in an array of in vitro biological and chemical experimentation where lipid membranes are necessary. DIBs are composed of aqueous droplets containing small unilameller vesicles (SUVs) immersed in oil solutions forming lipid monolayers at the interface. When these monolayers are pushed together they form a model membrane bilayer.1
The use of low cost plastics such as polymethyl methacrylate (PMMA) as a construction material for milli- and microfluidic devices is well established3–8 and a number of such devices have been recently proposed for the construction and manipulation of DIBs. In 2010, Sarles and Leo designed a PMMA chip to contain and stabilize DIBs with physical encapsulation; as an unintended consequence of durability testing it was discovered that by shaking at 20 Hz a DIB could be rendered into its individual droplets.9 This procedure was not shown to be reproducible, and it is therefore not suitable for rapid and on-demand DIB separation and re-formation, a requirement for serial high throughput analysis of lipid membranes. The controlled and repetitive formation of DIBs was demonstrated in a circular acrylic DIB well chip (DWC) by Tsuji et al.10 This “split-and-contact” method was employed to form, divide and re-form large (40 μL) DIBs rapidly for serial pore conductance experimentation with high statistical relevancy. The chip is size limited however, as it requires relatively large droplets in conjunction with a protective sheet between the droplets to control the interfacial area and stabilize the DIB. The resulting DIBs with large droplet volumes and small interfacial areas are not suitable for all applications, in particular membrane permeability assays require the opposite due to diffusion timescale limitations. A recent design by Czekalska et al. has the ability to repetitively and automatically form DIBs – one at a time – on a milled, polycarbonate chip for transmembrane electrical recordings, the droplets being subsequently separated after use.11 Despite their advantages of low-cost and ease of fabrication, the existing plastic devices can only house a limited number of DIBs and the statistical data of the membrane properties can only be accessed through repetitive membrane formation and serial data acquisition.
Nisisako et al. developed a chip for high-throughput passive permeability measurements on nL scale DIBs,12 the results of which demonstrate moderate statistical strength on improved time scales as compared to parallel artificial membrane permeability assay (PAMPA) techniques.13 The limitation, however, is the inability to simply vary the lipid or permeating solute without having to make changes to the microfluidic system inputs. Finally, this chip does not have the ability to reversibly return the droplets back into their original form.
This study in contrast applies a low cost, linear movable PMMA chamber array chip to allow for the simultaneous formation and analysis of sub-μL DIBs. The device capabilities are demonstrated by performing small molecule membrane permeability assays with different lipid compositions. By parallelising the DIB formation, rapid and high throughput analysis of the membrane transport properties could be achieved.
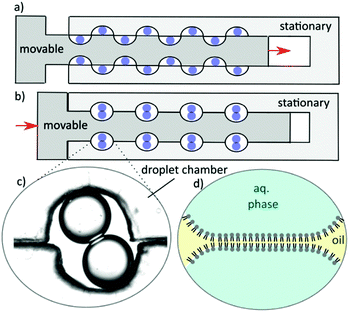
As shown in Fig. 1(a and b), multiple DIBs (c) can be formed in unison by mechanical action where lipid monolayer coated aqueous droplets in oil – manually pipetted into the device – come into contact to form a bilayer (d) without mechanical stabilization or area control.
Note that to form asymmetric membranes, DIBs can be formed with droplets of differing lipid composition.
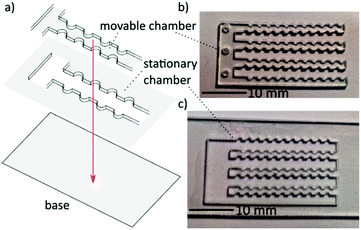
The chip is fashioned of three laser cut PMMA parts, a base, stationary chamber and a movable manifold as shown in Fig. 2. A photograph of the finished product with an array of 11 × 14 semi-circle chambers 1 mm in diameter is shown in Fig. 2(b and c). The base and stationary chambers were sealed together via acetone solvent bonding.14 The number of wells can be custom designed to accommodate a range of working areas depending upon the application. The movable chambers can be actuated by hand with tweezers, a fine instrument, or can be rigged with a screw adjustment mechanism. The free standing movable manifold is cut to fit with a tolerance of 100 μm. After use, the chip can be rinsed with ethanol, dried and reused. Details of fabrication are supplied in the ESI.†
To demonstrate the chip function, snapshots of the dynamic formation are shown in Fig. 3 where aqueous 1,2-diphytanoyl-sn-glycero-3-phosphocholine (DPhPC) lipid emulsion droplets in hexadecane are put into contact to form interfaces, and then reversibly rendered apart to return to their original droplet arrangement. From position (a) to position (b), after sufficient incubation time the nascent droplets form monolayers (approximately 2 minutes) and can be moved at an arbitrary rate. To prevent droplet coalescence, care must be taken when traversing from position (b) to (c). In a study by Schlicht and Zagnoni, it has been experimentally shown that for 1,2-dioleoyl-sn-glycero-3-phosphocholine (DOPC) and DPhPC lipids at 5 mg mL−1, the droplet approach velocities above 100 micrometres per second increase the chances of DIB coalescence substantially.15 This implies that the osculation process should take no less than approximately 10 seconds. From image (c) to (d) the bilayer interface forms, which takes typically less than a minute for this “lipid-in”1 system in hexadecane, but can vary on different surfaces. Image (e) demonstrates the effect of lateral shearing forces on the DIB without mechanical stabilization by an inner plate between the droplets as used in previous studies.10 The result of these forces show the interface shrink, and in image (f) the two droplets are separated. Bilayer formation process dynamics have been studied previously,16–18 and it has likewise been shown that the bilayer area can be modified on DIBs away from the thermodynamically favourable shape18 by mechanical forces19 or electric potential.20 The free energy landscape is however yet to be explored when the interface is sheared or indeed whether there are any associated hysteresis effects. As a proof of concept, it is shown that the movable chamber chip can be used for multiplexing DIB permeability assays. The DIB formation and assay were performed simultaneously with DOPC and DPhPC, both lipids shown previously to form stable DIBs.21 All lipids and lipid extrusion equipment were purchased from Avanti Polar Lipids. For DIB preparation, the lipids are first deposited from chloroform on a glass vial surface and emulsified in an aqueous phosphate (100 mM) buffer solution at 10 mg mL−1. The emulsions are prepared by lipid extrusion repeated 11× through a 100 nm pore to form a solution containing approximately 120 nm diameter SUVs. Then the fluorescent dye 7-hydroxy-3H-phenoxazin-3-one (resorufin), purchased from Sigma Aldrich, is prepared in solutions at 5 and 0.63 μM.
For the permeability assay, 200 nL droplets are deposited into the chambers, which consists of a set of DOPC DIBs and DPhPC DIBs loaded with 5 or 0.65 μM resorufin, which is within the linear region of fluorescence intensity with respect to concentration. Fluorescence images (excitation 556 nm and detection 580 nm) are obtained from laser scanning with a Typhoon FLA 9500 biomolecular imager – an example image is available in the ESI.† To measure permeability, a simple mathematic treatment of Fick's first law is employed similar to the approach taken by Cass et al.22 By standard solutions to first order differential equations, the rate of change of the fluorescence intensity [I]i in a droplet (i = 1, 2) is given by
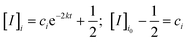 | (1) |
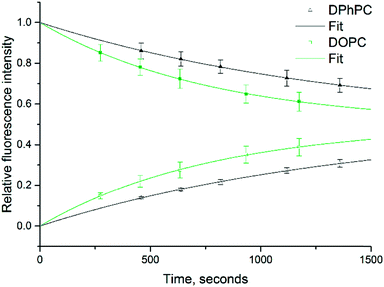
The relative fluorescence intensity dynamics of the DIB pair droplets [I]1 and [I]2 over a span of 25 minutes are plotted in Fig. 4 and are based on the measured fluorescence data taken using ImageJ pixel analysis. Additionally, the droplet volume and interfacial area measurements are taken from these images. To abate photobleaching, a minimal sample acquisition rate is chosen. Accordingly dynamic intensity data taken at five time points is used to solve for the rate constant k with a minimization of the root mean square error of the model. The permeability data shows the effect of the membrane type, specifically the DPhPC membranes are less permeable to resorufin than DOPC, with permeability values of 0.78 ± 0.17 and 1.26 ± 0.23 × 10−4 cm s−1 respectively. The higher permeability for DOPC confirms previous comparisons with norfloxacin.25 Note that this experiment was repeated 14 times for DOPC and 6 times for DPhPC, and the overall error comes from one standard deviation of experimental repeats on freshly made chips. Note also that the measured permeability value, due to the manifestation of an unstirred layer effect, is an underestimated value of the intrinsic permeability, particularly for higher permeability membranes, and should be taken as an unstirred, effective value.26 Despite this underestimate, a Welch's t-test shows an error rate of 0.001, which bolsters the supposition that there is a statistically significant difference between the two measurements. Furthermore, this result matches to an order of magnitude to an unstirred permeability measurement made by Bean et al. in 1968 with a similar small molecule, indole-3-ethanol (MW 161), across a planar brain lipid bilayer at 2.9 to 3.5 × 10−4 cm s−1.27 Additionally, as shown by the relatively rapid diffusion half-life (ca. 10 minutes) with sub-μL DIBs and interfacial areas on length scales comparable to the droplet volumes, it is apparent that for small molecule permeability assays, a low droplet volume to interfacial area ratio is necessary to attain reasonable timescales.28 This occludes the use of relatively large droplets for permeability studies, such as the 40 μL droplets with 250 μm membranes interfaces used by Tsuji.10
With the proposed low-cost PMMA-based movable chamber chip, high-throughput permeability assays can be performed at high statistical relevance. Indeed, compared to other PMMA-based devices, the design of our chip maximises the space available to DIBs so that a large number (theoretically ca. 100) of model lipid membranes could be simultaneously analysed. Unlike many previous PDMS and glass devices for on-chip multi-DIB formation and characterisation, requiring a clean-room for construction and a plasma source for bonding,29,30 our device was manufactured and assembled in a standard laboratory environment with simple procedures and equipment. This is expedient as it opens up our microfluidic approach for multiplexed DIB formation and membrane characterisation to a wider scientific community who do not have access to specialized facilities.
The ability to render the original droplets from a DIB pair extends the versatility and applicability of the device. For instance, the transport process of molecules across the membrane could be stopped when the pre-selected amount of fluorescent solute is transferred from the donor to the acceptor droplets, thereby enabling the control over the final droplet solute concentration. Additionally, if a permeating solute cannot be fluorescently labelled, a permeability assay could be designed where the diffusion could be halted by rendering DIB droplets at intervals and concentration measurements taken with FTIR, mass spectrometry or NMR among other techniques.
Microfluidic devices employing the parallel formation of several artificial membranes in the form of either planar lipid bilayers31,32 or DIBs30,33 can typically produce only identical copies of the same lipid system and hence they do not allow for the parallel analysis of membrane transport with different lipid types and permeating molecules. This is however desirable for pharmacological applications such as high throughput screening of drug candidates. Conversely, as demonstrated by the permeability assays, our chip enables the simultaneous characterisation of the properties of several membranes composed of different lipid types. By the same token, the effects of different droplet contents could also be tested at the same time.
Conclusions
It has been demonstrated that a laser cut PMMA chip can be deployed to manipulate DIBs for multiplexed formation and membrane transport analysis. Bilayer interface shearing and rendering of the original sub-μL droplets was also demonstrated. The device was manufactured and assembled with rapid, simple and inexpensive procedures, which do not require access to advanced fabrication facilities (i.e. cleanroom). The application of this low cost linear movable chamber chip to multiplexed membrane permeability assays with different lipid combinations shows the practicality of simultaneous DIB formation for membrane transport studies that require high statistical relevance. Classically, permeability is known to be a function of solute de-solvation, lipophilicity, molecular volume and dipole moment,34 however the effect of membrane properties such as asymmetry on permeability have experienced recent interest.35,36 Accordingly, this chip could be applied for rapid and quantitative analysis of membrane transport mediated by lipid asymmetry. Furthermore, it could be applied to high-throughput screening of drug–membrane and protein–membrane interactions as well as by membrane proteins including ion-channels,37 α-hemolysin38 or MscL.39 Finally, the droplet format would also enable the encapsulation of bio-synthetic systems with cell-like functions (e.g. gene expression, enzymatic cascades, etc.), thereby making our device a suitable platform for bottom-up synthetic biology studies.Acknowledgements
The research leading to these results has received funding from the People Programme (Marie Curie Actions) of the European Union's Seventh Framework Programme (FP7/2007-2013) under REA grant agreement no 607466. This research was funded by EPSRC grants: EP/J017566/1, EP/L015498/1, EP/J021199/1 and EP/K503733/1. The authors would like to thank Professor Ed Tate and Mr Roman Fedoryshchak for additional technical support and the Imperial College Advanced Hackspace for access to fabrication and prototyping facilities. All data created during this research are openly available from Imperial College London, please see contact details at http://www.imperial.ac.uk/membranebiophysics.Notes and references
- H. Bayley, B. Cronin, A. Heron, M. A. Holden, W. Hwang, R. Syeda, J. Thompson and M. Wallace, Mol. BioSyst., 2008, 4, 1191–1208 RSC.
- K. Funakoshi, H. Suzuki and S. Takeuchi, Anal. Chem., 2006, 78, 8169–8174 CrossRef CAS PubMed.
- J. M. Li, C. Liu, J. S. Liu, Z. Xu and L. D. Wang, J. Mater. Process. Technol., 2009, 209, 5487–5493 CrossRef CAS.
- T. Nisisako, T. Torii and T. Higuchi, in Micro Total Analysis Systems 2001: Proceedings of the μTAS 2001 Symposium, held in Monterey, CA, USA 21–25 October, 2001, ed. J. M. Ramsey and A. van den Berg, Springer Netherlands, Dordrecht, 2001, DOI:10.1007/978-94-010-1015-3_56, pp. 137–138.
- H. Suzuki, K. V. Tabata, H. Noji and S. Takeuchi, Langmuir, 2006, 22, 1937–1942 CrossRef CAS PubMed.
- T.-F. Hong, W.-J. Ju, M.-C. Wu, C.-H. Tai, C.-H. Tsai and L.-M. Fu, Microfluid. Nanofluid., 2010, 9, 1125–1133 CrossRef CAS.
- X. Li, D. Li, X. Liu and H. Chang, Sens. Actuators, B, 2016, 229, 466–475 CrossRef CAS.
- Y. Tsuji, R. Kawano, T. Osaki, K. Kamiya, N. Miki and S. Takeuchi, Lab Chip, 2013, 13, 1476–1481 RSC.
- S. A. Sarles and D. J. Leo, Lab Chip, 2010, 10, 710–717 RSC.
- Y. Tsuji, R. Kawano, T. Osaki, K. Kamiya, N. Miki and S. Takeuchi, Anal. Chem., 2013, 85, 10913–10919 CrossRef CAS PubMed.
- M. A. Czekalska, T. S. Kaminski, S. Jakiela, K. Tanuj Sapra, H. Bayley and P. Garstecki, Lab Chip, 2015, 15, 541–548 RSC.
- T. Nisisako, S. A. Portonovo and J. J. Schmidt, Analyst, 2013, 138, 6793–6800 RSC.
- M. Fujikawa, K. Nakao, R. Shimizu and M. Akamatsu, Bioorg. Med. Chem., 2007, 15, 3756–3767 CrossRef CAS PubMed.
- C.-W. Tsao and D. L. DeVoe, Microfluid. Nanofluid., 2009, 6, 1–16 CrossRef CAS.
- B. Schlicht and M. Zagnoni, Sci. Rep., 2015, 5, 9951 CrossRef CAS PubMed.
- J. N. Vargas, R. Seemann and J. B. Fleury, Soft Matter, 2014, 10, 9293–9299 RSC.
- P. Mruetusatorn, J. B. Boreyko, G. A. Venkatesan, S. A. Sarles, D. G. Hayes and C. P. Collier, Soft Matter, 2014, 10, 2530–2538 RSC.
- S. S. Dixit, A. Pincus, B. Guo and G. W. Faris, Langmuir, 2012, 28, 7442–7451 CrossRef CAS PubMed.
- L. C. Gross, A. J. Heron, S. C. Baca and M. I. Wallace, Langmuir, 2011, 27, 14335–14342 CrossRef CAS PubMed.
- G. J. Taylor, G. A. Venkatesan, C. P. Collier and S. A. Sarles, Soft Matter, 2015, 11, 7592–7605 RSC.
- G. A. Venkatesan, J. Lee, A. B. Farimani, M. Heiranian, C. P. Collier, N. R. Aluru and S. A. Sarles, Langmuir, 2015, 31(47), 12883–12893 CrossRef CAS PubMed.
- A. Cass and A. Finkelstein, J. Gen. Physiol., 1967, 50, 1765–1784 CrossRef CAS PubMed.
- I. N. Bojesen and H. S. Hansen, J. Lipid Res., 2006, 47, 561–570 CrossRef CAS PubMed.
- P. Pohl, S. M. Saparov and Y. N. Antonenko, Biophys. J., 1998, 75, 1403–1409 CrossRef CAS PubMed.
- S. Purushothaman, J. Cama and U. F. Keyser, Soft Matter, 2016, 12, 2135–2144 RSC.
- P. H. Barry and J. M. Diamond, Physiol. Rev., 1984, 64, 763–872 CAS.
- R. C. Bean, W. C. Shepherd and H. Chan, J. Gen. Physiol., 1968, 52, 495–508 CrossRef CAS PubMed.
- T. Tonooka, K. Sato, T. Osaki, R. Kawano and S. Takeuchi, Small, 2014, 10, 3275–3282 CrossRef CAS PubMed.
- Y. Elani, A. J. deMello, X. Niu and O. Ces, Lab Chip, 2012, 12, 3514–3520 RSC.
- T. Tonooka, K. Sato, T. Osaki, R. Kawano and S. Takeuchi, Small, 2014, 10, 3197–3197 CrossRef.
- S. Ota, H. Suzuki and S. Takeuchi, Lab Chip, 2011, 11, 2485–2487 RSC.
- R. Watanabe, N. Soga, D. Fujita, K. V. Tabata, L. Yamauchi, S. H. Kim, D. Asanuma, M. Kamiya, Y. Urano and H. Suga, Nat. Commun., 2014, 5, 4951 CrossRef PubMed.
- R. Watanabe, N. Soga, M. Hara and H. Noji, Lab Chip, 2016, 16, 3043–3048 RSC.
- C. W. Fong, J. Membr. Biol., 2015, 248, 651–669 CrossRef CAS PubMed.
- P. J. Milianta, M. Muzzio, J. Denver, G. Cawley and S. Lee, Langmuir, 2015, 31, 12187–12196 CrossRef CAS PubMed.
- Y. Elani, S. Purushothaman, P. J. Booth, J. M. Seddon, N. J. Brooks, R. V. Law and O. Ces, Chem. Commun., 2015, 51, 6976–6979 RSC.
- S. Leptihn, J. R. Thompson, J. C. Ellory, S. J. Tucker and M. I. Wallace, J. Am. Chem. Soc., 2011, 133, 9370–9375 CrossRef CAS PubMed.
- S. Leptihn, O. K. Castell, B. Cronin, E. H. Lee, L. C. Gross, D. P. Marshall, J. R. Thompson, M. Holden and M. I. Wallace, Nat. Protoc., 2013, 8, 1048–1057 CrossRef CAS PubMed.
- H. M. G. Barriga, P. Booth, S. Haylock, R. Bazin, R. H. Templer and O. Ces, J. R. Soc., Interface, 2014, 11, 98 CrossRef PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c6lc01011c |
| This journal is © The Royal Society of Chemistry 2016 |