Open Access Article
Open Access ArticleBacterial chemotaxis-enabled autonomous sorting of nanoparticles of comparable sizes†
SeungBeum
Suh‡
a,
Mahama A.
Traore‡§
a and
Bahareh
Behkam
*b
aDepartment of Mechanical Engineering, Virginia Tech, Blacksburg, Virginia 24061, USA
bDepartment of Mechanical Engineering, School of Biomedical Engineering and Sciences, Virginia Tech, Blacksburg, Virginia 24061, USA. E-mail: behkam@vt.edu
First published on 18th February 2016
Abstract
High throughput sorting of micro/nanoparticles of similar sizes is of significant interest in many biological and chemical applications. In this work, we report a simple and cost-effective sorting technique for separation of similarly-sized particles of dissimilar surface properties within a diffusion-based microfluidic platform using chemotaxis in Escherichia coli bacteria. Differences in surface chemistry of two groups of similarly-sized nanoparticles in a mixture were exploited to selectively assemble one particle group onto motile E. coli, through either specific or non-specific adhesion, and separate them from the remaining particle group via chemotaxis of the attached bacteria. To enable optimal operation of the sorting platform, the chemotaxis behavior of E. coli bacteria in response to casamino acids, the chemoeffector of choice was first characterized. The chemical concentration gradient range within which the bacteria exhibit a positive chemotactic response was found to be within 0.25 × 10−7–1.0 × 10−3 g ml−1 mm−1. We demonstrate that at the optimum concentration gradient of 5.0 × 10−4 g ml−1 mm−1, a sorting efficiency of up to 81% at a throughput of 2.4 × 105 particles per min can be achieved. Sensitivity of the sorting efficiency to the adhesion mechanism and particle size in the range of 320–1040 nm was investigated.
Introduction
Efficient and cost-effective micro/nanoparticle sorting and separation is fundamentally important in biological and chemical analyses such as cell separation, pathogen filtration and drug screening.1,2 At the micro/nanoscale, sorting is achieved through a variety of passive and active techniques or a combination of both.2 Active techniques are based on external recognition of particle properties (e.g. electrical charge) and subsequent force application (e.g. electrostatic force) to collect particles into a desired location.3 Examples of such techniques include fluidic, optical, dielectrophoretic and magnetic separation. Passive methods, which are based on one or more particle properties (e.g. size and density), result in particles exhibiting different behaviours when placed in a sorting platform, and are exploited for separation in the absence of any active recognition step.3 Examples of such methods include particle filtering and centrifugation. As such, the separation efficiency of active sorting techniques is usually higher than that of passive techniques. However, active sorting generally demands more complex set-ups operated by skilled users. On the other hand, conventional passive sorting methods are often inertia-based4–6 which leads to limitations of sorting particles of comparable sizes and density, as well as unintended aggregation and compromise of the stability of nanoparticle suspension. Recent progress in multiplex microfluidics has enabled miniaturization and increased parallelism of micro/nanoparticle manipulation and sorting. Active and passive micro-sorting devices based on several techniques including dielectrophoresis,7 acoustophoresis,8 hydrophoresis,9 hydrodynamic flow control,10 electrokinetic flow,10 and optical force switching11 have been developed. However, sorting particles of similar sizes and densities remains a challenge. Therefore, there is a substantial need for new, simple and cost-effective methodologies to be developed in order to sort particles of similar bulk physical properties (size, density, rigidity, etc.) at high efficiency.Over the past decade, whole-cell actuators have been implemented in microscale-engineered systems for applications such as load transport and mixing.12–14 It has been shown by us and others that flagellated bacteria can be used for controlled actuation,14–17 directed transport,18,19 or manipulation and assembly of microscale objects.20 To the best of our knowledge, for the first time, we present a separation method that exploits chemotaxis (i.e. directed migration in response to a chemoeffector gradient) and selective adhesion in flagellated swimming bacteria for autonomous (passive) sorting of similarly-sized nanoparticles of dissimilar surface properties. We have implemented an Escherichia coli chemotaxis-activated microfluidic particle sorter and evaluated its performance in separating polystyrene particles of comparable sizes in the range of 320–390 nm and 1000–1040 nm. Our method is based on selective adhesion of E. coli bacteria to one group of particles only and chemotactic transport of the adhered particles away from the mixture within a static body of fluid (Fig. 1). The simple hydrogel-based microfluidic sorting platform reported here is fabricated in a one-step direct photopolymerization process, is robust to variation in operational conditions, does not require continuous flow of the immersion medium, and obviates the need for additional external equipment (e.g. signal analyzers, function generator, etc.). This platform offers substantial flexibility compared with other microfluidic-based techniques such as dielectrophoresis, magnetic sorting and acoustic sorting, wherein strict requirements for particle properties and forces can add to the complexity of the task. In contrast to probe-based systems such as an atomic force microscope (AFM) or optical tweezers, this platform can easily achieve parallel operation as well as higher throughput autonomous separation and sorting.
Materials and methods
Bacterial culture
E. coli MG1655m, a derivative of E. coli MG1655 from the K-12 family with increased motility, was used in all the sorting experiments.19,21E. coli RP437, a chemotaxis model strain from the same family, was used in all the chemotaxis assays.22 To facilitate microscopy imaging, E. coli RP437 was transformed with a plasmid encoding GFP (pHC60; TetR, constitutive expression of green fluorescent protein)23 and E. coli MG1655m was transformed with a plasmid encoding RFP (p67TD1; AmpR, expression of red fluorescent protein in the presence of isopropyl-β-D-thiogalactopyranoside (IPTG)).24An E. coli RP437 culture from a single colony was incubated overnight in 10 ml of fresh T-broth (10 g l−1 tryptone, 5 g l−1 NaCl in deionized (DI) water) supplemented with 10 μg ml−1 tetracycline in a shaking incubator (30 °C, 180 rpm). A 100 μl aliquot of the overnight culture was inoculated in 10 ml of fresh T-broth supplemented with 0.04 g of the chemoattractant (casamino acids) to promote a pronounced chemotactic response of the bacteria cells in all the chemotaxis experiments.25 A 1 ml aliquot of the liquid culture at an OD600 of 0.5 was then centrifuged at a low speed (1700 × g) for 5 minutes at room temperature and suspended in 1 ml of freshly prepared chemotaxis buffer (150 mM NaCl, 2 mM Na2HPO4·7H2O, 1.9 mM KH2PO4, 0.1 mM EDTA, 0.01 mM L-methionine, and 10 mM DL-lactate)25 and was used in all the chemotaxis assays.
A similar culturing procedure was followed for E. coli MG1655m in 10 ml of L-broth (10 g l−1 tryptone, 5 g l−1 NaCl, and 5 g l−1 yeast extract in DI water) supplemented with 10 μg ml−1 ampicillin, 0.5 mM IPTG, and 0.04 g of casamino acids in a shaking incubator (30 °C, 150 rpm). A 1 ml aliquot of the liquid culture at an OD600 of 0.5 was centrifuged at a low speed (1700 × g) for 5 minutes at room temperature and suspended in 1 ml of a freshly prepared motility medium (0.01 M potassium phosphate, 0.067 M sodium chloride, 0.1 mM EDTA, 0.01 M glucose, and 0.002% Tween-20)14 and was used in all the sorting experiments.
Fabrication of the microfluidic platforms
The microfluidic chemotaxis assay device, shown in Fig. 2, was used to characterize the chemotaxis behavior of E. coli over a wide range of chemical gradients of the chemoeffector casamino acids. This device has been described in a previous work.26 Briefly, a solution of polyethylene glycol diacrylate (PEG-DA, MW = 700 Da, 10% (v/v) in PBS) hydrogel mixed with 0.5% (w/v) of the photoinitiator Irgacure® 2959 (Sigma-Aldrich, St. Louis, MO) was poured within a polydimethylsiloxane (PDMS) enclosure. A three-channel pattern was transferred into the hydrogel via UV photopolymerization (365 nm, 18 W cm−2, Omnicure S1000, Vanier, Quebec) for 15 seconds. A PDMS layer and two Plexiglas support layers were placed on top of the hydrogel device layer and one Plexiglas support layer was placed underneath the bottom glass slide. The top and bottom Plexiglas layers were clamped together to provide sufficient pressure to seal the device. Controllable, quasi-steady, and linear chemical concentration gradients were established by continuously flowing the buffer and the chemoeffector solution in the outer channels at a flow rate of 5 μl min−1 (PHD Ultra syringe pump, Harvard apparatus, Holliston, MA). The quasi-steady linear gradient was established in the center channel after 75 minutes as the chemoeffector diffused through the hydrogel wall into the buffer-filled center channel.Similar to the chemotaxis assay device, the sorting platform is composed of a chemoattractant channel, a center channel (contains the sorting work area), and a buffer channel, as is shown in Fig. 1. The device layer is made of the same PEG-DA gel (MW = 700 Da). The inlet ports of the center channel in the sorting platform are designed such that the nanoparticle suspension, containing similarly-sized particles, can be introduced in only one side of the work area. The chemoattractant channel is filled with the chemoeffector casamino acid at a concentration of 0.004 g ml−1 to establish the optimum chemotactic response-inducing gradient. A 500 μl aliquot of the mixture of the particle–bacteria assemblies and the unattached freely-diffusing particles was infused through the left side of the center channel while the motility buffer solution was simultaneously infused through the right side of the central channel at the same flow rate. The two parallel stream introduction ensures that the nanoparticle mixture remains on the left side of the central channel before the bacteria carrying nanoparticles start migrating away (towards the chemoattractant source) via chemotaxis.
Selective bacteria adhesion to nanoparticles
This sorting technique exploits surface property differences between two groups of similarly-sized nanoparticles to selectively attach bacteria to one group of particles. Specific attachment facilitated by biotin–streptavidin bonds and non-specific attachment facilitated by electrostatic interactions were explored. For the specific adhesion experiments, a mixture of streptavidin-coated 390 nm polystyrene particles (Bangs Laboratories, Fishers, IN) and 320 nm polystyrene particles (Bangs Laboratories, Fishers, IN) was prepared prior to bacterial assembly. Briefly, E. coli MG1655m bacteria were washed twice with the motility medium and incubated with 10 μg ml−1 biotin-conjugated goat polyclonal anti-Lipid A LPS antibody (Thermo Scientific, Waltham, MA) to enable attachment of the 390 nm streptavidin-coated nanoparticles on the bacterial cell membrane.27 The suspension was gyrated on a vortex shaker for one hour at 600 rpm to facilitate antibody attachment to the bacterial cell. The bacterial suspension was centrifuged at a low speed (1700 × g) for 5 minutes at room temperature to remove the unbound antibody from the solution and was then resuspended in 50 μL of motility buffer. The 320 nm and 390 nm particle mixture suspension was agitated with biotinylated antibody-coated bacteria at a 100![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 nanoparticle mixture to bacteria ratio for 30 minutes. Using the streptavidin–biotin complex, one of the strongest non-covalent bonds found in nature, bacteria–390 nm particle assemblies were formed and the 320 nm particles remained unattached (Fig. 5(C) and (D)). A similar procedure was followed to prepare a mixture of streptavidin-coated 1040 nm polystyrene particles (Bangs Laboratories, Fishers, IN) and 1000 nm polystyrene particles (Thermo Scientific, Waltham, MA). The microparticle mixture was agitated with biotinylated antibody-coated bacteria at a 2.5
1 nanoparticle mixture to bacteria ratio for 30 minutes. Using the streptavidin–biotin complex, one of the strongest non-covalent bonds found in nature, bacteria–390 nm particle assemblies were formed and the 320 nm particles remained unattached (Fig. 5(C) and (D)). A similar procedure was followed to prepare a mixture of streptavidin-coated 1040 nm polystyrene particles (Bangs Laboratories, Fishers, IN) and 1000 nm polystyrene particles (Thermo Scientific, Waltham, MA). The microparticle mixture was agitated with biotinylated antibody-coated bacteria at a 2.5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 bacteria to microparticle mixture ratio for 30 minutes. Bacteria–1040 nm particle assemblies were formed and the 1000 nm particles remained unattached (ESI,† Fig. S1(A)).
1 bacteria to microparticle mixture ratio for 30 minutes. Bacteria–1040 nm particle assemblies were formed and the 1000 nm particles remained unattached (ESI,† Fig. S1(A)).
For non-specific adhesion experiments, a mixture of positively charged 1000 nm polystyrene particles (Polysciences, Warrington, PA) and 1000 nm neutrally charged polystyrene particles (Thermo Scientific, Waltham, MA) was first prepared. E. coli MG1655m bacteria were washed twice with the motility medium and agitated with the particle suspension at a 2.5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 bacteria to particle mixture ratio for 30 minutes. The electrostatic interactions between the positively charged particles and the negatively charged bacteria facilitated their assembly and the neutrally charged particles remained unattached (ESI,† Fig. S1(B)).
1 bacteria to particle mixture ratio for 30 minutes. The electrostatic interactions between the positively charged particles and the negatively charged bacteria facilitated their assembly and the neutrally charged particles remained unattached (ESI,† Fig. S1(B)).
Imaging and data analysis
Spatiotemporal distribution of bacteria, nanoparticles and bacteria–nanoparticle assemblies within the work area of both microfluidic devices was captured using a Zeiss AxioObserver Z1 inverted microscope equipped with an AxioCam MRm camera and a 10× objective. The recorded images were converted to binary images using Zen software (Zeiss Microscopy, Oberkochen, Germany). The binary images were then imported in ImageJ (NIH, Bethesda, MD) to obtain the spatiotemporal distribution information across the center channel. The chemotactic behavior of the bacteria and bacteria–nanoparticle assemblies was quantified using the population-scale metrics of the chemotaxis partition coefficient (CPC) and the chemotaxis migration coefficient (CMC), which represent the direction and strength of the chemotaxis response, respectively.28 The coefficients are defined by the following equations:where Nr is the number of objects in the right side of the center channel, Nl is the number of objects in the left side of the center channel, N(x) is the number of objects at a given position from the middle of the center channel x = 0, and w is the width of the channel. Both coefficients range between −1 and 1, with 1 indicating the strongest attraction to a chemoeffector and −1 indicating the strongest repulsion. A coefficient value that is positive indicates that the cells respond positively to the chemoeffector present in the source channel.
Results and discussion
Quantification of the bacterial chemotactic response
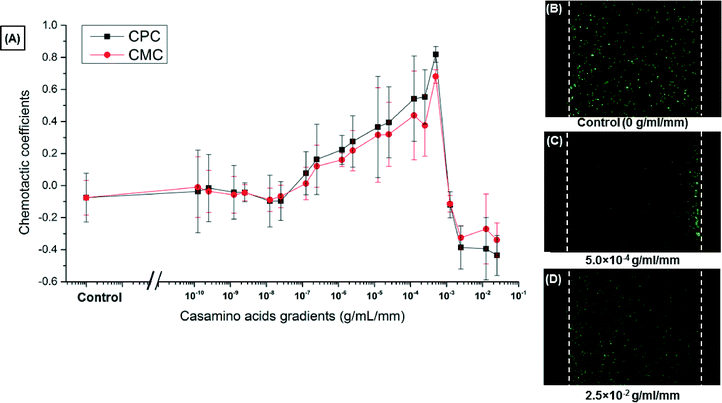
Various linear concentration gradients, from 1.25 × 10−10 g ml−1 mm−1 to 2.5 × 10−2 g ml−1 mm−1 of casamino acids, were generated inside the center channel of the microfluidic chemotaxis device to fully characterize the E. coli chemotaxis response in the presence of this chemoeffector and identify the optimal gradient that induces the strongest chemotaxis response. To this end, solutions of casamino acids (of various concentrations) in the right outer channel and a buffer solution in the left outer channel were continuously flowed. Since the three channels were separated by porous PEG-DA hydrogel walls, this resulted in a quasi-steady linear concentration gradient of casamino acids in the center channel. The chemotactic partition coefficient (CPC) and chemotactic migration coefficient (CMC) were computed and plotted as a function of the chemical concentration gradients generated in the center channel (Fig. 3(A)). In a control experiment, with chemotaxis buffer continuously flowed in both outer channels, the bacteria distribution did not show any bias over time (shown in Fig. 3(B)).The threshold chemical concentration that elicits a chemotactic response was found to be 0.25 × 10−7–1.25 × 10−7 g ml−1 mm−1, for which the chemotactic partition coefficient (CPC) value first became positive. The CPC value increased with the increase in concentration gradient slope and reached a maximum of 0.82 ± 0.05 at the chemical concentration gradient of 5.0 × 10−4 g ml−1 mm−1, as is shown in Fig. 3(C). At higher concentration gradients, the CPC begins to decrease. The bacteria exhibit a negative response towards the much higher gradient of 2.5 × 10−2 g ml−1 mm−1, as is shown in Fig. 3(D). These results can be explained by the adverse effects of high absolute chemical concentrations on bacterial motility and chemotaxis that prevent cells from performing chemotaxis towards a higher chemical concentration. This phenomenon where bacteria do not move up the gradient in a high chemical concentration environment has been discussed in previous studies.29,30 Earlier investigations have also shown that bacteria have the ability to sense a chemoeffector as an attractant at low chemical concentrations and as a repellent at higher chemical concentrations.28,31 Based on the chemotaxis characterization results, the optimum concentration gradient of 5.0 × 10−4 g ml−1 mm−1 was selected for the chemotaxis-enabled sorting of nanoparticles.
Sorting of nanoparticles using bacterial chemotaxis
Well-characterized and repeatable performance of the sorting platform requires operation under steady-state conditions. To estimate the amount of time required to achieve a quasi-steady linear gradient across the work area of the sorting platform, a computational model of the chemoattractant casamino acid transport through the device was carried out using the finite element analysis software package COMSOL®. For this model, the diffusion coefficient of casamino acids through the hydrogel was taken to be D = 1.5 × 10−6 cm2 sec−1, as was determined in a previous work using a Franz diffusion cell.26 The diffusion coefficient and flux for transport of casamino acids through PDMS were assumed to be zero given its non-permeable nature to casamino acids. The chemical concentrations in both outer channels were assumed to be constant. As is illustrated in Fig. 4, our results show that the quasi-steady optimal chemical gradient of 5.0 × 10−4 g ml−1 mm−1, can be reached within 75 min (4500 s). As is shown in Fig. 5 and S2,† this chemical attractant gradient across the work area of the sorting platform will prompt a strong chemotaxis response and chemotactic migration of the bacteria–particle assemblies towards the chemoattractant side of the work area, while the freely-diffusing particles will remain in a close proximity to their initial location within the work area.The diffusion length of the freely-diffusing particles due to Brownian motion can be calculated using  , where D = kBT/6πμR, kB is the Boltzmann constant, T is the absolute temperature, μ is the dynamic viscosity, R is the radius of the particle, and t is the time. Within the timeframe of the sorting experiment t = 45 min, the diffusion lengths of the 320 nm and 1000 nm particles are approximated as Ld = 120.61 μm and Ld = 72.19 μm, respectively, which are much smaller than the displacement of nanoparticles due to bacterial propulsion (∼500 μm), clearly demonstrating the feasibility of using the differential displacement of the diffusing particles and self-propelled particles for sorting.
, where D = kBT/6πμR, kB is the Boltzmann constant, T is the absolute temperature, μ is the dynamic viscosity, R is the radius of the particle, and t is the time. Within the timeframe of the sorting experiment t = 45 min, the diffusion lengths of the 320 nm and 1000 nm particles are approximated as Ld = 120.61 μm and Ld = 72.19 μm, respectively, which are much smaller than the displacement of nanoparticles due to bacterial propulsion (∼500 μm), clearly demonstrating the feasibility of using the differential displacement of the diffusing particles and self-propelled particles for sorting.
The sorting performance was quantified using the CPC metric by measuring the number of particles in each half of the work area every 5 minutes over a total duration of 60 minutes, as is shown in Fig. 6. The CPC values of 390 nm and 1040 nm streptavidin-coated particle–bacteria assemblies formed via specific adhesion increased from −0.58 ± 0.15 and −0.63 ± 0.01 to steady state values of 0.57 ± 0.10 and 0.62 ± 0.06, respectively, which were reached within 45 min. The CPC values of 320 nm and 1000 nm uncoated freely-diffusing particles remained negative and largely unchanged at −0.62 ± 0.06 and −0.66 ± 0.07. Control experiments in which a chemical attractant gradient is not present showed that bacteria-propelled particles could not be separated from unattached particles (data not shown). The CPC values of 0.57 and 0.62 for the 390 nm and 1040 nm bacteria-propelled particles indicate that about 79% and 81% of these particles have been transported to the right side of the work area, respectively, where the chemical concentration of casamino acids is the highest. In contrast, around 85% of 320 nm and 1000 nm unattached nanoparticles stayed near their initial location, on the buffer side of the work area. The sorting efficiency was not sensitive to the size of the nanoparticles for the size range we explored. The CPC of 1000 nm bacteria-propelled particles assembled via non-specific adhesion increased from −0.75 ± 0.07 to a steady state value of 0.49 ± 0.02, while the CPC of neutrally charged non-motile particles remained negative and largely unchanged at approximately −0.64 ± 0.10, as is shown in Fig. 6. The small reduction in the sorting efficiency of the non-specific attachment method can be attributed to weaker attachment between the bacteria and the particles that may result in reduced force transmission. The maximum CPC for the bacteria-propelled particles is somewhat smaller than the CPC obtained for the free swimming bacteria subjected to the same chemical attractant gradient value (Fig. 3). This can be attributed to disruption of flagellar bundling and bacteria motility in a small fraction of bacteria due to the random nanoparticle attachment.
Upon the completion of sorting, the two parallel streams, each containing one type of the particles from the mixture, can be purged into separate collectors on or off the chip. The separated particle–bacteria assemblies can be subjected to changes in temperature or pH in order to break the bonds between the bacteria and the nanoparticles.32–34 A density gradient centrifugation step can be implemented to retrieve nanoparticles from the suspension due to the size and density difference between the nanoparticles and the bacteria. The overall sorting yield of up to 80% can be improved through multiplexing and reintroducing unsorted particles propelled by bacteria in a similar sorting platform.
The strength of the sorting method presented here lies in its ability to separate micro/nanoscale objects with similar or even identical sizes and densities as long as their surface properties are different. Effective bacterial chemotaxis-enabled sorting requires careful selection of bacteria such that the surface energy difference between the bacteria and one set of particles is minimized and selective adhesion is achieved.35 We have previously demonstrated that the directed transport of 50 nm–10 μm particles can be achieved through bacterial motility and chemotaxis.18,19,27 Thus, this method will be suitable for sorting particles within the same size range. Furthermore, the current throughput of 2.4 × 105 particles per min can be enhanced by implementing bacteria with a higher motility speed, stronger chemoattractants and work areas with a smaller width and larger length (ESI,† section S.II). Moreover, different bacterial strains with a specific affinity to different chemoattractants can be used to sort more than two types of particles in terms of surface chemistry. Also, multiple sources of chemoattractant activated in a pre-designed time controlled manner can be used to establish a spatiotemporal varying chemical gradient and achieve multi-dimensional particle manipulation. A limitation of the proposed method is that for the single chemoattractant design shown here, the width of the work area (width of the center channel) cannot exceed 1500 μm, due to the limited bacterial-biased random walk distance (ESI,† section S.III). If a higher throughput is desired, a parallel array of microfluidic sorting devices could be implemented.
Conclusion
In this paper, we have introduced a passive sorting method for separating similarly-sized nanoparticles according to their surface properties. This bacteria-enabled sorting method is simple and cost-effective and the sorted particles need not be modified or stained. The required chemoattractant gradient can be established using gravity driven flow, thus eliminating the need for a syringe pump. The predominantly attractive feature of the proposed system revolves around the fact that the biological manipulators and the microfluidic platforms can be generated cost-effectively and swiftly while being highly scalable in nature. This bio-hybrid manipulation platform is fabricated using a simple one-step microfabrication process and it does not require electrical or magnetic energy sources generally required by active sorting systems. It mainly depends on a chemical energy source for actuation and chemical signalling for steering. Our previous efforts in bacteria-enabled propulsion of micro/nanoparticles suggest that the presented method can be applied to sort objects 50 nm–10 μm in size.The work presented here will serve as a stepping-stone for the development of inexpensive, self-directed, and chemically-based manipulation platforms which, in the long run, can contribute to reducing the complexity and costs associated with performing these tasks at reduced length scales. In the future, we seek to engineer bio-hybrid autonomous factories for transport and delivery, sorting, or bottom–up programmed self-assembly of micro/nanoscale objects. Effective development of such assembly and manipulation of workspaces could transform current practices and enable high throughput and high precision bottom–up assembly strategies.
Acknowledgements
The authors would like to thank our colleagues in the MicroN BASE laboratory at Virginia Tech, especially Ali Sahari. We also thank Birgit Scharf from the Department of Biological Sciences at Virginia Tech for generously providing E. coli MG1655m and assisting with the bacterial transformations. The RFP plasmid was kindly provided by John T. Singer at the University of Maine. This project was partially supported by the National Science Foundation (IIS-117519 and CAREER award, CBET-1454226).Notes and references
- D. R. Gossett, W. M. Weaver, A. J. Mach, S. C. Hur, H. T. K. Tse, W. Lee, H. Amini and D. Di Carlo, Anal. Bioanal. Chem., 2010, 397, 3249–3267 CrossRef CAS PubMed.
- P. Sajeesh and A. K. Sen, Microfluid. Nanofluid., 2013, 17, 1–52 CrossRef.
- P. Jákl, T. Čižmár, M. Šerý and P. Zemánek, Appl. Phys. Lett., 2008, 92, 161110 CrossRef.
- S. S. Kuntaegowdanahalli, A. A. S. Bhagat, G. Kumar and I. Papautsky, Lab Chip, 2009, 9, 2973–2980 RSC.
- M. Masaeli, E. Sollier, H. Amini, W. Mao, K. Camacho, N. Doshi, S. Mitragotri, A. Alexeev and D. Di Carlo, Phys. Rev. X, 2012, 2, 031017 Search PubMed.
- T. Morijiri, S. Sunahiro, M. Senaha, M. Yamada and M. Seki, Microfluid. Nanofluid., 2011, 11, 105–110 CrossRef.
- S. Fiedler, S. G. Shirley, T. Schnelle and G. Fuhr, Anal. Chem., 1998, 70, 1909–1915 CrossRef CAS PubMed.
- T. Laurell, F. Petersson and A. Nilsson, Chem. Soc. Rev., 2007, 36, 492–506 RSC.
- S. Choi and J.-K. Park, Lab Chip, 2007, 7, 890–897 RSC.
- A. Y. Fu, C. Spence, A. Scherer, F. H. Arnold and S. R. Quake, Nat. Biotechnol., 1999, 17, 1109–1111 CrossRef CAS PubMed.
- M. M. Wang, E. Tu, D. E. Raymond, J. M. Yang, H. Zhang, N. Hagen, B. Dees, E. M. Mercer, A. H. Forster, I. Kariv, P. J. Marchand and W. F. Butler, Nat. Biotechnol., 2004, 23, 83–87 CrossRef PubMed.
- J. Xi, J. Schmidt and C. Montemagno, Nat. Mater., 2005, 4, 180–184 CrossRef CAS PubMed.
- D. B. Weibel, P. Garstecki, D. Ryan, W. R. DiLuzio, M. Mayer, J. E. Seto and G. M. Whitesides, Proc. Natl. Acad. Sci. U. S. A., 2005, 102, 11963–11967 CrossRef CAS PubMed.
- N. Darnton, L. Turner, K. Breuer and H. Berg, Biophys. J., 2004, 86, 1863–1870 CrossRef CAS PubMed.
- B. Behkam and M. Sitti, Appl. Phys. Lett., 2007, 90, 023902 CrossRef.
- E. Steager, C. Kim, J. Patel, S. Bith, C. Naik, L. Reber and M. Kim, Appl. Phys. Lett., 2007, 90, 263901 CrossRef.
- S. Martel, C. Tremblay, S. Ngakeng and G. Langlois, Appl. Phys. Lett., 2006, 89, 233804 CrossRef.
- M. A. Traoré, A. Sahari and B. Behkam, Phys. Rev. E: Stat., Nonlinear, Soft Matter Phys., 2011, 84, 061908 CrossRef PubMed.
- A. Sahari, M. A. Traore, B. E. Scharf and B. Behkam, Biomed. Microdevices, 2014, 16, 717–725 CrossRef CAS PubMed.
- S. Martel and M. Mohammadi, in IEEE International Conference on Robotics and Automation, Anchorage, Alaska, 2010, pp. 500–505 Search PubMed.
- C. S. Barker, B. M. Prüss and P. Matsumura, J. Bacteriol., 2004, 186, 7529–7537 CrossRef CAS PubMed.
- J. S. Parkinson and S. E. Houts, J. Bacteriol., 1982, 151, 106–113 CAS.
- H. Cheng and G. C. Walker, J. Bacteriol., 1998, 180, 5183–5191 CAS.
- J. T. Singer, R. T. Phennicie, M. J. Sullivan, L. A. Porter, V. J. Shaffer and C. H. Kim, Appl. Environ. Microbiol., 2010, 76, 3467–3474 CrossRef CAS PubMed.
- D. L. Englert, M. D. Manson and A. Jayaraman, Nat. Protoc., 2010, 5, 864–872 CrossRef CAS PubMed.
- M. A. Traore and B. Behkam, J. Micromech. Microeng., 2013, 23, 085014 CrossRef.
- M. A. Traore, C. M. Damico and B. Behkam, Appl. Phys. Lett., 2014, 105, 173702 CrossRef.
- H. Mao, P. S. Cremer and M. D. Manson, Proc. Natl. Acad. Sci. U. S. A., 2003, 100, 5449–5454 CrossRef CAS PubMed.
- D. E. Koshland, Annu. Rev. Biochem., 1981, 50, 765–782 CrossRef CAS PubMed.
- R. Mesibov, G. W. Ordal and J. Adler, J. Gen. Physiol., 1973, 62, 203–223 CrossRef CAS PubMed.
- C. Li, A. J. Boileau, C. Kung and J. Adler, Proc. Natl. Acad. Sci. U. S. A., 1988, 85, 9451–9455 CrossRef CAS.
- A. Holmberg, A. Blomstergren, O. Nord, M. Lukacs, J. Lundeberg and M. Uhlén, Electrophoresis, 2005, 26, 501–510 CrossRef CAS PubMed.
- R. Faibish, M. Elimelech and Y. Cohen, J. Colloid Interface Sci., 1998, 204, 77–86 CrossRef CAS PubMed.
- U. A. Gurkan, S. Tasoglu, D. Akkaynak, O. Avci, S. Unluisler, S. Canikyan, N. Maccallum and U. Demirci, Adv. Healthcare Mater., 2012, 1, 661–668 CrossRef CAS PubMed.
- X. Zhang, Q. Zhang, T. Yan, Z. Jiang, X. Zhang and Y. Zuo, Environ. Sci. Technol., 2015, 49, 6164–6171 CrossRef CAS PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c6lc00059b |
| ‡ These authors contributed equally to this work. |
| § Current address: Department of Biomedical Engineering, Washington University in St. Louis, St. Louis, MO 63130, USA. |
| This journal is © The Royal Society of Chemistry 2016 |