Open Access Article
Open Access ArticleA microfluidic, dual-purpose sensor for in vitro detection of Enterobacteriaceae and biotinylated antibodies†
G.
Kokkinis
*a,
B.
Plochberger
b,
S.
Cardoso
c,
F.
Keplinger
a and
I.
Giouroudi
 ad
ad
aInstitute of Sensors and Actuators Systems, Vienna University of Technology, Gusshausstrasse 27-29, 1040 Vienna, Austria. E-mail: georgios.kokkinis@tuwien.ac.at
bInstitute of Applied Physics, Biophysics Group, Vienna University of Technology, Getreidemarkt 9, 1060 Vienna, Austria
cINESC Microsistemas e Nanotecnologias, Rua Alves Redol 9, 1000-029 Lisbon, Portugal
dInstitute for Biophysics, Department of Nanobiotechnology, BOKU – University of Natural Resources and Life Sciences, Muthgasse 11/II, 1190 Vienna, Austria
First published on 18th February 2016
Abstract
In this paper, we present a versatile, dual-purpose sensor for in vitro detection of Enterobacteriaceae (e.g. Escherichia coli) and biotinylated antibodies (e.g. IgG rabbit polyclonal antibodies), based on different detection principles for each bioanalyte. These bioanalytes are tagged individually with functionalized magnetic microparticles, suspended into a static fluid and injected into a microfluidic channel. Without the need for bulk or complicated pumping systems, the functionalized microparticles are set in motion by a magnetic force exerted on them by integrated microconductors. The fundamental detection principle is the decrease in the velocity of the microparticles that are loaded with the respective bioanalyte, due to factors inhibiting their motion. The velocity of the unloaded, bare microparticles is used as a reference. We discovered a novel mechanism on which the constrained particle motion is based; in the case of E. coli, the inhibiting factor is the enhanced Stokes' drag force due to the greater volume and altered hydrodynamic shape, whereas in the case of biotinylated antibodies, it is the increased friction force at the interface between the modified microparticle and the biosensor's surface. Friction force is for the first time employed in a scheme for resolving biomolecules. Integrated magnetic microsensors are used for the velocity measurements by detecting the microparticles' stray field. Moreover, we developed a biocompatible, easy to implement and reliable surface modification that practically diminishes the problem of bioadhesion on the sensor's surface.
1. Introduction
Enterobacteriaceae (e.g. E. coli) are considered to be the most common causative agents of food and waterborne diseases. Enterobacter infections are responsible for thousands of deaths annually,1 while prominent health institutions like the World Health Organization (WHO)2 are trying to tackle the increasing antimicrobial resistance (AMR), by means of the appropriate use of antimicrobials and adequate diagnostics. Rapid diagnostics can substantially address those issues.3Another analyte gaining considerable attention in diagnostics are the serum auto-antibodies due to their biomedical relevance. Systemic auto-antibody detection would facilitate the diagnostics of autoimmune disorders and evaluate their treatment and the sustained damage in organs.4 More interestingly, the presence of certain antibodies in the system is considered an increasingly important expression of diseases such as some types of cancer.5
Most of the existing laboratory techniques to identify suspected pathogens use culturing techniques to grow colonies large enough to identify. Other diagnostic methods such as flow cytometry, fluorescence probe detection and optical particle detection employ microscopy or fluorescence microscopy which is expensive and time consuming.6–8 Nevertheless, there exist methods which do not require a large amount of sample and provide rapid identification, without the disadvantages of microscopy such as immunological tests (e.g. ELISA immunoassays) and nucleic acid based diagnostics.9–14 Yet, these methods either require established laboratory infrastructure and well-trained personnel or are technologically complex and expensive to fabricate. Moreover, even though they are highly sensitive and specific, false positive and negative results may occur. These results can be caused by improper sample storage or treatment, improper washing methods or reagent deterioration. As far as antibody detection is concerned, ELISA is again the technique mostly used to detect and quantify those antibodies, manifesting the same limitations.
Therefore, the development of portable, sensitive and fully automated biosensors that directly translate the presence of certain analytes into an electronic signal gains increasing interest.15 All necessary sample handling and analysis steps are performed within the biosensor without the need for established laboratory infrastructure or well-trained personnel.
Microfluidic biosensing platforms are ideal for addressing the demand for such systems. Several biosensors utilizing different physical properties and detection principles have been proposed,16–22 with magnetic biosensors yielding promising results.23–27 However, the main disadvantages of these platforms are low flexibility, complicated microfluidic structures and pumping mechanics.
Our microfluidic biosensor, contrary to culture methods, can accurately and rapidly identify only a small number (e.g. a few tens) of pathogens due to magnetic labelling and the novel detection mechanism (section 4). Moreover, the detection procedure (using magnetic microsensors) eliminates the disadvantages of microscopy.
The main advantage of our approach over other cheap and fast diagnostic platforms lies on the fact that there is no specific molecular interaction that provides the results, such as that obtained by using ELISA, but instead the quantitative results are obtained by magnetically driven motion and its alternation (velocity change). Thus, our biosensor is insusceptible to common problems occurring in biosensing techniques such as wrong concentrations, buffer solutions or non-specific interactions.
Specifically, the presented microfluidic biosensor combines labeling of the analytes with magnetic microparticles (MPs),27 magnetophoretic manipulation of the tagged bioanalytes suspended in a static fluid (without flow) through integrated microconductors (MCs) and indirect evaluation of their velocity utilizing spin valve (giant magnetoresistance, GMR) sensors.28 Any difference in the evaluated velocity of the microparticles conjugated with the analytes (henceforth referred to as loaded microparticles, LMPs) due to factors hindering their motion with respect to a reference velocity, evaluated for unloaded MPs, yields positive results. The factors hindering the MPs' motion are different for each type of bioanalyte.
Capturing of E. coli is achieved using specific antibodies which are then conjugated to the MPs, while the detection of antibodies requires their biotinylation and immobilization on the MPs' surface through the streptavidin–biotin interaction. The conjugation process will be described in detail in section 3. When injected into the microfluidic channel and in the absence of flow, the MPs are manipulated by sequentially actuated MCs inducing a traveling magnetic field gradient, attracting the MPs and moving them along the channel. In the case of E. coli conjugated to the MPs (henceforth referred to as bacteria loaded magnetic particles, BLMPs), the greater hydrodynamic volume (while the magnetic volume remains constant) and the altered shape result in an enhanced Stokes' drag force29 and thus to a reduction in their velocity while in suspension. In the case of biotinylated antibodies conjugated to the MPs (henceforth referred to as antibody loaded magnetic particles, ALMPs), the overall volume of the compound also increases but to a negligible extent. The effect that slows down the ALMPs with respect to bare MPs in this case is the increased friction force between the ALMPs and the chip's surface. This effect is explained theoretically in section 2 and proven experimentally through atomic force microscopy (AFM) measurements in section 5.
Among others, the significance of the presented work lies on the fact that for the first time we present an alternative method for resolving biomolecules (i.e. biotinylated antibodies) based on the frictional interaction between tagged biomolecules and surfaces, a principle that could lead to the development of methods akin to chromatography and capillary electrophoresis.30,31
Friction forces are critical for the development of microelectromechanical systems (MEMS) and biosensors.32 A lot of the proposed systems are based on the manipulation of particles, biological entities etc. Several particle manipulation techniques bring the suspended particles in contact with the chip's surface, while exerting magnetic forces on them.33,34 Unfortunately, the most commonly used materials, such as silicon dioxide (SiO2) or silicon nitride (Si3N4), exhibit bioadhesion making them inappropriate for such applications. Another innovation of the presented work is the development of a biocompatible, easy to implement and reliable surface modification that practically diminishes the problem of biological entities being immobilized on the biosensor's surface which otherwise would render it inadequate for MPs' manipulation and multiple diagnostic tests.
2. Theoretical analysis
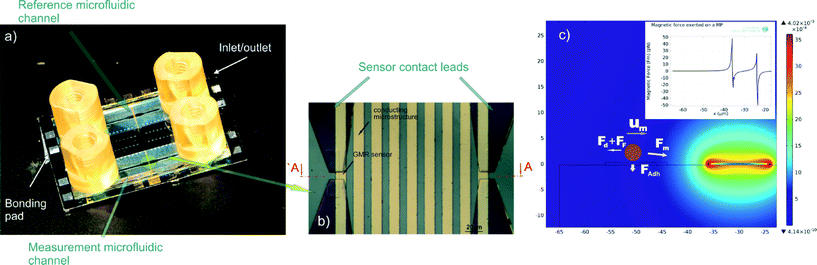
A better understanding of the detection principle is achieved by analysing the forces that act on a single particle (MP). Fig. 1c shows the forces exerted on a single particle by a single MC. The magnetic force is given by the following equation:35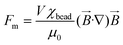 | (1) |
The main force opposing the movement of the MPs in suspension is the frictional Stokes' drag force, deriving from the Navier–Stokes differential equation for small Reynold's numbers and small spherical particles, acting on the interface between the fluid and the MP. It is given by the following equation:38
| Fd = 3πDηυ | (2) |
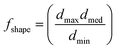 | (3) |
During the sliding of the MPs on the chip's surface, a friction force is exerted on them. Friction between two solid surfaces on the microscale and nanoscale involves complicated physical phenomena that are beyond the scope of this paper and are extensively reported in the literature.40–43 In the present work, we focus on providing the necessary elements for a qualitative analysis of the friction based on recently proposed models. The friction in the developed system can be modeled as two solid surfaces in sliding, relative motion with a molecularly thin film (the surface modification) confined between them, with low shear rates and smooth asperities. Such conditions classify the interfacial friction to the boundary tribological regime.44,45 As AFM experiments show (section 5), the two surfaces are in adhesive contact. Consequently, Amonton's law cannot sufficiently describe the friction; therefore, a more appropriate approach is given by the cobblestone model where friction force is evaluated as follows:46
| F = FF = ScA + μL, | (4) |
The surfaces involved in relative motion in the developed system are quite complex: on the one hand, a protein (streptavidin or antibodies) covered sphere and, on the other hand, a hydrogel flat surface. As it can easily be deducted, the definition of the intermolecular forces at the interface cannot be solely explained with the DLVO theory (named after Derjaguin and Landau, Verwey and Overbeek).47,48 For the qualitative study and the proof of concept of the biosensor, the determination of the adhesive nature of the interaction is sufficient and accurate. Further analysis of the additional forces involved which sum up to the principally adhesive forces, such as solvation, entropic, hydrophobic forces etc., is out of the scope of this paper.49
3. Material and methods
3.1 Device fabrication
Fig. 1c shows the developed biosensor with the integrated microfluidic channels and the integrated GMR sensors. The GMR sensors were fabricated as follows: a thin film spin valve stack was deposited in a Nordiko 3000 Ion Beam Deposition device. The stack had the following structure (thickness in nm, compositions in atomic percentage): Ta 3.0/Ni80Fe20 3.6/Mn76Ir24 8.5/Co80Fe20 2.3/Rn 0.8/Cu 3.0/Co80e20 3.0/Ni80Fe20 3.6/Ta 5.0. A 3 mT magnetic field was applied during the deposition in order to induce a parallel anisotropy for the free and pinned layer easy axis.50 The final structures as shown in Fig. 1 were defined using standard photolithography and an ion milling etching process carried out using a Nordiko 3600 device. 300 nm aluminum contact leads to the sensors were fabricated using again photolithography and an aluminum spattering process with the latter carried out using a Nordico 7000 device. The sensors were then passivated with a 15 nm thick TiW layer deposited in the aforementioned device. The MC fabrication and the finalization of the chips' surface have been reported in detail.28The microfluidic channels were fabricated as follows: the wafer was immersed in a 10% RBS 50 basic solution in deionized (DI) water and in an ultrasonic bath for 2 hours to remove potential contaminants present from the previous steps and dried for 30 min at 120 °C. Then, a 55 μm thick, negative type, dry-film, photoresist (Ordyl®) was laminated using a standard office laminator and exposed for 21 s using a Karl Suss MA-150 Mask Aligner; a 60 s post exposure bake at 85 °C on a hot plate followed. The exposed photoresist was developed for 1.5 min in 3 successive developer baths of increasing cleanness and rinsed with isopropanol and DI water. For the sealing of the channels, a cleaned (in the aforementioned solution) and cut glass slide was used, in which inlet and outlet holes were opened using a sand blasting device (BEGO Inc., Duostar®). The chip and the sealing slide were then bonded by applying a force of 60 N per cm2 of photoresist. The temperature was increased to 100 °C with a 5 °C min−1 ramp and maintained for 30 min. Then, the temperature was reduced to room temperature at 1 °C min−1. A DAD 3220 dicing saw with a 200 μm thick diamond blade was used to cut the bonded wafers into single chips. The inlet and outlet holes in the glass wafer were sealed with adhesive tape before dicing to prevent cooling water and debris from entering the chips. To free the contact pads, the glass wafer was partially removed in the areas above the pads. This was performed by dicing the chip halfway through, dicing only through the glass wafer and using the Ordyl® layer as a spacer that prevented the MCs and the contact pads from getting damaged.
3.2 Antibody loaded magnetic particle preparation
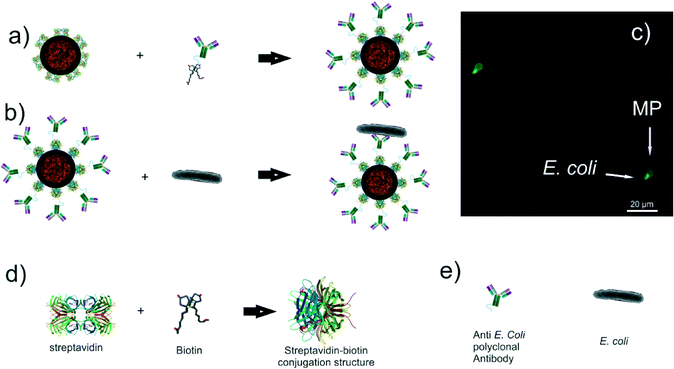
Life Technologies® Dynabeads® M-280 with covalently coupled recombinant streptavidin and a tosyl-activated, hydrophobic surface functionality have served as the basis for analyte (i.e. antibodies, E. coli) conjugation and sample preparation throughout the experiments. 100 μL of Dynabeads® M-280 (10 mg mL−1 concentration) was mixed with 900 μL of 0.01 M phosphate-buffered saline (PBS)–Tween (0.01% v/v) buffer, then washed 3 times with magnetic separation and resuspended in 1 mL of PBS–Tween. 7 μL of rabbit polyclonal antibodies to E. coli (Abcam® ab20640) covalently coupled with the N-hydroxysuccinimide ester of biotin was added and incubated for 30 min at room temperature. Then, the particles conjugated with the IgG were washed 5 times by magnetic separation and resuspended in 1 mL of 0.01 M PBS–BSA (0.1% w/v). 3 μL of donkey polyclonal secondary antibody to rabbit IgG–H&L (conjugated with the fluorophore Alexa Fluor® 488) was added and incubated for 30 min at room temperature for visualization purposes.3.3 Bacteria loaded magnetic particle preparation
One colony-forming unit (CFU) of Escherichia coli of the K12 “wild type” strain was suspended in 1 mL of 0.01 M PBS–Tween 20 (0.01% v/v), washed 3 times utilizing centrifugal separation and resuspended in the original volume. 7 mL of ab20640 antibodies with a concentration of 4 mg ml−1 was added and incubated for 30 min at room temperature, washed 5 times by centrifugal separation and resuspended in 100 μL of 0.01 M PBS–BSA (0.1% w/v). The Dynabeads® M-280 were washed using the same process as the one described in the ALMP preparation section and condensed in 100 μL of 0.01 M PBS–Tween 20 (0.01% v/v). 40 μL of the E. coli (conjugated with antibodies) solution was mixed with 1 μL of the Dynabeads® M-280 washed solution and was incubated for 30 min at room temperature. 200 μL of 0.01 M PBS–BSA (0.1% w/v) was added and then washed once by magnetic separation. 1 μL of donkey polyclonal secondary antibody to rabbit IgG–H&L (conjugated with the fluorophore Alexa Fluor® 488) was added and incubated for 30 min at room temperature for visualization purposes.3.4 Surface modification
A single chip was washed by three consecutive rinses with acetone, isopropanol and DI water to remove any contaminants. Then, it was left to dry on a hot plate for 30 min at 150 °C. Oxygen plasma was employed for the hydroxylation of the SiO2 surface (30 s oxygen plasma, 100 watt forward power, 30 sccm O2). Afterwards, the chip was dipped for 10 min in a 2 g L−1 branched, polyethyleneimine (Mw ∼ 24![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 by LS, Sigma-Aldrich®) solution in DI water and then rinsed with DI water. Another dip for 10 min in 2 g L−1 sodium alginate (alginic acid sodium salt, Sigma-Aldrich®) followed by a DI water rinse finalizes the antifouling surface modification.
000 by LS, Sigma-Aldrich®) solution in DI water and then rinsed with DI water. Another dip for 10 min in 2 g L−1 sodium alginate (alginic acid sodium salt, Sigma-Aldrich®) followed by a DI water rinse finalizes the antifouling surface modification.
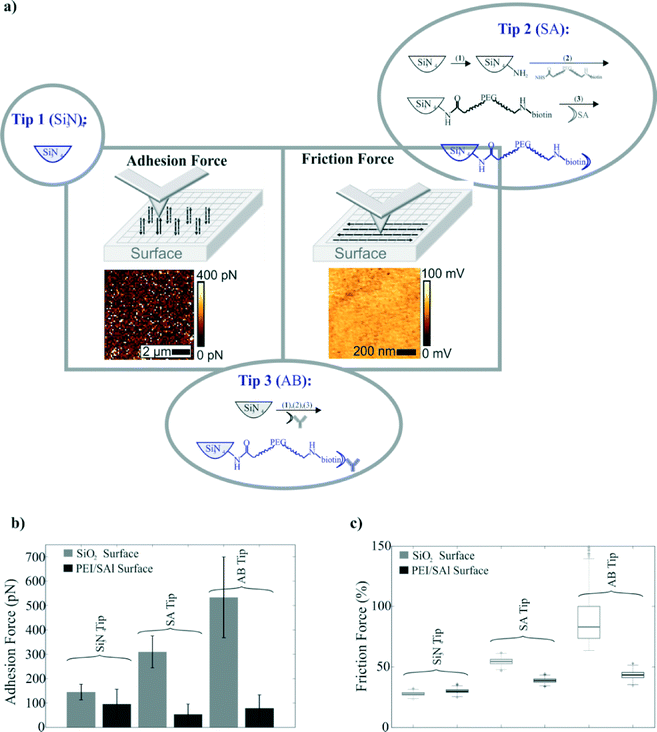
3.5 AFM tip functionalization
Si3N4 cantilever tips were stepwise chemically modified for friction and adhesion measurements. Commercially available silicon AFM cantilevers (MSNL-10, Veeco Instruments) were cleaned with chloroform (3 times), dried with N2 and stored in PBS buffer (tip 1: Si3N4 tip). The cleaned silicon AFM cantilevers were additionally amino-functionalized via gas-phase silanization with aminopropyltriethoxysilane (APTES) as described in ref. 51. A heterobifunctional linker (aldehyde–biotin) was prepared as described in ref. 52. Briefly, 3.3 mg of a linker (Nanocs, Biotin-PEG-NHS, MW 3400) was dissolved in 0.5 ml of chloroform and transferred into a small glass reaction chamber. 30 μl of trimethylamine was added, and the ethanolamine-coated AFM tips were immediately immersed for two hours. Subsequently, the tips were washed with chloroform and dried with N2 gas. After rinsing with chloroform and drying, the tips were immersed for 60 minutes in a 50 μg ml−1 streptavidin solution, then washed and stored in PBS buffer (tip 2: streptavidin (SA) tip). Finally, streptavidin coated tips were incubated for 60 minutes with biotinylated antibodies, washed and stored in PBS buffer (tip 3: antibody tip).3.6 Combined fluorescence microscopy and atomic force microscopy
The sample was sealed in a home-built chamber and rinsed with PBS. AFM measurements were performed using a NanoWizard 3 (JPK Instruments AG, Germany) system mounted on an Axiovert 200 inverted microscope (Carl Zeiss AG, Germany). The microscope is equipped with a 100× NA = 1.46 oil-immersion Plan-Apochromat TIRFM objective (Olympus, Japan). Samples were illuminated in objective-type, total internal reflection (TIR) configuration via the epiport using 488 nm (250 nW) and 647 nm (250 mW) light from a diode laser (Toptica 250 mW, Toptica Photonics, Germany) or 532 nm light from a solid state laser (Millennia X, Spectra Physics, USA), with intensities of 3–10 kW cm−2. After appropriate filtering, emitted signals were imaged using a back-illuminated, TE-cooled CCD camera (Andor iXon Du-897 BV, UK). For the precise control of the illumination timings, we used acousto-optical modulators (1205C, Isomet, USA). Timing protocols were generated by an in-house program package implemented in LABVIEW (National Instruments, USA). Illumination times were adjusted to values between 1 and 5 ms. The particle on the apex of the tip was first imaged using a fluorescence microscope to determine the lateral movement of the bead during scanning. Topographical and friction images were recorded in contact mode by keeping the vertical deflection constant and analyzed. Adhesion and elasticity measurements were recorded in quantitative imaging mode (QI mode) at room temperature in liquid (PBS) at a resolution of 128 × 128 pixels. The maximum force applied for adhesion measurements, determined by the vertical deflection of the cantilever, was set to 300 pN. Force distance cycles (scan rates) were controlled by the z length (250 nm), extension time (10 ms) and retraction time (50 ms). We used uncoated silicon cantilevers (MSNL-10, Bruker Corporation, USA) with a nominal spring constant in the range of 0.01–0.03 N m−1. The spring constant for each cantilever was calibrated using the thermal noise method.53–56 JPK data processing (JPK Instrument, Germany) software was used for image processing and estimation of the adhesion force and indentation. The height, adhesion and slope of the force curve were collected simultaneously in both trace and retrace directions. Height images were line-fitted as required. Isolated scan lines were occasionally removed. The Young's modulus was estimated from the force curve by using the JPK data processing software with the following settings: model (cantilever tip): cone; opening angle (cantilever tip): 35°, Poisson ratio: 0.5; method: Hertz model.4. Experimental
The microfluidic channel walls were fabricated utilizing standard photolithography techniques on a negative dry photoresist laminated on the biosensor's surface (Ordyl® 350Y) with 50 μm thickness while the length of the channel was 10 mm and the width varied between 90 μm and 500 μm. The maximum feature size of the microfluidic channel is dictated by the surface area of the developed photoresist so as to not inhibit or impede the bonding.57 As a final step, a glass slide with sand-blasted inlets and outlets was bonded on the photoresist sealing the channels. This fabrication technique provides the significant advantage of automatic, pump-less priming over polydimethylsiloxane (PDMS) channels, due to the hydrophilicity of the glass and the biosensor's surface. Thus, the channels are being filled solely due to capillary forces.58The commercially available magnetic microparticles, Dynabeads® M-280 which were used for the preliminary experiments, are uniform, superparamagnetic, porous polystyrene spheres with an even dispersion of magnetic material throughout the particle. The magnetic material within the Dynabeads is a mixture of two iron oxides, maghemite (gamma-Fe2O3) and magnetite (Fe3O4), which is encased in the bead matrix by an additional thin polymer shell (monodispersity: SD 0.04–0.05 lm and CV 1.6–1.8%, density: 1.6 g DS cm−3).59 These MPs, with a layer of covalently coupled recombinant streptavidin to the surface, are an ideal candidate for conjugation processes due to the high affinity of streptavidin to biotin (Kd = 10−15) while their superparamagnetic properties allow for high magnetophoretic mobility and rules out particle agglomeration.
The developed biosensor consists of two microfluidic channels; one is the measurement channel where the sample with the LMPs is injected and another is used as the reference channel, where the reference sample is injected. The reference sample is ALMPs when we want to detect E. coli and plain unloaded MPs for the detection of biotinylated antibodies. Once the samples are inserted into the channels, the magnetophoretic manipulation of the particles (MPs and LMPs) takes place. Superparamagnetic particles single or engulfed in a polymer matrix move towards an increasing magnetic flux density gradient.
This gradient is provided by integrated planar microconductors (MCs) fabricated perpendicular to the MPs' motion as shown in Fig. 1b. Since this gradient is especially sharp, as finite element method simulations suggest (Fig. 1c, inset), it can effectively move the MP over a limited distance of a few micrometers. Hence, an array of 9 conductors was fabricated to move the MPs over a distance of 180 μm, the manipulation area (MA), by switching “OFF” a current of 50 mA when the MPs reach the edge of one conductor and switching “ON” the consequent conductor. Four GMR spin valve microsensors fabricated underneath the MCs are situated as follows: two underneath the first MCs and two underneath the last MCs.
Sensing the stray fields of the MPs in both channels provides information about their presence or absence on top of the first and last MCs. For the detection, we utilized the double frequency modulation technique,60 substituting the lock-in amplifier with a spectral analysis algorithm described in ref. 61. Since we proved the successful performance of our microfluidic sensing system using uncoated MPs, we proceeded to determine the magnetophoretic behavior for the chemically modified ones.
In order to study the effects on MPs coated with bioanalytes (e.g. E. coli and antibodies) but also characterize the interaction forces between the sample and the surface, IgG rabbit polyclonal antibodies were conjugated to the MPs. These antibodies are specific for the “O” and “K” antigenic serotypes of E. coli gram negative bacteria and they are biotinylated through the amine-reactive cross-linker chemistry,62 (Fig. 2a). Subsequently, a part of the ALMPs was conjugated with E. coli K12 “wild type” gram negative bacteria. The common K12 strain was chosen for its equivalence to the pathogenic E. coli expressing the enterohemorrhagic serotype O157:H7, a profound food contaminant63 (Fig. 2b).
As stated in the introduction, biological entities on the surface of the particles exhibit adhesion with common materials used in MEMS technology. Preliminary experiments showed complete adhesion of the streptavidin coated particles on the biosensor's SiO2 surface. For that reason, a surface modification was developed, based on the layer-by-layer (LbL) electrostatic self-assembly (ESA) technique.64 Two biocompatible polyelectrolytes, polyethyleneimine (PEI) and sodium alginate (SAl), were sequentially immobilized on the biosensor's surface forming a bilayer. First, the SiO2 passivation layer of the biosensor was treated with oxygen plasma so as to enhance its negative charge; then PEI, a polycation, was attracted on the surface, and lastly, SAl, a polyanion, was electrostatically coupled with the PEI. With SAl having the properties of absorbing water molecules, the antifouling property of the final surface are greatly enhanced, while the overall thickness, as AFM measurements suggest, increases slightly (Fig. S1†).
4.1 Measurement process
For the measurement process, both the test and the reference samples are injected into the channels using pipets. Then, the excess liquid is removed from the inlets with fine swabs. This asserts that the level of liquid in the inlet and the outlet is equal and there is no gravity induced flow in the channel. With the injection of the samples, the MA is covered with scattered MPs and LMPs. In order to avoid false readings, the area has to be free of MPs. Therefore, we sequentially actuate the MCs from the last to the first with a time interval that allows even the slowest particle to move from one MC to the next. All the current switching sequences are operated by a programmable microcontroller. Subsequently, if the first spin valves in both channels detect one or more MPs, we again sequentially actuate the MCs this time in the opposite direction, from first to last (Fig. 3). The time each MC is “ON” is determined by the time the MPs in the reference channel (unloaded MPs) were required to move from one conductor to the next and has to be defined experimentally before the sensing procedure, as a calibration step. This certifies that the sensor underneath the last MC in the reference channel will give a signal change at the end of the sequential actuation. Simultaneously, in the measurement channel (in the presence of the analyte at the MPs' surface), the slower LMPs will fall behind or arrive with a delay at the vicinity of the sensor. That way, we deduct the test results.5. Results and discussion
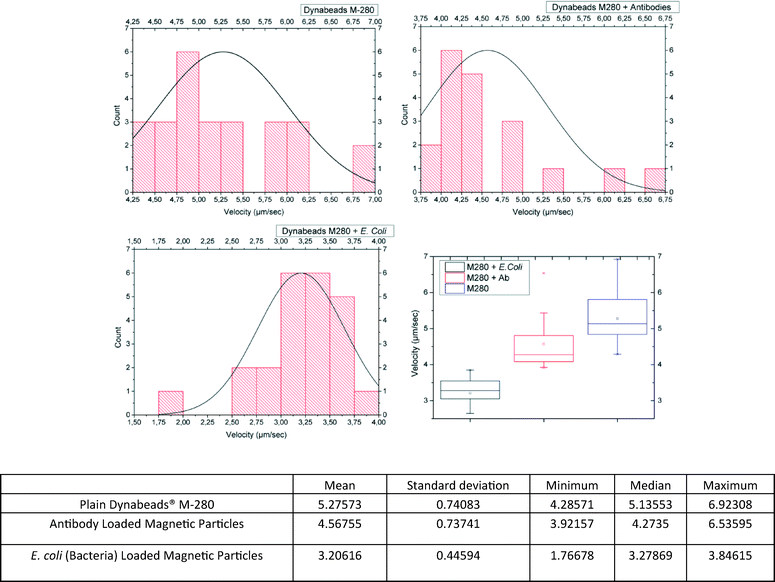
In order to prove the concept of the microfluidic biosensor, we had to show that the velocity of the MPs in suspension throughout the (on chip) magnetophoretic procedure was lower for ALMPs with respect to plain MPs (in the reference channel) and even lower when the antibody had captured an E. coli bacterium (e.g. BLMPs).To do so, the magnetophoretic procedure was filmed using a 50× microscope lens. The videos were then analyzed, and the velocities were deducted. The results are shown in Fig. 4. In order to cross-check the results and to test the biosensor's efficiency, the samples were again injected into the channels; this time the microcontroller was programmed to maintain each conductor “ON” for a period of time equal to the mean velocity value for each sample type, as the previous experiments suggested (values are presented in the table shown in Fig. 4).
The results were verified with the GMR sensors that could register a measurement at the end of the magnetophoretic procedure (Fig. 5). Such a registered signal by the GMR sensor means that at least one MP could travel with the mean velocity from one conductor to the other provided that they were carrying current (exerting magnetic field gradient) for the time interval corresponding to the velocity measured for each sample type. The reason for the decrease in velocity of the BLMPs has been sufficiently analyzed in ref. 28 and is based on the overall volumetric increase while the magnetic volume remains constant; the same does not apply for the ALMPs.
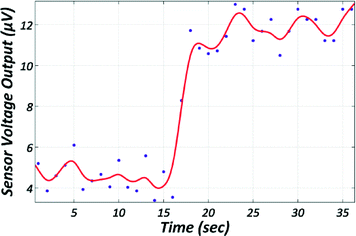 | ||
| Fig. 5 Graph acquired during the detection of a single MAP by the GMR sensor showing the sensor's voltage output over time. The red line is a second order polynomial fitting to the raw data. | ||
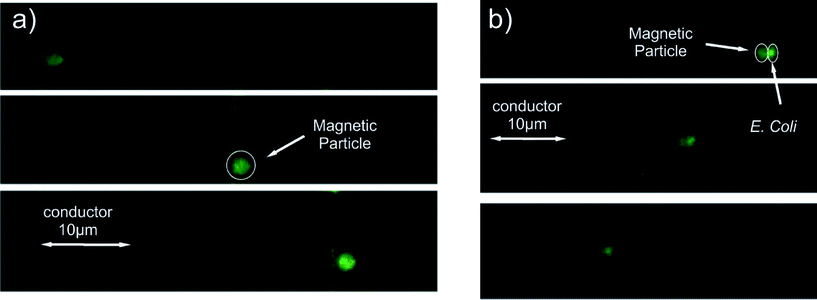
The ratio of gyration for the IgG antibodies conjugated with biotin molecules is of the order of a few nanometers. That can cause, considering the size of the MPs, an insignificant velocity decrease. Since it is not the Stokes' drag force that is significantly altered and the magnetic force remains constant, we conclude that the causes of this decrease in the velocity are the friction and the adhesion surface forces. As eqn (4) suggests, the friction and the adhesion are interconnected; thus, it is justified to assume that the change in velocity for the ALMPs with respect to plain, streptavidin coated MPs is due to enhanced friction forces. In order to experimentally prove this assumption, we conducted adhesion and friction force measurements utilizing an AFM device with a functionalized tip. As a first approach, we directly linked paramagnetic particles with different functionalities to the AFM tip apex. However, none of the used approaches, neither the linkage via biotin nor via antibody linkage, showed the expected performance. In both cases, we observed a visible lateral movement of the particle on the apex of the tip by scanning on a glass surface, which we imaged by the combined atomic force and fluorescence microscopy (Fig. S2†). Therefore, we changed the approach and directly chemically functionalized the AFM tip (Fig. 6a). In particular, we started by using a cleaned Si3N4 tip, followed by further functionalization processes as described in section 3 and measured the interaction between various functionalized tips and different surfaces.
Fig. 6b and c summarizes the adhesion and friction force study. Three different tip surfaces were tested on two different chip surfaces: a SiO2 surface (light grey) and the PEI/SAl modified surface (dark grey). It is apparent that the unmodified chip's surface could not be used to manipulate streptavidin coated (plain) MPs or ALMPs as the adhesion forces are an order of magnitude greater than the magnetophoretic force exerted on the MPs by the conductors. The study also suggests that the adhesion force is greater for the antibody coated tip than the streptavidin coated one. This implies, due to the first term of the friction force equation in section 2 (eqn (4)), that the friction force could be greater for the antibody coated tip as well. Indeed, friction force measurements showed that the friction force exerted on a moving tip (with a velocity of 5 μm s−1) on the modified PEI/SAl surface was significantly greater for the antibody coated tip than the streptavidin coated one.
The results lead to the assumption that frictional interactions between biomolecules and surfaces could be utilized to differentiate biomolecules with a distinct spatial structure, provided that this translates into different friction forces. Of course, the same principle could apply for differentiating the same protein but with different or incorrect folding.
6. Conclusions
The AFM friction force measurements proved the original assumption. The observed decrease in the mean velocity during the magnetophoretic manipulation of ALMPs with respect to plain, streptavidin coated MPs is due to the enhanced friction of the ALMPs. The actual nature of the enhanced friction is unclear but most probably linked to the 3D protein structure of the IgGs and their spatial orientation, protruding out of the MP surface.Thus, we demonstrated the potential efficiency of the developed microfluidic sensor in detecting E. coli, as a whole organism, by using the change in velocity measured indirectly with integrated GMR sensors, due to the enhanced Stokes' drag force exerted on the BLMPs because of their volumetric increase with respect to plain MPs. We also demonstrated the possibility of using the same biosensor for the detection of biotinylated antibodies. Again, a decrease in velocity is utilized for the detection scheme. However this time, the decrease is due to the enhanced friction exerted on the ALMPs.
Other members of the family Enterobacteriaceae (e.g. Salmonella and Klebsiella) are currently being measured with the presented microfluidic biosensor in order to prove that it can be used as a general pathogen sensor. In parallel, experiments are conducted with other molecules in order to demonstrate the possibility of using the same biosensor for the detection of smaller entities other than biotinylated antibodies, for example proteins and even non-organic substances as polymers.
Acknowledgements
The research at the Vienna University of Technology was supported by the Austrian Science Fund (FWF) with Project No. P 24372-N19. INESC-MN acknowledges FCT funding through the IN Associated Laboratory (Pest-OE/CTM/LA0024/2011) and Project EXCL/CTM-NAN/0441/2012.References
- W. Tian and E. Finehout, in Microfluidics for Biological Applications, 2008, pp. 271–322 Search PubMed.
- World Health Organization, Fact Sheet for Antimicrobial Resistance, http://www.who.int/mediacentre/factsheets/fs194/en/, 2015 Search PubMed.
- P. Yager, G. J. Domingo and J. Gerdes, Annu. Rev. Biomed. Eng., 2008, 10, 107–144 CrossRef CAS PubMed.
- A. Schlichtiger, P. Luppa, D. Neumeier and M. Thaler, Bioanal. Rev., 2012, 4, 75–86 CrossRef.
- D. M. Goldenberg, Am. J. Med., 1993, 94, 297–312 CrossRef CAS PubMed.
- BD Biosciences, Instrument Brochure, http://www.bdbiosciences.com/documents/BD_FACSMicroCount_Instrument_Brochure.pdf, 2015 Search PubMed.
- J. Moldenhauer, BioPharm Int., 2005, 11–20 Search PubMed.
- J. P. Jiang, Encyclopedia of rapid microbiological methods, 2005, vol. 3, pp. 121–141 Search PubMed.
- Alere Inc., Rapid diagnostic tests developer, http://www.alere.com, 2013 Search PubMed.
- Micronics Inc., Developer of near patient in vitro diagnostic products, www.micronics.net, 2013 Search PubMed.
- O. Lazcka, F. J. Del Campo and F. X. Muñoz, Biosens. Bioelectron., 2007, 22, 1205–1217 CrossRef CAS PubMed.
- B. Vuylsteke, Sex. Transm. Infect., 2004, 80, 333–334 CrossRef CAS PubMed.
- D. Nichols, FEMS Microbiol. Ecol., 2007, 60, 351–357 CrossRef CAS PubMed.
- R. M. Lequin, Clin. Chem., 2005, 51, 2415–2418 CAS.
- M. L. Y. Sin, K. E. Mach, P. K. Wong and J. C. Liao, Expert Rev. Mol. Diagn., 2014, 14, 225–244 CrossRef CAS PubMed.
- J. Loureiro, R. Ferreira, S. Cardoso, P. P. Freitas, J. Germano, C. Fermon, G. Arrias, M. Pannetier-Lecoeur, F. Rivadulla and J. Rivas, Appl. Phys. Lett., 2009, 95, 034104 CrossRef.
- A. K. Balasubramanian, A. Beskok and S. D. Pillai, J. Micromech. Microeng., 2007, 17, 1467–1478 CrossRef CAS.
- J. Chen, D. Chen, Y. Xie, T. Yuan and X. Chen, Nano-Micro Lett., 2013, 5, 66–80 CrossRef CAS.
- K.-S. Yun, D. Lee, H.-S. Kim and E. Yoon, Meas. Sci. Technol., 2006, 17, 3178–3183 CrossRef CAS.
- J. Mairhofer, K. Roppert and P. Ertl, Sensors, 2009, 9, 4804–4823 CrossRef CAS PubMed.
- K. S. Kim and J.-K. Park, Lab Chip, 2005, 5, 657–664 RSC.
- L. Y. Yeo, H.-C. Chang, P. P. Y. Chan and J. R. Friend, Small, 2011, 7, 12–48 CrossRef CAS PubMed.
- P. Kinnunen, I. Sinn, B. H. McNaughton, D. W. Newton, M. A. Burns and R. Kopelman, Biosens. Bioelectron., 2011, 26, 2751–2755 CrossRef CAS PubMed.
- M. Mujika, S. Arana, E. Castaño, M. Tijero, R. Vilares, J. M. Ruano-López, A. Cruz, L. Sainz and J. Berganza, Biosens. Bioelectron., 2009, 24, 1253–1258 CrossRef CAS PubMed.
- O. Laczka, J.-M. Maesa, N. Godino, J. del Campo, M. Fougt-Hansen, J. P. Kutter, D. Snakenborg, F.-X. Muñoz-Pascual and E. Baldrich, Biosens. Bioelectron., 2011, 26, 3633–3640 CrossRef CAS PubMed.
- A. Sandhu, Nat. Nanotechnol., 2007, 2, 746–748 CrossRef CAS PubMed.
- J. B. Haun, T.-J. Yoon, H. Lee and R. Weissleder, Wiley Interdiscip. Rev.: Nanomed. Nanobiotechnol., 2010, 2, 291–304 CrossRef CAS PubMed.
- G. Kokkinis, F. Keplinger and I. Giouroudi, Biomicrofluidics, 2013, 7, 054117 CrossRef CAS PubMed.
- M. Y. Okiishi, B. Munson and D. Young, Fundamentals of Fluid Mechanics, 2006 Search PubMed.
- G. R. Bartlett, J. Biol. Chem., 1959, 234, 466–468 CAS.
- J. W. Jorgenson and K. D. Lukacs, Science, 1983, 222, 4621 Search PubMed.
- B. Bhushan, Tribol. Int., 1995, 28, 85–96 CrossRef CAS.
- M. Suwa and H. Watarai, Anal. Chim. Acta, 2011, 690, 137–147 CrossRef CAS PubMed.
- R. Ganguly and I. K. Puri, Wiley Interdiscip. Rev.: Nanomed. Nanobiotechnol., 2010, 2, 382–399 CrossRef CAS PubMed.
- N. Pamme, Lab Chip, 2006, 6, 24–38 RSC.
- T. Hung, D. Kim, B. Rao and C. Kim, in State of the Art in Biosensors – General Aspects, 2013, pp. 197–239 Search PubMed.
- K. N. Tu, J. Appl. Phys., 2003, 94, 5451 CrossRef CAS.
- S. S. Shevkoplyas, A. C. Siegel, R. M. Westervelt, M. G. Prentiss and G. M. Whitesides, Lab Chip, 2007, 7, 1294–1302 RSC.
- E. Loth, Powder Technol., 2008, 182, 342–353 CrossRef CAS.
- E. Meyer, Nanoscience: friction and rheology on the nanometer scale, World Scientific, 1998 Search PubMed.
- J. Van Alsten and S. Granick, Phys. Rev. Lett., 1988, 61, 2570 CrossRef CAS PubMed.
- R. W. Carpick and M. Salmeron, Chem. Rev., 1997, 97, 1163–1194 CrossRef CAS PubMed.
- B. Bhushan, J. N. Israelachvili and U. Landman, Nature, 1995, 374, 607–616 CrossRef CAS.
- S. Granick, Science, 1991, 253, 1374–1379 CrossRef CAS PubMed.
- M. L. Gee, P. M. McGuiggan, J. N. Israelachvili and A. M. Homola, J. Chem. Phys., 1990, 93, 1895–1906 CrossRef CAS.
- M. Ruths and J. N. Israelachvili, Nanotribology and Nanomechanics II, ed. B. Bhushan, Springer Berlin Heidelberg, Berlin, Heidelberg, 2011 Search PubMed.
- B. V. Derjaguin, Acta Physicochim. USSR, 1941, 14, 633–662 Search PubMed.
- E. J. W. Verwey and J. T. G. Overbeek, Theory of the stability of lyophobic colloids, Courier Corporation, 1999 Search PubMed.
- J. N. Israelachvili, Intermolecular and surface forces: revised third edition, Academic Press, 2011 Search PubMed.
- V. Gehanno, P. P. Freitas, A. Veloso, J. Ferrira, B. Almeida, J. B. Soasa, A. Kling, J. C. Soares and M. F. da Silva, IEEE Trans. Magn., 1999, 35, 4361–4367 CrossRef CAS.
- A. Ebner, P. Hinterdorfer and H. J. Gruber, Ultramicroscopy, 2007, 107, 922–927 CrossRef CAS PubMed.
- L. Wildling, B. Unterauer, R. Zhu, A. Rupprecht, T. Haselgrübler, C. Rankl, A. Ebner, D. Vater, P. Pollheimer, E. E. Pohl, P. Hinterdorfer and H. J. Gruber, Bioconjugate Chem., 2011, 22, 1239–1248 CrossRef CAS PubMed.
- R. Levy and M. Maaloum, Nanotechnology, 2002, 13, 33 CrossRef.
- R. W. Stark, T. Drobek and W. M. Heckl, Ultramicroscopy, 2001, 86, 207–215 CrossRef CAS PubMed.
- M.-S. Kim, J.-H. Choi, J.-H. Kim and Y.-K. Park, Measurement, 2010, 43, 520–526 CrossRef.
- J. L. Hutter and J. Bechhoefer, Rev. Sci. Instrum., 1993, 64, 1868–1873 CrossRef CAS.
- T. Huesgen, G. Lenk, B. Albrecht, P. Vulto, T. Lemke and P. Woias, Sens. Actuators, A, 2010, 162, 137–144 CrossRef CAS.
- D. Puchberger-Enengl, S. van den Driesche, C. Krutzler, F. Keplinger and M. J. Vellekoop, Biomicrofluidics, 2015, 9, 14127 Search PubMed.
- ThermoFisher Scientific, Dynabeads® M-280 Streptavidin, https://www.thermofisher.com/order/catalog/product/11205D, 2015 Search PubMed.
- F. Li and J. Kosel, IEEE Trans. Magn., 2013, 49, 3492–3495 CrossRef.
- G. Kokkinis, M. Jamalieh, F. Cardoso, S. Cardoso, F. Keplinger and I. Giouroudi, J. Appl. Phys., 2015, 117, 17B731 CrossRef.
- G. Hermanson, Bioconjugate techniques, 1996 Search PubMed.
- P. M. Griffin and R. V. Tauxe, Epidemiol. Rev., 1991, 13, 60–98 CAS.
- Q. Zhao, J. Qian, Q. An and B. Du, J. Mater. Chem., 2009, 19, 8448 RSC.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c6lc00008h |
| This journal is © The Royal Society of Chemistry 2016 |