Open Access Article
Open Access ArticleChemical communication between bacteria and cell-free gene expression systems within linear chains of emulsion droplets†
M.
Schwarz-Schilling‡
a,
L.
Aufinger‡
a,
A.
Mückl
a and
F. C.
Simmel
*ab
aTechnical University of Munich, Physics Department E14 and ZNN/WSI, Am Coulombwall 4a, 85748 Garching, Germany. E-mail: simmel@tum.de
bNanosystems Initiative Munich, Schellingst. 4, 80539 München, Germany
First published on 12th January 2016
Abstract
Position-dependent gene expression in gradients of morphogens is one of the key processes involved in cellular differentiation during development. Here, we study a simple artificial differentiation process, which is based on the diffusion of genetic inducers within one-dimensional arrangements of 50 μm large water-in-oil droplets. The droplets are filled with either bacteria or cell-free gene expression systems, both equipped with genetic constructs that produce inducers or respond to them via expression of a fluorescent protein. We quantitatively study the coupled diffusion-gene expression process and demonstrate that gene expression can be made position-dependent both within bacteria-containing and cell-free droplets. By generating diffusing quorum sensing signals in situ, we also establish communication between artificial cell-free sender cells and bacterial receivers, and vice versa.
Insight, innovation, integrationThe creation of synthetic multicellular structures composed of artificial and chemical cells, which have the ability to respond to their environment is a topic of increasing interest in synthetic biology. Such systems could be used to create soft, adaptive structures and materials that differentiate in response to environmental cues or morphogens. Here we demonstrate a simple form of spatial differentiation within linear chains of emulsion droplets that are filled either with bacteria or with cell-free gene expression systems. Confinement of genetic inducers to diffuse in only one dimension enables strong coupling of neighboring droplet cells. As an application, we establish chemical communication between cell-free systems and bacteria acting as senders and receivers, and vice versa. |
Introduction
Over the past decade, the investigation of bacterial growth, gene expression or population dynamics in artificially structured microenvironments have become increasingly popular,1 as they allow researchers to follow the dynamics of individual cells within a population over time, and also to precisely control their spatial, temporal, and chemical boundary conditions. In this context, the combination of fluorescence microscopy methods and microfluidic techniques has been shown to be particularly versatile.2 Microfabricated bacterial traps have been applied to monitor gene expression dynamics over extended periods of time,3,4 and study, e.g. stochastic effects in gene expression,5–7 or the synchronization of bacterial oscillators.8 Microfluidic environments have also been used to study the growth of bacteria9,10 or perform evolutionary experiments.11,12 Other experiments were designed to control bacterial shape,13 or study biofilm formation and bacterial adhesion properties.14 Microfluidic techniques have been also frequently used for compartmentalization studies, in which single or small numbers of cells were isolated from their environment and also from each other. This has been recently utilized in single cell genomic studies,15 where the genomic content of a single cell is amplified directly within the compartment using droplet PCR.16,17In previous work, we used picolitre-sized water-in-oil emulsion droplets to compartmentalize small bacterial consortia and study their response to small diffusible inducer molecules.18 This was based on the finding that inducers such as IPTG or the quorum sensing signal N-(3-oxo-hexanoyl)-L-homoserine lactone (abbreviated 3-oxo-C6-HSL or simply AHL) could permeate through the separating oil phase and thus diffuse from one microcompartment into another. In this system, engineered “sender bacteria” in one droplet could communicate with “receiver bacteria” in neighbouring compartments.19,20 Gene expression in bacteria equipped with a genetic “AND gate” was switched on only in the presence of both IPTG and AHL containing reservoir droplets.
Compartmentalized bacterial consortia are of particular interest for applications in synthetic biology. For instance, compartmentalization could be used to create systems, in which several bacterial species interact and cooperate, which might be incompatible in co-culture (due to different growth conditions or predation of one species on the other). Furthermore, hybrid systems could be created, in which some droplets contain cell-free gene expression systems, while others contain bacteria, or simply nutrients or other chemicals. One vision for such systems could be the creation of a semi-synthetic super-organism composed of spatially arranged “droplet cells”, which have the ability of position-dependent gene expression, and thus spatial differentiation and pattern formation.21–23
In the present work we study spatially distributed gene expression in strictly linear arrangements of microdroplet compartments. A quasi one-dimensional geometry allows for a better control of boundary conditions, and also facilitates a straightforward analysis of the experiments. One of the most important aspects for our present study is the stronger mutual coupling of neighbouring compartments in a 1D geometry.
We not only studied microdroplets containing bacteria, but also emulsion droplets filled with cell-free gene expression systems24–29 as well as hybrid systems. In experiments with compartmentalized bacteria, gene expression dynamics is strongly affected by bacterial cell growth in the droplets. By contrast, cell-free systems display different gene expression dynamics,30 as the gene products are not diluted by cell growth. An obvious advantage of cell-free systems is the presence of the whole transcription and translation machinery and the simultaneous lack of an own genetic agenda. Cell-free systems can thus be programmed by simply feeding synthetic DNA without interference with an existing genetic background.24,26,31 The scope of cell-free systems can be easily expanded by the addition of supplementary components such as enzymes and their substrates, crowding agents,32 or other chemicals. Encapsulation of cell-free systems thus results in flexible biochemical compartments, which inherit some of their characteristics from bacteria. We demonstrate that in the context of our linear microdroplet geometries, cell-free and bacterial systems can be even made to communicate with each other.
Results and discussion
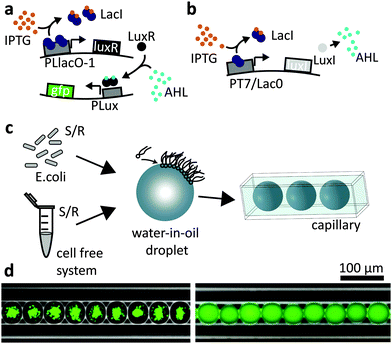
Our experiments are based on two simple synthetic gene circuits. As explained in Fig. 1a, expression of green fluorescent protein (GFPmut3*) is activated only in the presence of both inducer molecules IPTG and AHL. In this respect, the gene construct on the reporter plasmid approximates the function of a logical AND gate. The second gene construct is termed “sender” since it can synthesize AHL in the presence of IPTG as shown in Fig. 1b. Operating the AND gate circuit within gradients of inducer molecules can be used to generate spatially differentiated gene expression. In order to study this effect, we produced picolitre-sized emulsion droplets (diameter d ≈ 40–50 μm), containing either the cell free transcription/translation mix or bacteria using a microfluidic droplet generation system (see Experimental section). In order to create a linear arrangement of such compartments, the droplets were loaded into a squared capillary with a side length of 50 μm (Fig. 1c and d). Hence, diffusion of inducer molecules was effectively confined to the dimension along the long axis of the capillary.Inducer response in the cell free system
We first studied the bulk response of the AND gate plasmid in the cell-free system to the inducer molecules IPTG and AHL in bulk in titration experiments (for the characterization in E. coli cf.ref. 18). To this end, we fitted the maximum rate of GFP expression as a function of the inducer concentrations with a Hill curve. We found that the response of the AND gate to varying concentrations of IPTG (for constant [AHL]) did not follow a typical induction behaviour. This may be caused by batch-to-batch variations of the cell-free system,33 and is further related to the fact that in the cell-free system the concentration of the lac repressor LacI relative to the plasmid concentration (7.5 nM) is not sufficient26 (Fig. S2, ESI†). By contrast, the AND gate displayed a proper, slightly cooperative response to AHL (Hill exponent n ≈ 1.8) with an induction threshold of Ka ≈ 9.9 nM (Fig. S1, ESI†). These values are within the range previously found for AHL induction in bacteria.18 In the cell-free context, further analysis therefore mainly focused on the response of the AND gate to AHL.1D droplet chains connected to an inducer reservoir
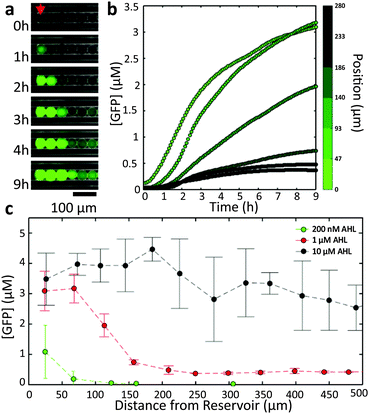
In order to characterize the diffusion of the inducers within our 1D geometry, we studied the spatial variation of gene expression in the droplet chains as a function of only one of the two inducer molecules, i.e., the AND gate droplets were only used as sensors or “receiver” droplets. The dispersed phase was first saturated with one of the inducers (either 200 nM AHL or 10 mM IPTG), and then a fixed concentration of the other inducer was established at one end of a droplet chain. This was accomplished by placing a droplet-filled capillary into a reservoir solution containing both the saturated and the second inducer molecule. The volume of the reservoir was created much larger than the interior volume of the capillary (Vreservoir/Vcapillary ≈ 103). As the diffusion coefficient of the inducer inside the aqueous reservoir is much larger than the (apparent) diffusivity in the water-in-oil emulsion (Dreservoir/Dcapillary > 10–100, cf. discussion below), the inducer concentration at the opening of the capillary can be assumed constant. Furthermore, due to the length of the capillaries (l ≥ 1 cm) and the time scale of our observations (t ≤ 15 h) one can safely assume zero inducer concentration sufficiently far inside the capillary. This will result in the formation of a gradient in the inducer concentration along the aligned droplets, which can be observed via GFP expression using an epifluorescence microscope.A representative image time series recorded from cell-free receiver droplets inside a capillary connected to a reservoir with [AHL] = 1 μM is shown in Fig. 2a. As AHL diffuses into the capillary, GFP expression is sequentially induced in the droplets, starting with the compartments closest to the reservoir. The temporal progress of GFP production as a function of distance to the reservoir (Fig. 2b) reflects the AHL dependence of GFP expression expected from the bulk experiments – larger distances correspond to lower AHL concentrations and thus slower expression kinetics. As the encapsulated cell-free system has a limited run-time of ≈5–6 hours, droplets at a distance too far to be reached by AHL within this time were not induced at all.
In fact, the number of droplets which are induced within a given time span can be simply controlled by the inducer concentration in the reservoir (Fig. 2c). Larger concentrations lead to larger diffusion fluxes, resulting in the induction of an increasing number of droplets. For large fluxes, however, the spatial differentiation of neighbouring droplets is less pronounced (Fig. S6, ESI†).
We found similar behaviour as in Fig. 2 also for linear chains of droplets filled with bacterial AHL receivers (Fig. S7, ESI†), but with slightly different dynamics, mainly due to bacterial growth within the droplets (cf.ref. 18).
Experiments with IPTG reservoirs of varying concentrations generally did not lead to a clear spatial differentiation (see Fig. S8, ESI† for [IPTG] = 10 mM), which presumably is caused by the leakiness of the lac promoter and the lack of sufficient LacI for the cell-free system (cf. Fig. S2, ESI†).
Quantitative considerations
For programmed pattern formation, it is of interest to tune the apparent diffusion coefficient of the “morphogens” and thus the corresponding patterning length scale. To this end a quantitative understanding of the diffusion properties of AHL within a heterogeneous medium such as a water-in-oil emulsion is necessary. We used the GFP expression strength in cell-free droplets as reporter for the local AHL concentration and estimated its apparent diffusion coefficient Da. Surprisingly, for the reservoirs with [AHL] = 200 nM and [AHL] = 1 μM we obtain values around Da ≈ 0.1 μm2 s−1, while for the reservoir with [AHL] = 10 μM we found a higher Da of about ≈25 μm2 s−1 (Fig. S3 and S4, ESI†). This discrepancy indicates that gene activation proceeds through different mechanisms for low and high reservoir concentrations.In general, transport of molecules within a water-in-oil emulsion based on our specific surfactant and fluorocarbon oil is dominated by partitioning effects (the droplets are not expected to form adhesive bilayers at their interface,34 for a discussion see ESI†). Thus, inducer molecules partition from the reservoir into the oil phase, diffuse through the oil and then enter droplets in the capillaries. An alternative mechanism involves diffusive transport of inducer via surfactant micelles.35 Whereas the diffusion coefficient through oil is expected to be reduced compared to the aqueous phase (Daqueous ≈ 100–1000 μm2 s−1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 36–38) by a factor of ≈4 (due to its higher viscosity), diffusion via micelles has been measured to proceed with a diffusion coefficient of Dmicelle ≈ 1 μm2 s−1.35
36–38) by a factor of ≈4 (due to its higher viscosity), diffusion via micelles has been measured to proceed with a diffusion coefficient of Dmicelle ≈ 1 μm2 s−1.35
Apparently, for the lower reservoir concentrations ([AHL] = 200 nM, 1 μM) droplet induction is dominated by micelle diffusion. By contrast, for the [AHL] = 10 μM reservoir, a large enough amount of AHL partitions into the oil phase to elicit a “fast” and more global response in the capillaries (for a discussion see ESI†).
It is also important to consider effects, which are caused by the geometry of our experimental set-up. The linear arrangement of droplets in glass capillaries confines the inducer molecules to effectively diffuse in 1D and thus enforces their sequential diffusion through neighbouring droplets. In the absence of a bulk phase – acting as diffusion sink – this creates a relatively strong coupling between neighbouring droplets compared to other geometries. For instance, in a close-packed geometry an inducer-filled droplet is surrounded by two nearest neighbours in 1D, while it has six and even twelve neighbours in 2D and 3D, respectively, which would result in a correspondingly faster dilution. We can thus observe spatial effects over much longer distances in 1D than in our previous work based on a 2D arrangement of droplets in contact with a bulk oil phase.18
Inducer reservoir droplets
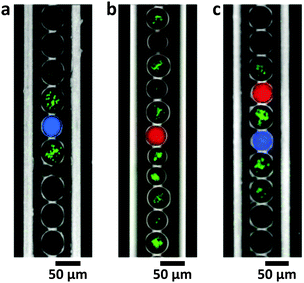
In the following set of experiments, we localized GFP expression by introducing inducer-filled reservoir droplets rather than using a macroscopic reservoir. To this end, one inducer was distributed globally to all droplets, while the second inducer was encapsulated only in dedicated droplets. In the presence of AHL sender droplets ([AHL] = 200 nM), GFP expression was induced only in nearest neighbour droplets containing bacteria and IPTG (Fig. 3a). In stark contrast, in the case of IPTG reservoir droplets ([IPTG] = 10 mM) GFP expression in droplets containing AND gate bacteria with saturating AHL concentration was not restricted to neighbouring droplets (Fig. 3b). However, in the absence of IPTG reservoir droplets in a capillary, no GFP production is observed at all. Using a combination of AHL and IPTG sender droplets, it was possible to spatially confine gene expression only to AND gate bacteria close to the reservoir droplets (Fig. 3c).Communication between bacteria and artificial cellular compartments
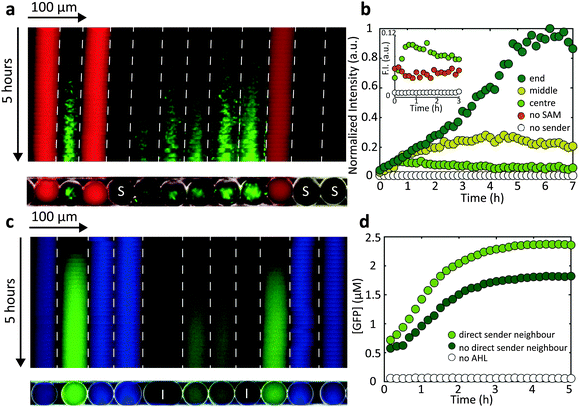
We finally studied communication between the cell-free and bacterial droplets. As shown in Fig. 4a, cell-free sender droplets equipped with a LuxI encoding sender plasmid generate enough AHL to elicit a gene expression response in neighbouring droplets containing AND gate bacteria. To this end, the cell-free medium actually had to be supplemented with S-adenosyl methionine (SAM), which is a precursor for AHL not contained in sufficient quantities in the conventional cell extract. In order to further increase the “sender strength” of the cell-free droplets, additional T7 RNA polymerase was added, which resulted in an enhanced production of LuxI (Fig. 4b and Fig. S1b, ESI†). The kymograph representation of Fig. 4a indicates that – as expected – the response is stronger and starts earlier in droplets in the direct vicinity of the cell free sender droplets.We also found, that the intensity of the GFP signal was higher towards the end of the capillary compared with the centre (Fig. 4b). We assume that the growth of bacteria within the emulsion droplets is actually limited by the availability of oxygen. Since oxygen can dissolve and transfer through fluorinated oil, bacterial growth is prolonged towards the end of the capillaries, resulting in a a stronger overall GFP production. This effect was not observed for cell-free gene expression in droplets.
The direction of communication can also be reversed by encapsulating LuxI producing sender bacteria that generate AHL and thus induce gene expression in cell-free receiver droplets (Fig. 4c). For “bacterial-to-cell-free” droplet communication, a clear difference between gene expression in nearest neighbour receivers and receivers more remote from the sender droplets could be resolved (Fig. 4d).
Experimental
Preparation of the cell-free gene expression system
Crude S30 cell extract was obtained by beat beating a BL21-Rosetta2(DE3) mid-log phase culture with 0.1 mm glass beads in a Minilys device (Peqlab, Germany) as described in ref. 39. Instead of 3-phosphoglyceric acid (3-PGA), phospho-enolpyruvate (PEP) was utilized as an energy source40 in the composite buffer (50 mM Hepes pH 8, 1.5 mM ATP and GTP, 0.9 mM CTP and UTP, 0.2 mg ml−1 tRNA, 26 μM coenzyme A, 0.33 mM NAD, 0.75 mM cAMP, 68 μM folinic acid, 1 mM spermidine, 30 mM PEP, 1 mM DTT, 2% PEG-8000). All components were stored at −80 °C before thawing on ice. The composition of a single cell-free reaction was: 33% (v/v) S30 cell extract mixed with 42% (v/v) buffer and 25% (v/v) DNA plus additives. Each final reaction mix was supplemented with 13.3 mM maltose for ATP regeneration.41 300 μM of S-(5′-adenosyl)-L-methionine chloride dihydrochloride (Sigma-Aldrich, #A7007) and 1 U μl−1 of T7 polymerase (Epicentre, TM910K) was added to the cell free sender droplets. LacI was His-tagged and purified by gravity-flow chromatography with Ni-NTA Agarose Beads (Qiagen).Bacterial strains and culture media
The receiver (AND gate) plasmid was cloned into Escherichia coli DH5αZi (ExpressSys) and the sender plasmid into E.coli BL21(DE3)pLysS (Promega) as described previously.18 As culture media, Luria-Bertani (LB) medium (Carl Roth, #X968.1) and M9 minimal supplemented with 20 mM glucose and 300 μg ml−1 thiamine hydrochloride (Sigma-Aldrich, #T1270) were used. For IPTG reservoir experiments with AND gate bacteria, M9 minimal medium was supplemented with 0.4% (w/v) glycerol instead of glucose. Cells from glycerol stock were grown overnight in Falcon tubes with 5 ml LB medium containing 100 μg ml−1 carbenicillin (Carl Roth) and 30 μg ml−1 chloramphenicol (Carl Roth) at 37 °C shaken at 250 rpm. The overnight cultures were diluted to an initial optical density (OD600nm) of 0.01 in Falcon tubes containing fresh LB or M9 minimal medium (Sigma-Aldrich, #M6030) for sender or receiver cells, respectively (LB medium is autofluorescent in the GFP channel) with 50% reduced antibiotic concentrations. Before encapsulation of bacteria into emulsion droplets, the cultures were incubated for 2–3 h until an OD600nm of 0.1–0.2 was reached. Sender bacteria droplets were grown in LB medium supplemented with 1 mM IPTG, two hours before encapsulation.Microfluidics
For the generation of droplets, we utilized a microfluidic flow-focusing geometry, which was defined using soft lithographic techniques and the elastomer poldimethysiloxane (PDMS). PDMS devices were micromolded from silicon masters containing the channel structures, which were defined using the negative photoresist Epocore 20 (micro resist technology, Germany). The cured microfluidic device was bonded onto a glass cover slide after oxygen plasma treatment and baked at 200 °C for 3 hours.42 To avoid cross contamination, each droplet species was produced in a separate device. Fluid flows were generated with a pressure controller OB-1K (Elveflow, France) and appropriate PTFE-tubings (inner diameter: 0.8 mm). Pressures were set between 150–300 mbar. The droplet device was loaded with the carrier oil Fluorinert FC-40 (Sigma-Aldrich, Germany) blended with 2% (w/w) surfactant (EA, RainDance Technologies, USA) and focused with the appropriate aqueous phase. The critical micelle concentration of the surfactant is ≈0.02–0.04%.35 The reservoirs and the collection tubes for the cell free system were placed on ice during droplet production. Different species of droplets were collected separately and then mixed in a new test tube in defined volume ratios with a pipette. Filling of the capillaries with droplets is described in detail in the ESI† (Fig. S9). For capillary measurements with inducer reservoirs, the capillary ends were placed in a chamber containing the reservoir solution. The chamber was built with two stripes of melted parafilm on a glass slide, immediately covered with another glass slide. The chambers were sealed with silicone vacuum grease and nail polisher.Bulk characterization
Cell-free expression of GFP was characterized via plate reader measurements (BMG FLUOstar Optima) using 15 μl reaction volumes in 384-well plates.Microscopy
Video microscopy of droplets containing only bacteria was performed with an inverted epifluorescence microscope IX81 (Olympus, Japan) using a 10× magnification objective, an EMCCD camera (iXon3, Andor, UK), and a mercury fluorescence excitation light source (X-Cite 120Q, Excelitas Technologies, USA). The microscope was equipped with an incubator box (Okolab, Italy) to maintain a temperature of T = 37 °C. All time-lapse microscopy measurements containing the cell-free system, also in combination with bacteria, were conducted on an IX71 microscope (Olympus, Japan) using a 10× objective, a CCD camera (LucaR, Andor, UK) and a 4-wavelength fluorescence LED source (Thorlabs, USA). The samples were thermostatted at 30 °C using a heating plate (Tokai Hit Co. Ltd, Japan).Data analysis
Image analysis is performed using a customized automated droplet tracking software programmed in MATLAB.43 Extracted fluorescence and position data was further processed as described in detail in S7, ESI.†Conclusions
We have demonstrated spatially differentiated gene expression in linear chains of emulsion droplets containing either bacteria or cell-free gene expression systems. Spatial effects can be either achieved by establishing concentration gradients of genetic inducers such as AHL and IPTG via macroscopic reservoirs or local sender droplets. For low inducer concentrations, the apparent diffusion coefficient of AHL is reduced by several orders of magnitude, consistent with micelle-mediated transport between neighbouring droplets. Confinement to an effective 1D geometry leads to a stronger chemical coupling of neighbouring droplets and an extended spatial range of the inducer molecules. This also enabled us to establish communication between cell-free sender droplets and neighbouring bacterial receiver droplets and, conversely, between sender bacteria and cell-free receivers.Our experiments represent a step towards artificial multicellular hybrid systems, in which the “cells” are constituted either by encapsulated bacterial consortia, artificial cells containing cell-free gene expression systems, or supplementary cells with chemical supplies, nutrients, etc. The 1D geometry utilized in this work could prove particularly useful to achieve spatial differentiation in such systems, to generate interesting spatiotemporal effects, and thus emulate simple developmental processes in an artificial context.
Acknowledgements
We gratefully acknowledge financial support by the DFG Research Training Group GRK2062 “Molecular Principles of Synthetic Biology” and the DFG Cluster of Excellence NIM. We wish to thank Michael Heymann and Yannick Rondelez for useful experimental tips.References
- F. J. H. Hol and C. Dekker, Science, 2014, 346, 1251821 CrossRef PubMed.
- J. C. W. Locke and M. B. Elowitz, Nat. Rev. Microbiol., 2009, 7, 383–392 CrossRef CAS PubMed.
- F. K. Balagadde, L. C. You, C. L. Hansen, F. H. Arnold and S. R. Quake, Science, 2005, 309, 137–140 CrossRef CAS PubMed.
- M. R. Bennett and J. Hasty, Nat. Rev. Genet., 2009, 10, 628–638 CrossRef CAS PubMed.
- M. B. Elowitz, A. J. Levine, E. D. Siggia and P. S. Swain, Science, 2002, 297, 1183–1186 CrossRef CAS PubMed.
- I. Golding, J. Paulsson, S. Zawilski and E. Cox, Cell, 2005, 123, 1025–1036 CrossRef CAS PubMed.
- L. S. Tsimring, Rep. Prog. Phys., 2014, 77, 026601 CrossRef PubMed.
- T. Danino, O. Mondragón-Palomino, L. Tsimring and J. Hasty, Nature, 2010, 463, 326–330 CrossRef CAS PubMed.
- P. Wang, L. Robert, J. Pelletier, W. L. Dang, F. Taddei, A. Wright and S. Jun, Curr. Biol., 2010, 20, 1099–1103 CrossRef CAS PubMed.
- L. Boitard, D. Cottinet, C. Kleinschmitt, N. Bremond, J. Baudry, G. Yvert and J. Bibette, Proc. Natl. Acad. Sci. U. S. A., 2012, 109, 7181–7186 CrossRef CAS PubMed.
- J. E. Keymer, P. Galajda, C. Muldoon, S. Park and R. H. Austin, Proc. Natl. Acad. Sci. U. S. A., 2006, 103, 17290–17295 CrossRef CAS PubMed.
- J. E. Keymer, P. Galajda, G. Lambert, D. Liao and R. H. Austin, Proc. Natl. Acad. Sci. U. S. A., 2008, 105, 20269–20273 CrossRef CAS PubMed.
- J. Männik, R. Driessen, P. Galajda, J. E. Keymer and C. Dekker, Proc. Natl. Acad. Sci. U. S. A., 2009, 106, 14861–14866 CrossRef PubMed.
- K. Drescher, Y. Shen, B. L. Bassler and H. A. Stone, Proc. Natl. Acad. Sci. U. S. A., 2013, 110, 4345–4350 CrossRef CAS PubMed.
- T. Kalisky and S. Quake, Nat. Methods, 2011, 8, 311–314 CrossRef CAS PubMed.
- M. Nakano, J. Komatsu, S. Matsuura and K. Takashima, et al. , J. Biotechnol., 2003, 102, 117–124 CrossRef CAS PubMed.
- R. Williams, S. G. Peisajovich, O. J. Miller, S. Magdassi, D. S. Tawfik and A. D. Griffiths, Nat. Methods, 2006, 3, 545–550 CrossRef CAS PubMed.
- M. Weitz, A. Mückl, K. Kapsner, R. Berg, A. Meyer and F. C. Simmel, J. Am. Chem. Soc., 2014, 136, 72–75 CrossRef CAS PubMed.
- R. Weiss, T. F. Knight, A. E. Condon and G. Rozenberg, DNA Computing, 6th International Workshop on DNA-Based Computers, DNA6, 2000, 2054, 1–16.
- R. Weiss, S. Basu, S. Hooshangi, A. Kalmbach, D. Karig, R. Mehreja and I. Netravali, Nat. Comput., 2003, 2, 47–84 CrossRef.
- M. Isalan, C. Lemerle and L. Serrano, PLoS Biol., 2005, 3, 488–496 CAS.
- S. Basu, Y. Gerchman, C. H. Collins, F. H. Arnold and R. Weiss, Nature, 2005, 434, 1130–1134 CrossRef CAS PubMed.
- J. J. Tabor, H. M. Salis, Z. B. Simpson, A. A. Chevalier, A. Levskaya, E. M. Marcotte, C. A. Voigt and A. D. Ellington, Cell, 2009, 137, 1272–1281 CrossRef PubMed.
- Y. Shimizu, A. Inoue, Y. Tomari, T. Suzuki, T. Yokogawa, K. Nishikawa and T. Ueda, Nat. Biotechnol., 2001, 19, 751–755 CrossRef CAS PubMed.
- V. Noireaux, R. Bar-Ziv and A. Libchaber, Proc. Natl. Acad. Sci. U. S. A., 2003, 100, 12672–12677 CrossRef CAS PubMed.
- J. Shin and V. Noireaux, ACS Synth. Biol., 2012, 1, 29–41 CrossRef CAS PubMed.
- Z. Z. Sun, C. A. Hayes, J. Shin, F. Caschera, R. M. Murray and V. Noireaux, J. Visualized Exp., 2013, 1–15, DOI:10.3791/50762.
- E. Sokolova, E. Spruijt, M. M. K. Hansen, E. Dubuc, J. Groen, V. Chokkalingam, A. Piruska, H. A. Heus and W. T. S. Huck, Proc. Natl. Acad. Sci. U. S. A., 2013, 110, 11692–11697 CrossRef CAS PubMed.
- P. Torre, C. D. Keating and S. S. Mansy, Langmuir, 2014, 30, 5695–5699 CrossRef CAS PubMed.
- H. Niederholtmeyer, V. Stepanova and S. J. Maerkl, Proc. Natl. Acad. Sci. U. S. A., 2013, 110, 15985–15990 CrossRef CAS PubMed.
- D. C. Harris and M. C. Jewett, Curr. Opin. Biotechnol., 2012, 23, 672–678 CrossRef CAS PubMed.
- C. Tan, S. Saurabh, M. P. Bruchez, R. Schwartz and P. Leduc, Nat. Nanotechnol., 2013, 8, 602–608 CrossRef CAS PubMed.
- M. K. Takahashi, J. Chappell, C. A. Hayes, Z. Z. Sun, J. Kim, V. Singhal, K. J. Spring, S. Al-Khabouri, C. P. Fall, V. Noireaux, R. M. Murray and J. B. Lucks, ACS Synth. Biol., 2015, 4, 503–515 CrossRef CAS PubMed.
- A. R. Thiam, N. Bremond and J. Bibette, Langmuir, 2012, 28, 6291–6298 CrossRef CAS PubMed.
- Y. Skhiri, P. Gruner, B. Semin, Q. Brosseau, D. Pekin, L. Mazutis, V. Goust, F. Kleinschmidt, A. El Harrak, J. B. Hutchison, E. Mayot, J.-F. Bartolo, A. D. Griffiths, V. Taly and J.-C. Baret, Soft Matter, 2012, 8, 10618 RSC.
- W. S. Choi, D. Ha, S. Park and T. Kim, Biomaterials, 2011, 32, 2500–2507 CrossRef CAS PubMed.
- G. Dilanji, J. Langebrake, P. De Leenheer and S. Hagen, J. Am. Chem. Soc., 2012, 134, 5618–5626 CrossRef CAS PubMed.
- B. A. Hense, J. Müller, C. Kuttler and A. Hartmann, Sensors, 2012, 12, 4156–4171 CrossRef PubMed.
- Z. Z. Sun, C. A. Hayes, J. Shin, F. Caschera, R. M. Murray and V. Noireaux, J. Visualized Exp., 2013, e50762, DOI:10.3791/50762.
- D.-M. Kim and J. R. Swartz, Biotechnol. Bioeng., 2001, 74, 309–316 CrossRef CAS PubMed.
- F. Caschera and V. Noireaux, Biochimie, 2014, 99, 162–168 CrossRef CAS PubMed.
- S. Kaneda, K. Ono, T. Fukuba, T. Nojima, T. Yamamoto and T. Fujii, Anal. Sci., 2012, 28, 39 CrossRef CAS PubMed.
- K. Kapsner and F. C. Simmel, ACS Synth. Biol., 2015, 4, 1136–1143 CrossRef CAS PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available: Supplementary methods, data, and modeling. See DOI: 10.1039/c5ib00301f |
| ‡ Authors have contributed equally to this work. |
| This journal is © The Royal Society of Chemistry 2016 |