Open Access Article
Open Access ArticleA chemical–biological similarity-based grouping of complex substances as a prototype approach for evaluating chemical alternatives†
Fabian A.
Grimm
a,
Yasuhiro
Iwata
a,
Oksana
Sirenko
b,
Grace A.
Chappell
a,
Fred A.
Wright
c,
David M.
Reif
c,
John
Braisted
d,
David L.
Gerhold
d,
Joanne M.
Yeakley
e,
Peter
Shepard
e,
Bruce
Seligmann
e,
Tim
Roy
f,
Peter J.
Boogaard
g,
Hans B.
Ketelslegers
h,
Arlean M.
Rohde
i and
Ivan
Rusyn
*a
aDepartment of Veterinary Integrative Biosciences, Texas A&M University, College Station, TX, USA. E-mail: irusyn@cvm.tamu.edu; Fax: +(979) 458-9866; Tel: +(979) 458-9866
bMolecular Devices LLC, Sunnyvale, CA, USA
cBioinformatics Research Center, North Carolina State University, Raleigh, NC, USA
dNational Institutes of Health, National Center for Advancing Translational Sciences, Bethesda, MD, USA
eBioSpyder Technologies Inc., Carlsbad, CA, USA
fDepartment of Natural Science, University of South Carolina, Beaufort, SC, USA
gSHELL International BV, The Hague, NL, USA
hEuropean Petroleum Refiners Association, Concawe Division, Brussels, BE, USA
iExxonMobil Chemical Co., Spring, TX, USA
First published on 16th May 2016
Abstract
Comparative assessment of potential human health impacts is a critical step in evaluating both chemical alternatives and existing products on the market. Most alternatives assessments are conducted on a chemical-by-chemical basis and it is seldom acknowledged that humans are exposed to complex products, not individual substances. Indeed, substances of Unknown or Variable composition, Complex reaction products, and Biological materials (UVCBs) are ubiquitous in commerce yet they present a major challenge for registration and health assessments. Here, we present a comprehensive experimental and computational approach to categorize UVCBs according to global similarities in their bioactivity using a suite of in vitro models. We used petroleum substances, an important group of UVCBs which are grouped for regulatory approval and read-across primarily on physico-chemical properties and the manufacturing process, and only partially based on toxicity data, as a case study. We exposed induced pluripotent stem cell-derived cardiomyocytes and hepatocytes to DMSO-soluble extracts of 21 petroleum substances from five product groups. Concentration-response data from high-content imaging in cardiomyocytes and hepatocytes, as well as targeted high-throughput transcriptomic analysis of the hepatocytes, revealed distinct groups of petroleum substances. Data integration showed that bioactivity profiling affords clustering of petroleum substances in a manner similar to the manufacturing process-based categories. Moreover, we observed a high degree of correlation between bioactivity profiles and physico-chemical properties, as well as improved groupings when chemical and biological data were combined. Altogether, we demonstrate how novel in vitro screening approaches can be effectively utilized in combination with physico-chemical characteristics to group complex substances and enable read-across. This approach allows for rapid and scientifically-informed evaluation of health impacts of both existing substances and their chemical alternatives.
Introduction
Comparative analysis of potential human health effects and physicochemical properties, combined with valuation of exposure scenarios, environmental impacts and other factors, is a critical step in evaluating the safety of both existing products and potential chemical alternatives. However, most complex substances and chemical alternatives lack traditional animal study-derived data that can be used to comprehensively evaluate their safety. Recent National Research Council (NRC) report1 “A Framework to Guide Selection of Chemical Alternatives” argued for the transition towards using data from novel high throughput and in silico approaches and posited that “it is critical that the scientific community embrace the challenge and advantages of using novel data streams in the alternatives assessment process.”In response to the NRC's call for developing principles and tools that support the benchmarking and integration of high-throughput data on chemical effects, especially in the context of different regulatory requirements, we pursued a case study of very complex high-production volume (HPV) substances that fall into a broad category of Unknown or Variable composition, Complex reaction products and Biological materials (UVCB). Petroleum products are a prototypical example of UVCB. Given their chemical complexity and multi-constituent nature, which is further complicated by batch- or manufacturer-dependent compositional variations, UVCBs continue to represent a major challenge to regulatory agencies and registrants alike.2,3 All petroleum products were registered in the European Union for the 2010 submission deadline under REACH (≥1000 tonnes registration band), comprising more than 8000 individual registrations.2 A number of these submissions were accompanied by testing proposals to fill data gaps in specific toxicity endpoints.4 To minimize the need for testing in vertebrate animals, the majority of data gaps were addressed by using read-across to similar substances for which the required data were available.
Read-across of petroleum substances within the REACH framework is typically done by grouping the individual substances into product categories with similar manufacturing processes, physico-chemical (including refining history and boiling point/carbon number ranges) and chemical properties (such as hydrocarbon classes).3,4 Several recent studies have shown that analytical–chemical properties, specifically the aromatic ring class (ARC) profiles of certain high-boiling petroleum substances, correlate well with reproductive and developmental toxicity and mutagenicity of petroleum substances.5–7 However, category read-across approaches for UVCBs that are based solely on such broad similarity parameters may not always be considered sufficient. Adding a biological data-based read-across component,8i.e. categorizing substances according to similarity in their biological responses in addition to the physico-chemical and manufacturing characteristics, may represent an enhanced strategy and provide complementary experimental evidence to support distinctive product categories for petroleum substances.3,4
Recent advances in in vitro high-content screening (HCS) technologies have improved their potential for multidimensional bioactivity profiling in a rapid and relatively cost-efficient way.9–12 Importantly, HCS can be used in conjunction with induced pluripotent stem cell (iPSC)-derived organotypic cell culture models, including iPSC-derived cardiomyocytes and hepatocytes. Such iPSCs derived from non-embryonic human stem cells are a particularly attractive and physiologically relevant in vitro model that mimics and maintains the phenotypic characteristics of their respective somatic counterparts.13,14
Collectively, the need for increased confidence in read-across of complex UVCBs and the advantages afforded by novel in vitro model systems and high-dimensional bioactivity data readouts create the opportunity for the biological data-assisted categorization of UVCBs. Thus, we hypothesized that modern bioactivity profiling may be used to support categorization and read-across of UVCBs using a case study of complex petroleum substances. Herein, we describe a comprehensive experimental and computational approach based on HCS screening of 21 petroleum substances from five distinct product groups and use these data to categorize them into groups for read-across. In particular, we determined bioactivity-based concentration-response profiles for these substances using multidimensional HCS of iPSC derived cardiomyocytes and hepatocytes. Concentration-response profiling allowed derivation of quantitative estimates of bioactivity for each parameter, data that were integrated and visualized into aggregate bioactivity profiles using ToxPi approach.9,15 Similarities in bioactivity profiles were then used for biological and chemical–biological data-integrative groupings of substances, an approach that allows for rapid and scientifically-informed evaluation of health impacts of both existing substances and their chemical alternatives.
Experimental
Chemicals and biologicals
iCell cardiomyocytes (Catalogue #: CMC-100-010-001) and hepatocytes (Catalogue #: PHC-100-020-001), including their respective plating and maintenance media were obtained from Cellular Dynamics International (Madison, WI). EarlyTox Cardiotoxicity kits were purchased from Molecular Devices LLC (Sunnyvale, CA). Reference standard compounds (isoproterenol, sotalol, and propranolol) were included in these kits. Hank's Balanced Salt Solution, RPMI 1640 medium, B-27 medium supplement, gentamicin (50 mg ml−1), penicillin/streptomycin solution, Hoechst 33342, and MitoTracker Orange CMTMRos reagent were all purchased from Life Technologies (Grand Island, NY). Cisapride monohydrate, tetraoctyl ammonium bromide, and formaldehyde solution were purchased from Sigma-Aldrich (St. Louis, MO). Dimethyl sulfoxide (DMSO), dexamethazone, hydrogen peroxide (3%), and recombinant oncostatin M were obtained from Fisher Scientific (Waltham, MA).Sample preparation
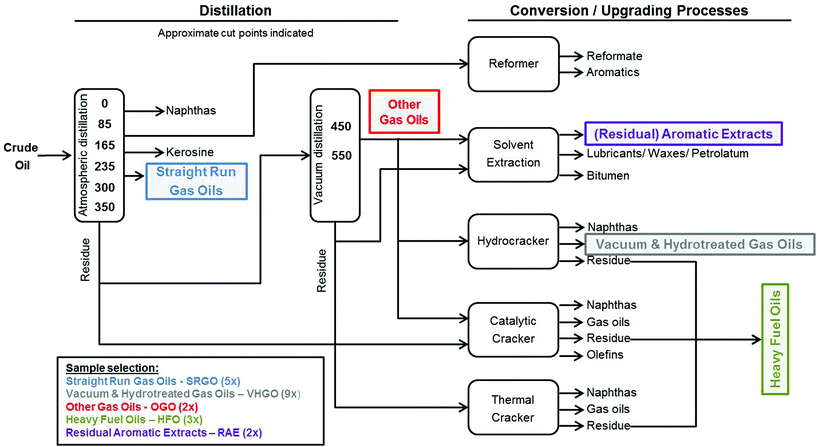
DMSO-soluble extracts of petroleum substances from five distinct product categories (SRGO – Straight Run Gas Oils, OGO – Other Gas Oils, VHGO – Vacuum & Hydrotreated Gas Oils, RAE – Residual Aromatic Extracts, and HFO – Heavy Fuel Oils) were provided by Concawe (Brussels, Belgium) (Fig. 1, Table 1). Samples were prepared using previously published extraction procedures for routine isolation of complex polycyclic aromatic compounds (PAC) in petroleum substances.16,17 The DMSO extraction procedure used herein is designed to concentrate the ‘biologically active’ fraction (i.e., aromatics) of the refinery streams; the extracts obtained using this method are used routinely for safety testing (e.g., mutagenicity) and chemical characterization of the refinery streams.18 Briefly, 4 grams of each petroleum substance was dissolved in 10 ml of cyclohexane, 10 ml of DMSO was added and the mixture was vigorously shaken for several minutes. The DMSO layer was removed and the cyclohexane was re-extracted with an additional 10 ml of DMSO. Both PAC-enriched DMSO layers were combined and diluted 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 with two volumes of 4% (w/v) sodium chloride solution. Following subsequent extraction with 20 ml and 10 ml cyclohexane to isolate the PAC fraction, the organic layers were washed twice with distilled water and filtered through anhydrous sodium sulfate. The solvent was evaporated to almost dryness (rotary evaporator at 15–20 Torr) at 40 °C, followed by final evaporation at 80 °C for 30 min. Prior to in vitro experiments dried PAC extracts were weighed and solubilized in up to 6 ml DMSO.
1 with two volumes of 4% (w/v) sodium chloride solution. Following subsequent extraction with 20 ml and 10 ml cyclohexane to isolate the PAC fraction, the organic layers were washed twice with distilled water and filtered through anhydrous sodium sulfate. The solvent was evaporated to almost dryness (rotary evaporator at 15–20 Torr) at 40 °C, followed by final evaporation at 80 °C for 30 min. Prior to in vitro experiments dried PAC extracts were weighed and solubilized in up to 6 ml DMSO.
| # | Sample | Category | CAS | ToxPi (biological) | ToxPi (volatility) | ToxPi (chem-biol) |
|---|---|---|---|---|---|---|
| a Straight run gas oil. b Other gas oil. c Vacuum & hydrotreated gas oil. d Residual aromatic extract. e Heavy fuel oil. | ||||||
| 1 | CON-01 | SRGOa | 64741-43-1 | 9.0 | 9.0 | 2.8 |
| 2 | CON-02 | SRGOa | 68814-87-9 | 8.2 | 9.6 | 2.7 |
| 3 | CON-03 | SRGOa | 68814-87-9 | 5.9 | 10.1 | 2.1 |
| 4 | CON-04 | SRGOa | 68915-96-8 | 9.5 | 8.3 | 2.7 |
| 5 | CON-05 | SRGOa | 64741-43-1 | 7.9 | 8.6 | 2.4 |
| 6 | CON-07 | OGOb | 64742-46-7 | 4.1 | 11.1 | 1.8 |
| 7 | CON-09 | OGOb | 64742-80-9 | 5.6 | 10.5 | 2.0 |
| 8 | CON-12 | VHGOc | 64741-49-7 | 10.8 | 9.5 | 3.2 |
| 9 | CON-13 | VHGOc | 64741-58-8 | 11.5 | 7.8 | 3.0 |
| 10 | CON-14 | VHGOc | 64741-77-1 | 8.7 | 9.7 | 2.7 |
| 11 | CON-15 | VHGOc | 64742-87-6 | 9.9 | 9.6 | 3.1 |
| 12 | CON-16i | VHGOc | 68334-30-5 | 8.8 | 12.7 | 2.9 |
| 13 | CON-16ii | VHGOc | 68334-30-5 | 7.9 | 10.6 | 2.8 |
| 14 | CON-17 | VHGOc | 68476-30-2 | 6.3 | 11.3 | 2.5 |
| 15 | CON-18 | VHGOc | 68476-31-3 | 10.2 | 8.5 | 3.2 |
| 16 | CON-20 | VHGOc | 92045-24-4 | 10.5 | 8.1 | 2.8 |
| 17 | CON-26 | RAEd | 64742-10-5 | 1.2 | 4.3 | 0.4 |
| 18 | CON-27 | RAEd | 91995-70-9 | 2.7 | 4.4 | 0.7 |
| 19 | A083/13 | HFOe | 68476-33-5 | 6.6 | — | 1.3 |
| 20 | A087/13 | HFOe | 68476-33-5 | 3.6 | — | 0.8 |
| 21 | A092/13 | HFOe | 68476-33-5 | 5.4 | — | 1.2 |
Cells were exposed to the extracts in descending logarithmic order of concentrations (100%, 10%, 1%, 0.1%, 0.01% and 0.001% dilutions of the DMSO-solubilized dried extracts from each petroleum substance). Serial dilutions were originally prepared in 100% cell-culture grade DMSO and then further diluted fifty-fold in corresponding cell culture media to yield 5× working solutions in 5% DMSO. The final concentration of DMSO in assay wells following addition of test substances was 1% (v/v), an amount of DMSO consistent with previous reports which by itself had no effect on cardiomyocyte- or hepatocyte-derived phenotypes.12
Cell culture
iPSC cardiomyocytes and hepatocytes were cultured according to the manufacturer's (Cellular Dynamics International, Madison, WI) recommendations and as described in more detail elsewhere.9,10,12,19 Briefly, frozen vials of cardiomyocytes were thawed for 4 min at 37 °C before cells were resuspended in 10 ml plating medium. Cell density was determined by microscopic analysis using disposable hemocytometers and the concentration of the suspension was adjusted to yield a target cell density of 2 × 105 viable cells per ml. Cells were seeded at a density of 5000 cells per well in pre-gelatinized (2 hours at 37 °C with 25 μl 0.1% gelatin in water) 384-well plates and rested for 30 min at room temperature before being incubated at 37 °C and 5% CO2. After 48 hours, the plating medium was replaced with 30 μl per well of fresh maintenance medium containing 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 500 penicillin/streptomycin solution. After approximately five days in culture, cardiomyocyte-specific synchronous contractions were observable. Cells were maintained in culture for a total of 10 days before experiments were conducted.
500 penicillin/streptomycin solution. After approximately five days in culture, cardiomyocyte-specific synchronous contractions were observable. Cells were maintained in culture for a total of 10 days before experiments were conducted.
Similarly, vials of hepatocytes were thawed for 3 min at 37 °C in a water bath and subsequently resuspended in RPMI medium containing 2% (v/v) iCell hepatocyte medium supplement, 0.1 μM dexamethasone, 2% (v/v) B27 supplement, 25 μg per ml Gentamicin, and 20 ng ml−1 Oncostatin-M. Following microscopic evaluation of the cell density, the suspension was further diluted to a final concentration of 1 × 106 cells per ml. 25 μl of this suspension was then added to each sample well on collagen I coated 384-well plates, yielding a final cell density of 25![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 cells per well. Plates were initially kept at room temperature for 30 min and then transferred to an incubator set at 37 °C and 5% CO2. After four hours of incubation, the plating medium was replaced with 25 μl fresh medium, a step that was repeated daily for four days. On day five, the plating medium was exchanged with 25 μl per well maintenance medium, consisting of RPMI containing 2% (v/v) iCell hepatocyte medium supplement, 0.1 μM dexamethasone, 2% (v/v) B27 supplement, and 25 μg per ml gentamicin. Maintenance medium was exchanged daily for the duration of the experiment.
000 cells per well. Plates were initially kept at room temperature for 30 min and then transferred to an incubator set at 37 °C and 5% CO2. After four hours of incubation, the plating medium was replaced with 25 μl fresh medium, a step that was repeated daily for four days. On day five, the plating medium was exchanged with 25 μl per well maintenance medium, consisting of RPMI containing 2% (v/v) iCell hepatocyte medium supplement, 0.1 μM dexamethasone, 2% (v/v) B27 supplement, and 25 μg per ml gentamicin. Maintenance medium was exchanged daily for the duration of the experiment.
Calcium flux assay
Intracellular calcium flux in iPSC cardiomyocytes exposed to the test solutions for 120 min was measured using FLIPR tetra (Molecular Devices) instrument using the EarlyTox Cardiotoxicity Kit as described previously.9,12 Cardiomyocytes were incubated for 2 hours at 37 °C following the addition of one volume of pre-equilibrated calcium-dye reagent. Prior to exposure of iPSC cardiomyocytes to test solutions, baseline calcium flux measurements were recorded at 515–575 nm following excitation at 470–495 nm and at a frequency of 8 Hz for 100 seconds. The internal instrument temperature was regulated at 37 °C. Cells were then simultaneously exposed to test solutions using the internal fluidics handling system. 120 min post-exposure, the beating of iPSC cardiomyocytes was monitored as specified above. Between measurements, cells were incubated under cell culture conditions at 37 °C and 5% CO2. Recorded data were processed in Screenworks 4.0 software (Molecular Devices LLC., Sunnyvale, CA) and statistical parameters were exported as Microsoft Excel files for concentration-response assessment.Cellular imaging
Cytotoxicity and mitochondrial integrity were assessed in both cardiomyocytes and hepatocytes by high-content live cell imaging 24 h and 72 h following exposure to test solutions, respectively. Cells were stained with one volume of 2× concentrated Hoechst 33342 (4 μg ml−1), Calcein AM Green (2 μM), and MitoTracker Orange (0.4 μM) for 30 minutes at 37 °C and 5% CO2 prior to image acquisition. Images were acquired using the ImageXpress Micro XL system (Molecular Devices) using the DAPI (Hoechst 3342), FITC (Calcein AM Green), and TRITC (MitoTracker Orange) filters at 20× magnification. Acquired images were processed using the multi-wavelength cell scoring applications module in MetaXpress image processing software (Molecular Devices) and quantitative data were extracted for concentration-response profiling.Assay quality controls
The qualitative integrity of the screening assays in this study was evaluated using previously established conditions.12 In addition to sample wells, tissue culture plates included wells containing untreated and vehicle-treated (1% DMSO in media) cells. Phenotypic outputs from vehicle-treated cells did not differ significantly from untreated cells, a finding that is consistent with previous reports.12 In addition, cardiomyocytes and hepatocytes were treated with 46 μM tetra-octyl ammonium bromide (TAB), which serves as a positive control for cytotoxicity. Cardiomyocytes were also treated with reference compounds: 0.1 μM isoproterenol (positive chronotrope), 0.15 μM cisapride (long QT) and 10 μM propranolol (negative chronotrope), and expected drug-associated phenotypes were observed.Coefficients of variation (%CV), herein defined as the standard deviation of the means of vehicle-treated controls, correlated well with previously published values (Fig. S2†).12 The consistency of replicates in high-content screening assays was determined using correlation analysis for duplicate determinations of all 21 petroleum substance extracts and each assay parameter (Fig. S3†). Pearson (r = 0.79–0.98) and Spearman (ρ = 0.75–0.98) correlations indicated good to excellent intra-plate replicability in all applied assays. The inter-plate assay reproducibility was evaluated through comparisons of %CV values. %CV values were found to be almost exclusively below 10% and to be consistent between different plates (Pearson r = 0.97, Spearman ρ = 0.78) for all sixteen assays/assay parameters, an indicator of the excellent robustness of the utilized screening assays.
Data processing and concentration-response profiling
Quantitative outputs from screening assays were normalized to the mean of responses measured in wells containing vehicle-treated (1% DMSO) cardiomyocytes or hepatocytes (Fig. S1†). Concentration-response curves, converted from % dilution to μg ml−1 based on the starting weight of the extracted substance (see Sample preparation section above) and subsequent dilutions in DMSO and media, were then fitted to a nonlinear logistic function in R and used to derive point-of-departure (POD) values as described previously.19 POD values are herein defined as concentrations at which the fitted curve exceeds one standard deviation above or below the mean of vehicle-treated controls. Calculated POD values based on percent dilution were converted to μg ml−1 concentrations in order to correct for differences in the concentration ranges of the initial stock extracts.Data integration in ToxPi
For data integration and visualization in Toxicological Priority Index Graphical User Interface (ToxPi GUI),15 we selected a total of fifteen experimental phenotypes, eight cardiophysiology parameters (peak frequency, peak rise/decay time, peak baseline, peak width, peak width at 10% amplitude, peak spacing, and peak amplitude), three cardiomyocyte parameters (total number of cardiomyocytes, cardiomyocytes with intact mitochondria, and cardiomyocyte viability), and five hepatocyte parameters (hepatocyte viability, total area of live hepatocytes, % hepatocytes with intact mitochondria, hepatocyte mitochondrial integrated intensity, hepatocyte nuclear mean area). POD values for each phenotype were inversely normalized on a 0–1 scale with 0 representing the highest POD value in a given data set (i.e. the lowest observed bioactivity) and 1 representing the lowest measured POD values (i.e. the highest observed bioactivity) (Fig. S1†). These normalized POD values were then used as quantitative inputs for bioactivity profiling in ToxPi.Differential gene expression analysis
Global changes in gene expression patterns were analyzed using a targeted RNA sequencing technology, TempO-Seq (BioSpyder Technologies, Inc., Carlsbad, CA) measuring a set of genes selected by an effort organized by NIEHS to represent a surrogate whole transcriptome assay, the S1500 (https://federalregister.gov/a/2015-08529). For TempO-Seq analysis, we exposed iPSC hepatocytes to five representative petroleum substance extracts (two SRGOs, two VHGOs, and one HFO) in concentration-response for 48 hours as described above. Following cell lysis in the TempO-Seq lysis buffer, mRNA targets present in the cell lysates were hybridized with a detector oligo mix, followed by nuclease digestion of excess oligos and ligation to generate a pool of amplification templates that share PCR primer landing sites. During product amplification, each sample well was assigned a specific, “barcoded” primer pair, allowing for proper identification and matching of mRNAs and samples following sequencing. Sample amplicons were pooled and cleaned up using a PCR clean-up kit (Clontech, Mountain View, CA). Libraries were sequenced at Texas A&M University Genomics & Bioinformatics Services using a HiSeq 2500 Ultra-High-Throughput Sequencing System (Illumina, San Diego, CA). Sequencing readouts were demultiplexed to generate FASTQ files, and passed all internal quality controls. Genes that were differentially expressed in each of the tested petroleum substances relative to the DMSO controls were identified using the DESeq2 R package.20 DESeq2 employs a normalization method that accounts for compositional bias in sequenced libraries and individual library size. This method calculates a size factor for each sample as the median ratio of the read count relative to the corresponding row geometric average (i.e. that gene for all samples in a group), and then divides the raw counts by that associated size factor. The size factor is an estimate of the necessary correction factor for all read counts of the corresponding column, which is then applied to enable appropriate sample-to-sample comparisons. The adjusted counts undergo a Wald test followed by Benjamini-Hochberg false discovery rate (FDR) correction for multiple testing to identify significantly differentially expressed transcripts (FDR q-value of <0.1).21 Model based clustering was performed using the hclust package in R, with number of components chosen using a maximum median Bayesian Information Criterion.22Results and discussion
The process of safety evaluation for existing or novel products and materials is predicated on the availability of comprehensive datasets that include extensive animal testing. While the requirements for the information needed for approval of pharmaceuticals and pesticides are very proscriptive and largely harmonized across the globe, the regulatory regimes for most chemicals in commerce not only vary from country to country, but are also evolving rapidly.23,24 Identification of “safer alternatives” to products already on the market is yet another challenge whereby no uniform process or standard exists to establish what “safer” means.1 Human health considerations are frequently behind a desire for an alternative, and weigh heavily into the overall safety considerations for the choice among the alternatives. Regardless of the exact criteria to be applied to satisfy decisions in a comparative alternatives assessment, the principles of grouping and read-across25 can aid in filling data gaps or selecting among options. While demonstrating the similarity among individual chemicals is already a formidable challenge in read-across,26 real-life exposures are to complex substances. Little methodological work has been done beyond chemical–physical characterization on how to establish with confidence their similarity in terms of human health impacts. Thus, there is a need to create experimental and data integration approaches that can aid selection of safer alternative chemicals and contribute to pollution prevention. This study tested a hypothesis that multidimensional in vitro bioactivity profiling may be used to support categorization and read-across of complex substances; a case study of petroleum substances as prototypical UVCB was investigated.Sample selection
The process of crude oil refining yields a variety of complex products which fall into specific refinery streams based on their physico-chemical and structural properties to meet product specifications. We therefore selected 21 petroleum substances comprising five of these refinery streams, i.e. manufacturing process-defined product categories (Fig. 1, Table 1).27 For example, included in our experiments were a number of gas oils (five SRGOs, nine VHGOs, and two OGOs) which are considered fairly chemically similar because they boil off in the comparable range of temperatures (∼150–500 °C) during the refining process. These are composed mainly of straight, branched and cyclic alkanes, and aromatics of wide, but largely overlapping range of carbon chain lengths (C9–C25 for SRGOs, C9–C36 for OGOs and C9–C30 for VHGOs).27 In addition, we selected three HFOs and two RAEs, all of which differ significantly in their physico-chemical characteristics from gas oils.27 HFOs comprise complex residual fractions from (vacuum) distillations of crude oil or catalytically or hydro-cracked oil products. RAEs, by contrast, are the products of solvent extractions of various petroleum residues. As such, HFOs and RAEs are composed of higher-molecular weight constituents than gas oils; typically ranging between carbon numbers C14–C98 (HFO) and C25–C95 (RAE) and a wider range of boiling points (265–715 °C for HFOs and 403–702 °C for RAEs).High-content screening of iPSC cardiomyocytes and hepatocytes
Phenotypic screening for concentration-response assessment of DMSO-soluble extracts of all 21 petroleum substances in iPSC cardiomyocytes and hepatocytes was performed using previously established high-content screening assays.9,10,12 Cells were exposed to serial dilutions of test substances in concentration-response over 6 (cardiomyocytes) or 5 (hepatocytes) logs. The respective highest concentrations were in the range between 100 and 1000 μg ml−1.Prior to exposure and after two hours of incubation in the presence of test substances, cardiomyocyte contractility was monitored by fluorescent visualization of internal calcium flux. Subsequently, cell viability was evaluated by live cell imaging following staining with Calcein AM (cell viability), Hoechst 33258 (nuclei), and MitoTracker Orange (mitochondria). iPSC hepatocytes were incubated in presence of test substances for 72 hours before the cells were stained with the same probe combination as cardiomyocytes in preparation for high-content live cell imaging. Processing of the acquired images and calcium flux-traces resulted in quantitative outputs for a variety of cell type-specific and generic phenotypes (see Experimental section).
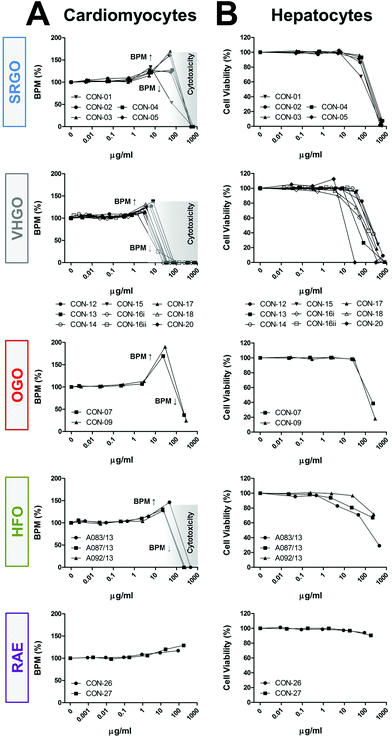
Some of the petroleum substances exhibited bioactivity in cardiomyocytes (Fig. 2A and S4†), primarily increasing the beat frequency and decreasing peak amplitudes, and affected cell viability of both cardiomyocytes and hepatocytes (Fig. 2B). These effects were concentration-dependent. Substances defined under the three VHGO and SRGO categories generally showed higher potency than OGOs and the heavier HFOs and RAEs. Gas oil and heavy fuel oil extracts increased cardiomyocyte beat frequency at sub-cytotoxic concentrations, resulting in a biphasic appearance of the respective concentration-response plots, i.e. initial increases in the beat rate (approx. at concentrations ranging between 1 and 100 μg ml−1) were followed by decreasing beat rate indicating the threshold for cytotoxicity. The only exceptions were the two OGOs, in which the highest tested concentrations had no effect on cell viability, but they were capable of inhibiting the cardiomyocyte contractility by approximately 70% (Fig. 2). While HFOs still elicited a variety of phenotypic responses, albeit at lower potencies than gas oil extracts, RAE were generally associated with the lowest bioactivity in the current study. These findings are concordant with previous studies of petroleum substances in fish.28,29
Bioactivity trends observed in cardiomyocyte-derived phenotypes correlated well with cell viability measurements in exposed hepatocytes (Fig. 2); SRGOs, VHGOs, and OGOs constituted the most bioactive cluster of tested petroleum substances. However, it should be noted that while treatment of hepatocytes with HFOs and RAEs resulted in moderate to low cytotoxicity, it induced pronounced effects on the mitochondria, i.e. decreased numbers of cells with intact mitochondria and increases in mitochondrial intensity.
The results of these experiments, especially a remarkable similarity in concentration-response curves within a product category, but differences between categories (Fig. 2), provided evidence that these data can be utilized to build confidence in the groupings of the petroleum substances according to parallels in their bioactivity profiles.
Computational bioactivity profiling and grouping of petroleum substances
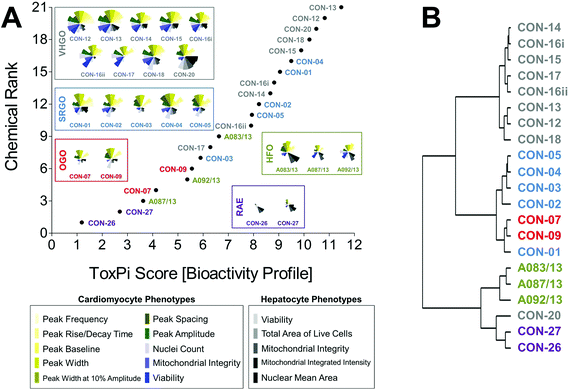
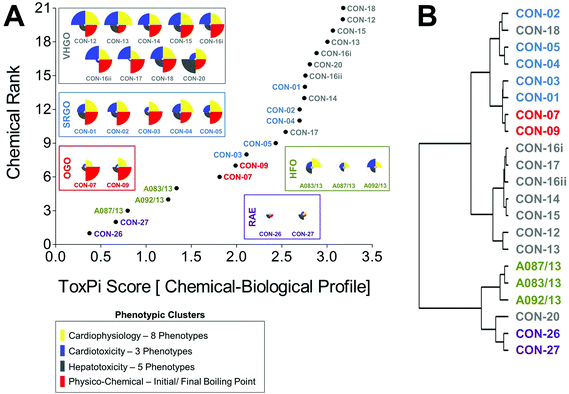
Curve-fitting to concentration-response data derived from in vitro screening assays yielded quantitative measures of bioactivity that were used as inputs for data integration using ToxPi (Fig. S1†). In order to conduct an unbiased data analysis, each assay parameter was assigned an individual ToxPi slice with equally distributed weighting (Fig. 3A). The cumulative ToxPi score for each petroleum substance, reflecting the sum of normalized input scores for each slice of the respective bioactivity profile, was then used as a score to rank substances according to their overall bioactivity.ToxPi ranking using quantitative bioactivity data allowed clustering of petroleum substances into groups. A high-degree of similarity was found in bioactivity profiles of substances within a product group, as well as pronounced differences between substances from different groups. VHGOs and SRGO shared some overlap and contributed the most bioactive petroleum substances in the ranking with ToxPi scores ranging between 6.3 and 11.5 (VHGO), and 5.9 and 9.5 (SRGO) (Fig. 3A, Table 1). HFOs (ToxPi scores = 3.6–6.6) and OGOs (ToxPi scores = 4.1–5.6) were moderately bioactive, with RAEs (ToxPi scores = 1.2–2.8) showing very little bioactivity other than mitochondrial effects in hepatocytes. More importantly, trends for chemical categories observed from the global ranking were consistent with similarities in bioactivity profiles of individual substances of distinct product groups. In fact, key characteristics of bioactivity profiles within a product group were remarkably similar, with a single notable exception: CON-20 differed from other VHGOs through its pronounced effects on hepatocytes (Fig. 3A).
Corroborating evidence for the integrative groupings based on ToxPi scores was provided by cluster analysis of the original POD data set (Fig. 3B). The depicted dendrogram clearly distinguishes two major branches separating the three gas oil groups from HFOs and RAEs. Due to its comparably higher potency with regard to bioactivity parameters in hepatocytes, CON-20 was most correlated with the RAE group. Within each major branch, two sub-clusters further distinguished between petroleum product groups, separating VHGOs from SRGOs and OGOs, and HFOs from RAEs. Clustering of SRGOs and OGO was reflective of qualitative similarities in their bioactivity profiles, which indicated lower levels of bioactivity in cardiomyocytes as compared to VHGOs.
Thus, computational bioactivity profiling in ToxPi and clustering analysis of POD values allowed for groupings of petroleum substances that were closely similar within product categories. This outcome indicates the utility of in vitro HCS data to categorize highly complex substances and to potentially increase confidence in read-across analyses.
Physico-chemical data-based integrative grouping of petroleum substances
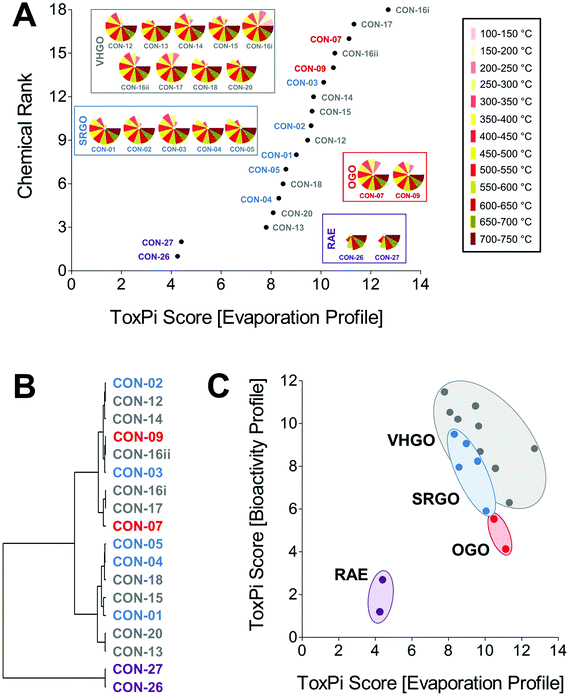
Petroleum substances are very complex and variable in chemical composition with many different chemical components. In addition to the manufacturing process, physico-chemical properties and analytical–chemical information are used to group them for read-across. Here, we used one such parameter, mass % evaporation data (in 50 °C increments), to probe similarities and differences among petroleum samples from four groups, SRGO, VHGO, OGO, and RAE. Mass % evaporation data were integrated in ToxPi to yield evaporation profiles, as well as a volatility-based ToxPi score (Fig. 4A). Global ranking of petroleum substances according to their relative volatility was approximately proportional to their respective boiling point ranges. Most notably, the three gas oil groups formed a large cluster of significantly more volatile (ToxPi score ranges for SRGO = 8.3–10.1, VHGO = 7.8–12.7, OGO = 10.5–11.1) substances as compared to the RAEs (ToxPi score range = 4.3–4.4) and could not be distinguished on the basis of volatility data alone (Table 1). A challenge in confident grouping of different gas oil categories through comparison of their volatility was further demonstrated through cluster analysis (Fig. 4B). We found two major clusters, gas oils and RAEs. Within the gas oils cluster there were additional sub-clusters, but no distinct separation of SRGOs, OGOs, or VHGOs.In order to test the initial hypothesis that in vitro HCS data can improve and refine physico-chemical data based groupings of petroleum substances we correlated evaporation profile and bioactivity profile-derived ToxPi scores (Fig. 4C).8,30 While both data sets were not significantly correlated (Pearson r = 0.35; Spearman ρ = −0.21), the inclusion of biological data indicated a clear trend towards clustering of petroleum substances within each of the four included substance groups with RAEs and OGOs being defined as separate clusters, whereas SRGO and VHGO clusters were slightly overlapping.
Chemical–biological data-integrative categorization of petroleum substances
Combined chemical–biological data integration in ToxPi was approached using four equally (25% each) weighted data clusters combining eight cardiophysiology phenotypes, three cardiomyocyte bioactivity and five hepatocyte bioactivity parameters, and two physico-chemical (initial and final boiling point) descriptors (Fig. 5A). Integrated ToxPi profiles showed a high degree of similarity within each petroleum product group, with CON-20 as an exception due to its previously discussed higher activity in hepatocyte assays. Indeed, inclusion of physico-chemical descriptors and balanced weighting of data types in ToxPi provided a more refined grouping of petroleum substances than was achieved using the global ranking of just the biological data-integrative ToxPi score. RAEs (ToxPi score = 0.4–0.7), HFOs (ToxPi score = 0.8–1.3), and OGOs (ToxPi score = 1.8–2.0) formed three clearly isolated groups, with some overlap being present between SRGOs (ToxPi score = 2.1–2.8) and VHGOs (ToxPi score = 2.5–3.2) (Table 1).ToxPi-derived groupings were consistent with the clustering analysis (Fig. 5B). As previously described for the biological data-based analysis, two major branches separated gas oils from HFOs and RAEs, which in turn were sharply separated within their sub-branches. Within the gas oil branch, most VHGOs, except for CON-18, were distinctly clustered. However, OGOs and SRGO were still clustered within the tertiary branch, with the OGOs only separating at the quaternary level.
Gene expression profiling as an unbiased source of bioactivity data for grouping and read-across
Gene expression profiling provides additional high-dimensional comprehensive data stream for biological read-across.8,31,32 However, full-genome transcriptomic analyses are rather low-throughput and costly. Therefore, to explore the utility of high-throughput transcriptomics for bio profiling and biological read-across of UVCBs, gene expression analysis was conducted using a highly multiplexed templated oligo targeted sequencing assay, TempO-Seq, which uses ligation of detector oligos hybridized to RNA targets, sample barcoding during amplification and pooling prior to sequencing. Unlike other ligation-based assays that have been described,33,34 TempO-Seq can be performed without capture of the target RNA which makes the assay very simple, robust and easy to fully automate.The feasibility of TempO-Seq to provide mechanistic insight into petroleum substance-mediated differential gene expression patterns was tested following 48 hours of exposure of iPSC hepatocytes to a select set of petroleum substances from three distinct product classes (SRGO, VHGO, HFO) in concentration-response. Samples were selected from concentrations that had minimal effect on cell viability (>80% cell viability).
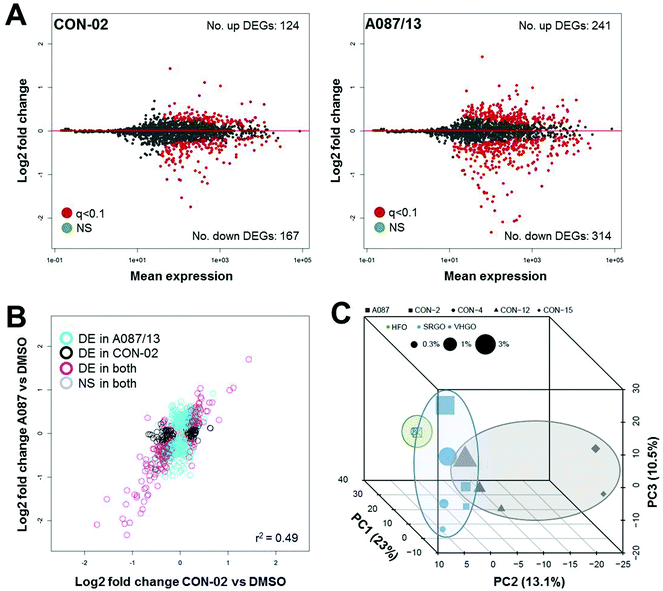
The number of differentially expressed genes varied for each of the substances (Fig. 6A and S5†), with the heavy fuel oil (A087/13) showing the highest number of transcriptional changes. Overall, changes in gene expression were moderately substance-specific, with only 10 genes (out of 1767 queried) commonly differentially expressed (q-value <0.1) after exposure to any of the five petroleum substances at any concentration; however, considerable similarity in the genes that were significantly differentially expressed was observed among all tested substances (Fig. 6B and S6†). In addition, three distinct clusters were observed (Fig. 6C), with the heavy fuel oil separating significantly from the other substances (p = 0.001). In addition, for the combined data, principal component 3 was significantly associated with tested concentration (p = 0.0002, Spearman ρ = 0.86). Commonalities in gene expression signatures among the substances suggest that the overall treatment effect may be greater than substance-specific transcriptomic effects. It is likely that inclusion of additional test substances in future studies will improve the separation of petroleum substance groups and provide further insight into group-specific signatures of differential gene expression patterns.
When data from the highest tested concentrations was analyzed, a total of 66 genes were found to be commonly differentially expressed in cells treated with any of the five petroleum substances (Table S1†). However, despite the above described clustering of petroleum substances by class according to transcriptional profiling (Fig. 6C), most profoundly altered genes were similar across classes, and even across all of the petroleum substances evaluated (Tables S2 and S3†). For example, expression of COL4A1 and COL4A2 (collagen, type IV, alpha 1 and alpha 2, respectively) was reduced in cells treated with any of the petroleum substances, a phenomenon associated with cell death.35 Two isoforms of the UDP glucuronosyltransferase gene family (UGT1A1 and UGT1A6), which are involved in the glucuronidation of small lipophilic molecules into more polar glucuronic acid conjugates, were induced in cells treated with each of the five tested petroleum substances, an observation that indicates the involvement of these genes in the metabolism and clearance of petroleum substances. TIPARP (TCDD-Inducible Poly(ADP-Ribose) Polymerase), an aryl hydrocarbon receptor transactivation suppressor,36 had the largest fold difference change in expression, as well as the smallest corrected p-values of all genes for every sample. CDKN1A (Cyclin-Dependent Kinase Inhibitor 1A) was also found to be commonly up-regulated, which indicates a signature of cell cycle arrest caused by cellular damage. Among the top-most down regulated genes, UBE2C (Ubiquitin-Conjugating Enzyme E2C) and cyclin B1 and B2 were the most significantly suppressed, genes that are involved in the progression of mitosis.37,38 Altogether, genes that code for proteins involved in metabolism and cell cycle are those that were the most significantly changes in expression, and these genes were observed to be differentially expressed across all tested petroleum substances.
Relevance of the biological read-across to human health assessments
The inherent complexity and qualitative variability, largely a result of the manufacturing process-dependent differences in oil refining, create a challenge for traditional, chemical structure-based groupings of petroleum products for read-across. The uncertainties in grouping UVCBs, combined with a lack of a standardized framework for the application of read-across for UVCBs by the regulatory agencies, creates a need for novel approaches to increase the confidence in using novel toxicological data streams, such as in vitro assays and transcriptomics, and to improve transparent communication of complex multi-dimensional datasets. Considering the complexities of the chemical exposure-associated biological processes, confident application of a similarity principle in regulatory decision-making will undoubtedly benefit from the inclusion of multi-dimensional bioactivity data in addition to chemical information. Indeed, knowledge-based chemical design principles increasingly rely on both physico-chemical properties of molecules, as well as the use of higher throughput in vitro and molecular assays to evaluate both UVCBs and chemical alternatives in existing products.1 It has also been suggested that “hybrid” approaches to read-across, models that use both biological (e.g., -omics data or in vitro toxicity screening profiles) and chemical features increase accuracy, expand domain of applicability of the models, and improve transparency and interpretability.8,30–32 Another inherent challenge for data-integrative safety assessments in a regulatory context is the appropriate communication of large chemical–biological data sets to regulators and the general public. Thus, in order to fully implement novel in vitro screening approaches into science-based decision-making, we will inevitably need to increasingly rely on computational toxicology and bioinformatics for data processing and visualization.The data presented in this study indicate that multi-parametric in vitro screenings utilizing iPSC-derived cell types coupled to computational analysis using ToxPi and clustering visualizations can indeed improve the data-assisted confidence in conventional, manufacturing class-derived petroleum substance groupings. Importantly, in vitro screening data independently established the groupings of petroleum substances that closely match the manufacturing process and physico-chemical property-based groupings already in use for registration (Fig. 3). As such, bioactivity profiling provided an increased resolution and increased confidence as compared to physico-chemical property-derived groupings, in which the gas oil groups were indistinguishable, but well separated from RAEs and HFOs (Fig. 4A and B). Moreover, correlation of biological and volatility ToxPi scores indicated improved clustering of petroleum substance groups as compared to volatility alone (Fig. 4C). Chemical–biological data-integrative ToxPi analysis may represent the best of both worlds, indicating sharp segregation of all petroleum substance groups, except for SRGOs and VHGOs. However, based on similarities in their physico-chemical characteristics, this overlap was in fact anticipated.27 It should be noted that while global rankings provided reasonable groupings, a qualitative comparison of the actual bioactivity profiles should be taken into account, providing a supporting basis for clustering UVCBs together. Clustering based on bioactivity profiles has the potential to segregate agents that may share a similar ToxPi score, but differ significantly in their biological effect from other correlated substances, as was the case for the VHGO CON-20.
The transcriptomic analysis of hepatocytes treated with five petroleum extracts from three product groups revealed that principal component analysis of global patterns in differential gene expression can provide corroborating evidence for the groupings determined by phenotypic screenings, and thus indicate its potential utility in data-informed decision making. In addition, targeted high-throughput transcriptomics analysis may eventually provide additional mechanistic insights into toxicological effects through pathway analysis; however, more studies need to be conducted in order to fully evaluate the mechanistic signatures obtained through gene expression-based pathway analyses.
Conclusions
In summary, we demonstrate an experimental approach to grouping complex substances for read across using chemical–biological data integration. This study directly follows from the NRC recommendations1 to demonstrate how novel data streams could be used as primary data in human health assessments or to fill data gaps across a broad range of domains, including health. We show that petroleum substances, prototypical high-production volume UVCBs, can be categorized using global similarities in their bioactivity profiles using multi-parametric HCS of iPSC-derived cardiomyocytes and hepatocytes. In combination with high-throughput transcriptomics analysis, interpretation of these multidimensional data sets is not limited to biological property-derived groupings for regulatory applications, but may eventually also be informative for mechanistic toxicity evaluations. Together, our findings strengthen the argument that high-content in vitro screens combined with computational data integration and visualization possess the potential to improve chemical–biological read-across applications, particularly with regard to the regulatory challenge that is represented by UVCBs, and as such may represent feasible alternatives to minimize the need for unwarranted traditional toxicity testing in animals in regulatory submissions, especially in situations of chemical alternatives assessment.8,12,39,40Author contributions
I. Rusyn, F. Grimm, Y. Iwata, and O. Sirenko designed the experiments with programmatic support from H. Ketelslegers, P. J. Boogaard, and A. M. Rohde. In vitro screenings were conducted by F. Grimm, Y. Iwata, and O. Sirenko. T. Roy prepared the petroleum extracts. F. Wright and D. Reif provided statistical support for the analysis of transcriptomics and in vitro data. J. M. Yeakley, P. Shepard., and B. Seligmann developed the TempO-Seq assay and mapped the genes of the transcriptomics data set. D. L. Gerhold, J. Braisted, and G. Chappell evaluated the transcriptomics data. F. Grimm and I. Rusyn prepared the manuscript.Conflict of interest
O. Sirenko is employed by Molecular Devices LLC, the manufacturer of the FLIPR tetra and ImageXpress instruments used for the in vitro screenings. J. M. Yeakley, P. Shepard, and B. Seligmann are owners/employees of BioSpyder Technologies Inc., the manufacturer of the TempO-Seq assay. P. J. Boogaard, H. Ketelslegers, and A. M. Rohde are employees of Shell International BV, Concawe, and ExxonMobil Chemical Co, the producers and suppliers of the petroleum substances used in this study.Acknowledgements
This work was funded by grants from the United States Environmental Protection Agency (STAR RD83516601 and RD83574701) and the National Institutes of Health (P42 ES005948) and was supported, in part, by the Intramural Research Program of the NIH, National Center for Advancing Translational Sciences. F. Grimm was the recipient of the 2015 Society of Toxicology-Colgate Palmolive postdoctoral fellowship in in vitro toxicology. The authors appreciate useful discussions and technical support from Carole Crittenden (Molecular Devices LLC., Sunnyvale, CA), Joel McComb (BioSpyder Technologies Inc., Carlsbad, CA), and Yi-Hui Zhou (North Carolina State University).References
- National Research Council, A Framework to Guide Selection of Chemical Alternatives, The National Academies Press, Washington, DC, 2014, DOI:10.17226/18872.
- ECHA, European Chemicals Agency, Helsinki, 2012, http://echa.europa.eu/documents/10162/13643/substance_id_en.pdf.
- P. Tsitou, M. Heneweer and P. J. Boogaard, Toxicol. In Vitro, 2015, 29, 299–307 CrossRef CAS PubMed.
- A. R. Scialli, Regul. Toxicol. Pharmacol., 2008, 51, 244–250 CrossRef CAS PubMed.
- F. J. Murray, T. M. Gray, L. G. Roberts, R. N. Roth, M. J. Nicolich and B. J. Simpson, Regul. Toxicol. Pharmacol., 2013, 67, S60–S74 CrossRef PubMed.
- F. J. Murray, R. N. Roth, M. J. Nicolich, T. M. Gray and B. J. Simpson, Regul. Toxicol. Pharmacol., 2013, 67, S46–S59 CrossRef CAS PubMed.
- R. N. Roth, B. J. Simpson, M. J. Nicolich, F. J. Murray and T. M. Gray, Regul. Toxicol. Pharmacol., 2013, 67, S30–S45 CrossRef CAS PubMed.
- Y. Low, A. Sedykh, D. Fourches, A. Golbraikh, M. Whelan, I. Rusyn and A. Tropsha, Chem. Res. Toxicol., 2013, 26, 1199–1208 CrossRef CAS PubMed.
- O. Sirenko, C. Crittenden, N. Callamaras, J. Hesley, Y. W. Chen, C. Funes, I. Rusyn, B. Anson and E. F. Cromwell, J. Biomol. Screening, 2013, 18, 39–53 CrossRef CAS PubMed.
- O. Sirenko, J. Hesley, I. Rusyn and E. F. Cromwell, Assay Drug Dev. Technol., 2014, 12, 43–54 CrossRef CAS PubMed.
- O. Sirenko, J. Hesley, I. Rusyn and E. F. Cromwell, Assay Drug Dev. Technol., 2014, 12, 536–547 CrossRef CAS PubMed.
- F. A. Grimm, Y. Iwata, O. Sirenko, M. Bittner and I. Rusyn, Assay Drug Dev. Technol., 2015, 13, 529–546 CrossRef CAS PubMed.
- M. Mercola, A. Colas and E. Willems, Circ. Res., 2013, 112, 534–548 CrossRef CAS PubMed.
- I. Acimovic, A. Vilotic, M. Pesl, A. Lacampagne, P. Dvorak, V. Rotrekl and A. C. Meli, BioMed. Res. Int., 2014, 512831 Search PubMed.
- D. M. Reif, M. Sypa, E. F. Lock, F. A. Wright, A. Wilson, T. Cathey, R. R. Judson and I. Rusyn, Bioinformatics, 2013, 29, 402–403 CrossRef CAS PubMed.
- R. H. McKee, M. Nicolich, T. Roy, R. White and W. C. Daughtrey, Int. J. Toxicol., 2014, 33, 17S–27S CrossRef CAS PubMed.
- T. A. Roy, S. W. Johnson, G. R. Blackburn and C. R. Mackerer, Fundam. Appl. Toxicol., 1988, 10, 466–476 CrossRef CAS PubMed.
- ASTM International, Standard Test Method for Determining Carcinogenic Potential of Virgin Base Oils in Metalworking Fluids, ASTM International, West Conshohocken, PA, 2010 Search PubMed.
- O. Sirenko, E. F. Cromwell, C. Crittenden, J. A. Wignall, F. A. Wright and I. Rusyn, Toxicol. Appl. Pharmacol., 2013, 273, 500–507 CrossRef CAS PubMed.
- M. I. Love, W. Huber and S. Anders, Genome Biol., 2014, 15, 550 CrossRef PubMed.
- Y. Benjamini and Y. Hochberg, J. R. Stat. Soc., 1995, B57, 289–300 Search PubMed.
- C. Fraley and A. E. Raftery, J. Am. Stat. Assoc., 2002, 97, 611–631 CrossRef.
- E. Berggren, P. Amcoff, R. Benigni, K. Blackburn, E. Carney, M. Cronin, H. Deluyker, F. Gautier, R. S. Judson, G. E. Kass, D. Keller, D. Knight, W. Lilienblum, C. Mahony, I. Rusyn, T. Schultz, M. Schwarz, G. Schuurmann, A. White, J. Burton, A. M. Lostia, S. Munn and A. Worth, Environ. Health Perspect., 2015, 123, 1232–1240 CrossRef PubMed.
- N. Burden, C. Mahony, B. P. Muller, C. Terry, C. Westmoreland and I. Kimber, Toxicology, 2015, 330, 62–66 CrossRef CAS PubMed.
- G. Patlewicz, N. Ball, P. J. Boogaard, R. A. Becker and B. Hubesch, Regul. Toxicol. Pharmacol., 2015, 72, 117–133 CrossRef CAS PubMed.
- N. Ball, M. Bartels, R. Budinsky, J. Klapacz, S. Hays, C. Kirman and G. Patlewicz, Regul. Toxicol. Pharmacol., 2014, 68, 212–221 CrossRef CAS PubMed.
- CONCAWE REACH – Analytical characterisation of petroleum UVCB substances, Brussels, Belgium, 2012 Search PubMed.
- F. Brette, B. Machado, C. Cros, J. P. Incardona, N. L. Scholz and B. A. Block, Science, 2014, 343, 772–776 CrossRef CAS PubMed.
- J. H. Jung, C. E. Hicken, D. Boyd, B. F. Anulacion, M. G. Carls, W. J. Shim and J. P. Incardona, Chemosphere, 2013, 91, 1146–1155 CrossRef CAS PubMed.
- I. Rusyn, A. Sedykh, Y. Low, K. Z. Guyton and A. Tropsha, Toxicol. Sci., 2012, 127, 1–9 CrossRef CAS PubMed.
- Y. S. Low, A. Y. Sedykh, I. Rusyn and A. Tropsha, Curr. Top. Med. Chem., 2014, 14, 1356–1364 CrossRef CAS PubMed.
- Y. Low, T. Uehara, Y. Minowa, H. Yamada, Y. Ohno, T. Urushidani, A. Sedykh, E. Muratov, V. Kuz'min, D. Fourches, H. Zhu, I. Rusyn and A. Tropsha, Chem. Res. Toxicol., 2011, 24, 1251–1262 CrossRef CAS PubMed.
- H. B. Larman, E. R. Scott, M. Wogan, G. Oliveira, A. Torkamani and P. G. Schultz, Nucleic Acids Res., 2014, 42, 9146–9157 CrossRef PubMed.
- H. Li, J. Qiu and X. D. Fu, Curr. Protoc. Mol. Biol., 2012 Search PubMed , Chapter 4, Unit 4–13, 11–19.
- C. W. Brown, A. S. Brodsky and R. N. Freiman, Mol. Cancer Res., 2015, 13, 78–85 CrossRef CAS PubMed.
- L. MacPherson, S. Ahmed, L. Tamblyn, J. Krutmann, I. Forster, H. Weighardt and J. Matthews, Int. J. Mol. Sci., 2014, 15, 7939–7957 CrossRef PubMed.
- M. Le Breton, P. Cormier, R. Belle, O. Mulner-Lorillon and J. Morales, Biochimie, 2005, 87, 805–811 CrossRef CAS PubMed.
- Z. Hao, H. Zhang and J. Cowell, Tumor Biol., 2012, 33, 723–730 CrossRef CAS PubMed.
- G. Patlewicz, T. Simon, K. Goyak, R. D. Phillips, J. C. Rowlands, S. D. Seidel and R. A. Becker, Regul. Toxicol. Pharmacol., 2013, 65, 259–268 CrossRef CAS PubMed.
- G. Patlewicz, N. Ball, R. A. Becker, E. D. Booth, M. T. Cronin, D. Kroese, D. Steup, B. van Ravenzwaay and T. Hartung, Altex Chromatogram, 2014, 31, 387–396 CrossRef.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c6gc01147k |
| This journal is © The Royal Society of Chemistry 2016 |