Open Access Article
Open Access ArticleImproving lipid recovery from Scenedesmus wet biomass by surfactant-assisted disruption†
YenJung Sean
Lai
*a,
Federica De
Francesco
b,
Alyssa
Aguinaga
a,
Prathap
Parameswaran
*c and
Bruce E.
Rittmann
a
aSwette Center for Environmental Biotechnology, The Biodesign Institute at Arizona State University, P.O. Box 875701, Tempe, AZ 85287-5701, USA. E-mail: ylai30@asu.edu; prathapp@ksu.edu
bDepartment of Applied Science and Technology, Politecnico di torino, Corso Duca degli Abruzzi, 24-10129 Torino, Italy
cDepartment of Civil Engineering, Kansas State University, 2123 Fiedler Hall, Manhattan, KS 66506, USA
First published on 20th October 2015
Abstract
Microalgae-derived lipids are good sources of biofuel, but extracting them involves high cost, energy expenditure, and environmental risk. Surfactant treatment to disrupt Scenedesmus biomass was evaluated as a means to make solvent extraction more efficient. Surfactant treatment increased the recovery of fatty acid methyl ester (FAME) by as much as 16-fold vs. untreated biomass using isopropanol extraction, and nearly 100% FAME recovery was possible without any Folch solvent, which is toxic and expensive. Surfactant treatment caused cell disruption and morphological changes to the cell membrane, as documented by transmission electron microscopy and flow cytometry. Surfactant treatment made it possible to extract wet biomass at room temperature, which avoids the expense and energy cost associated with heating and drying of biomass during the extraction process. The best FAME recovery was obtained from high-lipid biomass treated with Myristyltrimethylammonium bromide (MTAB)- and 3-(decyldimethylammonio)-propanesulfonate inner salt (3_DAPS)-surfactants using a mixed solvent (hexane![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) isopropanol = 1
isopropanol = 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, v/v) vortexed for just 1 min; this was as much as 160-fold higher than untreated biomass. The critical micelle concentration of the surfactants played a major role in dictating extraction performance, but the growth stage of the biomass had an even larger impact on how well the surfactants disrupted the cells and improved lipid extraction. Surfactant treatment had minimal impact on extracted-FAME profiles and, consequently, fuel-feedstock quality. This work shows that surfactant treatment is a promising strategy for more efficient, sustainable, and economical extraction of fuel feedstock from microalgae.
1, v/v) vortexed for just 1 min; this was as much as 160-fold higher than untreated biomass. The critical micelle concentration of the surfactants played a major role in dictating extraction performance, but the growth stage of the biomass had an even larger impact on how well the surfactants disrupted the cells and improved lipid extraction. Surfactant treatment had minimal impact on extracted-FAME profiles and, consequently, fuel-feedstock quality. This work shows that surfactant treatment is a promising strategy for more efficient, sustainable, and economical extraction of fuel feedstock from microalgae.
Introduction
Microalgae-derived lipids have promise to be carbon-neutral substitutes for fossil fuels.1–4 For example, Scenedesmus, Chlorella, Nannochloropsis, and Chlamydamonas can contain a high fraction of high-density lipid inclusions (30–60% lipids as dry weight) that are good biofuel feedstock.5–8 Microalgae-derived lipids are mainly triacylglycerols (TAGs) that are enclosed within intracellular compartments.9,10 Efficient extraction of lipids requires cell disruption and solvents able to penetrate into the intracellular inclusions.10–14Cells can be disrupted by several pre-treatment techniques – e.g., mechanical, thermal, ultrasound, microwave, osmotic shock, enzymatic, and pulsed electric fields.11–15 All require intensive capital investments and incur operating costs for energy, chemicals, or both. Furthermore, their scalability for disrupting microalgae is yet to be demonstrated.10,11,13,16
Lipid extraction also depends upon having solvents that are able to penetrate the cell membrane and dissolve the lipids.17 Examples of proven solvent mixtures include Folch (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 chloroform
1 chloroform![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) methanol)18 and Bligh and Dyer (2
methanol)18 and Bligh and Dyer (2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1
1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0.8 chloroform
0.8 chloroform![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) methanol
methanol![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) H2O).19 However, these solvents are hazardous and expensive, making them inappropriate for large-scale application.11,13 More sustainable “green” solvents, such as ethyl acetate and super-critical CO2, have been developed,13,20 but their scalability has not been confirmed.11 Hexane has been applied at the industrial scale, but needs intensive pre-treatment to enhance recovery.21,22 Moreover, conventional lipid-extraction techniques require dewatering, which consumes much or all of energy than what could be gained from the microalgal feedstock.11,23,24 Wet-biomass extraction is a new alternative, but it has not been embraced for large-scale microalgae-to-fuel initiatives.11,22–24
H2O).19 However, these solvents are hazardous and expensive, making them inappropriate for large-scale application.11,13 More sustainable “green” solvents, such as ethyl acetate and super-critical CO2, have been developed,13,20 but their scalability has not been confirmed.11 Hexane has been applied at the industrial scale, but needs intensive pre-treatment to enhance recovery.21,22 Moreover, conventional lipid-extraction techniques require dewatering, which consumes much or all of energy than what could be gained from the microalgal feedstock.11,23,24 Wet-biomass extraction is a new alternative, but it has not been embraced for large-scale microalgae-to-fuel initiatives.11,22–24
Surfactants are well established for isolating proteins by binding the hydrophobic parts of a cell, which releases proteins from the disrupted membranes.25–27 Anionic, cationic, non-ionic, and zwitterionic surfactants are commercially available for achieving different purposes. Surfactants are distinguished by their hydrophilic and hydrophobic constituents; the hydrophobic components can insert into outer membranes and, thus, lyse the cells.28,29 Recent studies30,31 showed that cationic surfactants could easily bind with microalga membranes that have a negative charge, bringing about effective cell disruption.30 Furthermore, the affinity of binding may not just rely on the charge, but also depend on the affinities of other components: i.e., the hydrophilic–lipophilic interplay31 between microalgae and surfactant could also be associated with cell disruption. Many surfactants are biodegradable and of little or no toxicity,32,33 and thus they are widely used in many environmentally sensitive applications.34,35
Given their proven ability to disrupt membranes, surfactants appear to be an ideal aid for enhanced lipid extraction from microalgae. Despite this potential, the literature offers minimal reports30,31 of surfactant-assisted lipid extraction. Fundamental understanding is absent concerning the parameters that need to be considered in the choice of an effective surfactant and how to employ it. In general, the efficacy of a surfactant for cell lysis can be evaluated based on its critical micelle concentration (CMC), which is defined as the threshold concentration of a surfactant to form a mixed lipid-surfactant micelle and/or surfactant solubilized protein units that lead to cell membrane fragmentation or solubilization.26 However, the interaction between surfactant and cell membrane leading to lysis and disruption also may be achieved at surfactant concentrations below the CMC value, depending on the composition and structure of the surfactant.26
The composition of the cell membrane not only changes with microalgae species, but also varies with physiological state for a single strain.36 For instance, the microalga Botryococcus braunii race A possesses a resistant non-hydrolyzable polymer, called algaenan, with a trilaminar structure (TLS) and located in the outer layer of the cell wall, but the polymer composition can change, depending on growth stage.37Chlorella emersonii, having a TLS structure, was more recalcitrant to detergent action than Chlorella vulgaris without a TLS structure.38 Presumably, a change in cell-wall structure will affect the efficacy of cell disruption by a surfactant. Therefore, it is important to understand how growth stages affect surfactant-based disruption.
We first screened anionic, cationic, non-ionic, and zwitterionic surfactants for their ability to disrupt cells of Scenedesmus and increase the efficiency of lipid extraction using a green solvent, isopropanol. We documented that an effective surfactant eliminates the need for toxic Folch solvent, and we used Transmission Electron Microscopy (TEM) and flow cytometry to document the disruptive impact on cell morphology. Second, we demonstrated that surfactants work well for wet-biomass extraction when using an isopropanol and hexane solvent mixture, and we assessed the kinetics of lipid extraction for different surfactant incubation times. Third, we correlated efficient lipid extraction with the surfactant's CMC and also the biomass growth stage.
Experimental
Microalgal biomass/elemental analysis
We obtained freshly harvested Scenedesmus biomass at three growth stages – protein-rich, intermediate-lipid, and high-lipid – from a pilot-scale photobioreactor at the Arizona Center for Algae Technology and Innovation (AzCATI), located at the Polytechnic campus of Arizona State University (Mesa, AZ). We measured dry weight as total suspended solids (TSS) and the organic fraction of the dry weight as volatile suspended solids (VSS) according to Standard Methods.39 We performed elemental analysis by a CHN elemental analyzer (CE-440, Exeter Analytical Inc., USA) to quantify the individual element composition and summarized the characteristics of each growth stage in Table S1.†Screening surfactants for lipid extraction via dried biomass extraction
The seven surfactants we evaluated were obtained from Sigma-Aldrich (St. Louis, MO), and their properties were listed in Table 1. For the cell-disruption screening test, high-lipid biomass (45 mL) was soaked with 50 mM of the respective surfactants and gently mixed using a rocker (Lab-line, TX, USA) for 18–20 hours at room temperature (∼24 °C). After overnight incubation, the surfactant-treated biomass was washed twice by distilled water and pelleted with a centrifuge (Eppendorf 5810R, NY, USA) at 4000 rpm for 20 min, and then the biomass was lyophilized using FreeZone Benchtop instrument (Labconco, MO, USA).| Type | Surfactant | CMCa (mM) | Total FAME lossb (%) |
|---|---|---|---|
| (20–25 °C) | |||
| a The values obtained from the manufacture, Sigma-Aldrich. b The loss was due to washing process within the dried biomass extraction procedure. | |||
| Anionic | Sodium dodecyl sulfate (SDS) | 7–10 mM | 3 ± 1 |
| Anionic | N-Lauroylsarcosine sodium salt (N_LS) | 14.6 mM | 0 ± 1 |
| Non-ionic | Tween-20 | 0.06 mM | −2 ± 11 |
| Non-ionic | Triton-100 | 0.2–0.9 mM | 19 ± 3 |
| Zwitterionic | 3-(N,N-Dimethylmyristylammonio)propanesulfonate (3_DMAPS) | 0.1–0.4 mM | 18 ± 4 |
| Zwitterionic | 3-(Decyldimethylammonio)propanesulfonate inner salt (3_DAPS) | 25–40 mM | 26 ± 2 |
| Cationic | Myristyltrimethylammonium bromide (MTAB) | 4–5 mM | 27 ± 3 |
The assay for lipid extraction from dried biomass was adapted from previous work.38 In brief, Folch (chloroform![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) methanol = 2
methanol = 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, V/V) and isopropanol (IPA) were selected for lipid extraction. The solvent-to-dried biomass ratio was 3 mL
1, V/V) and isopropanol (IPA) were selected for lipid extraction. The solvent-to-dried biomass ratio was 3 mL![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 15 mg for all the cases. We vortexed the mixtures for 3 hours using a vortex mixer (Scientific Industries, NY, USA) at room temperature and filtered the solvent mixtures through a 0.2 μm PVDF membrane (Pall Science, NY, USA) to remove the biomass debris. Afterward, the crude lipids in the filtrate were dried in a Nitrogen evaporator (Labconco RapVap, MO, USA), and their weight was obtained by subtracting the weight of the empty tubes and the weight of any breakthrough materials released from the syringe filter.
15 mg for all the cases. We vortexed the mixtures for 3 hours using a vortex mixer (Scientific Industries, NY, USA) at room temperature and filtered the solvent mixtures through a 0.2 μm PVDF membrane (Pall Science, NY, USA) to remove the biomass debris. Afterward, the crude lipids in the filtrate were dried in a Nitrogen evaporator (Labconco RapVap, MO, USA), and their weight was obtained by subtracting the weight of the empty tubes and the weight of any breakthrough materials released from the syringe filter.
Surfactant treatment released lipids from the biomass solids to the aqueous phase, and this led to a loss of lipid mass from the biomass when it was washed, as described above. In contrast, direct transesterification (DT) yields the maximum extractable FAMEs, commonly referred as total FAME, for a dried biomass sample. We performed direct transesterification by adding 2 ml of 3-N-methanolic HCl (Sigma-Aldrich, MO, USA) to the entire dried biomass (15 mg) in a test tube and incubating the mixture at 85 °C in the oven for 2.5 h.40 Thus, we evaluated lipid loss by comparing the total fatty acid methyl ester (FAME) in surfactant-treated biomass versus control biomass without any treatment using direct transesterification.
Wet biomass extraction and the kinetics of FAME recovery
We evaluated wet-biomass extraction using the three best-performing surfactants from the dried-biomass study: 3_DAPS, MTAB, and SDS (50 mM in each case). Because we hypothesized that the growth stages could change the compositions of cell membranes and affect the cell-disruption efficiency, we evaluated Scenedesmus biomass from the three growth phases (protein-rich, intermediate-lipid, and high-lipid). First, we replaced the supernatant from the centrifuged whole algae cells with surfactant solutions (dissolved in tap water) at desired concentrations. Centrifugation (Eppendorf 5810R, NY, USA) was at 4000 rpm for 15 min. Incubation was carried out within an incubator shaker at 210 rpm (New Brunswick Scientific, Enfield, CT) and kept at room temperature (∼24 °C).Four incubation times were evaluated: 0, 1, 2, and 3 days. At the specified time, duplicate 1 mL samples were withdrawn from the suspensions and mixed with a hexane and isopropanol solvent mixture (3 mL, HEX![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) IPA = 1
IPA = 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) in the test tube. Since solutions contained biomass, surfactant, and solvent, the selected solvent had to be compatible with all three components. A pure alcohol usually is not an effective solvent for recovering lipids from the aqueous phase due to its naturally strong binding to the membrane-associated lipid complex and high water solubility.11 Therefore, an alcohol usually is combined with a non-polar solvent, e.g., hexane, to recover the desired neutral lipids. Thus, we chose the mixed solvent (HEX
1) in the test tube. Since solutions contained biomass, surfactant, and solvent, the selected solvent had to be compatible with all three components. A pure alcohol usually is not an effective solvent for recovering lipids from the aqueous phase due to its naturally strong binding to the membrane-associated lipid complex and high water solubility.11 Therefore, an alcohol usually is combined with a non-polar solvent, e.g., hexane, to recover the desired neutral lipids. Thus, we chose the mixed solvent (HEX![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) IPA). In addition, an emulsion was not formed with any of the surfactants, a condition favoring effective separation of the extracted lipids.
IPA). In addition, an emulsion was not formed with any of the surfactants, a condition favoring effective separation of the extracted lipids.
The mixtures of solvent and surfactant-treated biomass were vortexed for 1 minute and then incubated without agitation for 20 minutes. Afterwards, 1.0 mL of supernatant was withdrawn from the clear separated solvent phase for FAME quantification. At the same time as sampling for wet extraction, the control and surfactant-treated samples were also evaluated by DT to compare with wet biomass extraction. The extractable FAME (%) is defined as FAME extracted from solvent extraction normalized to the total FAME obtained from 1 mL sample via DT.
We quantified the FAME components using a gas chromatograph (Shimadzu GC 2010, Japan) equipped with a Supelco SP-2380 capillary column (30 m × 0.25 mm × 0.20 μm) and flame ionization detector (FID)40 calibrated with a 37-component FAME Mix standard (Supelco, PA, USA). FAME profiles for the untreated control biomass were compared to profiles for biomass immediately after surfactant amendment in order to detect any background shifts in the FAME profiles due to the presence of surfactants.
Effects of CMC on FAME recovery
For the three selected surfactants, we evaluated the effects of CMC on FAME recoveries. We maintained a constant biomass concentration (2%, shown in Table S1†) for each biomass growth phase, but set the surfactant concentration at 0.5, 5, 25, or 50 mM in 15 mL centrifuge tubes (VWR, PA, USA). The incubation time of the biomass and surfactant suspension was 18 to 20 hours within an incubator shaker at 210 rpm (New Brunswick Scientific, Enfield, CT) and kept at room temperature (∼24 °C). Then, duplicate 1 mL biomass samples were extracted using a mixture of HEX![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) IPA = 1
IPA = 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 (3 mL, v/v) with a 1 min vortex time and 20 min static incubation, followed by the extraction assay described above. The total FAME obtained from DT for control and surfactant treated biomass was quantified using the same assay described above.
1 (3 mL, v/v) with a 1 min vortex time and 20 min static incubation, followed by the extraction assay described above. The total FAME obtained from DT for control and surfactant treated biomass was quantified using the same assay described above.
Characterization of cell disruption by transmission electron microscopy (TEM) and flow cytometer
We followed previous methods14,40 to characterize surfactant disruption of Scenedesmus cells using transmission electron microscopy (TEM) and flow cytometry (FC). For TEM, we initially fixed control and surfactant-treated (3_DAPS, MTAB, and SDS surfactants) cells in 2% glutyraldehyde in 50 mM NaPO4 at pH 7.2 and then post-fixed them with 1% osmium tetroxide in the same buffer. After sequential acetone dehydration steps, we infiltrated and embedded the cells in Spurr's epoxy resin polymerized at 60 °C for 36 h. We cut 60 nm sections and post-stained them in uranyl acetate and lead citrate. We then generated images using a Philips CM12 TEM operated at 80 kV with a Gatan model 791 camera.In parallel when evaluating the high-lipid biomass, we performed FC of SYTOX-green-stained samples using a BD FACSCalibur (BD Biosciences, CA, and USA) flow cytometer. When cell walls were disrupted by surfactants, SYTOX molecules were able to penetrate the cell membrane and exhibit their characteristic green fluorescence by binding to DNA. SYTOX was applied according to manufacturer guidelines (Invitrogen, Carlsbad, CA). Excitation was with an air-cooled 20 mW argon ion laser at 488 nm, and the fluorescence emission of SYTOX was detected using a 510–550 nm FITC filter with readings counted for 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 events from each sample.
000 events from each sample.
Results and discussion
Screening surfactants for lipid extraction via dried biomass extraction
Fig. 1 summarizes how surfactants improved FAME recovery with Folch and isopropanol (IPA) solvents. All samples of surfactant-treated biomass exhibited better FAME recoveries than control biomass for both solvents. The most effective surfactants were 3_DAPS, MTAB, and SDS, which yielded extractable FAME up to 70% of DT with the IPA solvent, and they had 16-fold higher efficiency compared to control biomass when using IPA solvent. A significant advantage of using 3_DAPS surfactant is that it could give FAME-extraction efficiency equivalent to Folch with only IPA. A majority of the surfactant-treated biomass samples could achieve 90–100% extractable FAME when Folch was applied as an extraction solvent, compared to only 64% obtained from control biomass (Fig. 1).Improved FAME recovery was correlated with the type of surfactant; in most cases, charged surfactants worked better than non-ionic surfactants (Fig. 1), a trend that could be associated with better solubility of ionic surfactants.26 Lipid extraction from dried biomass involved multiple washes with deionized water, which could lead to additional loss of FAME in the washing supernatant. Correspondingly, the FAME loss from surfactant-treated biomass (compared to untreated biomass) should match well with the degree of cell disruption by the surfactants to release the lipids: i.e., 3_ DAPS and MTAB, which showed a higher FAME loss of 26 and 27%, also showed higher extractable FAME of 72 and 54% with IPA solvent, respectively (shown in Table 1, Fig. 1 and S1†). On the other hand, surfactants such as Triton, Tween, and 3_DMAPS yielded negligible FAME recoveries with IPA solvent, probably due to an absence of charged groups, low solubility, and poor matching of structures between the surfactant and components of the membrane.26
Most surfactant amendments did not change the FAME profiles when using either Folch or isopropanol for extraction (Fig. S2†). However, Tween and NL_S surfactants, which contain C12 fatty acids in their hydrophobic ends, may insert within cell membranes, and they affected the FAME profile. Selection of surfactants that contain fatty acids that could not insert across the microalgae cell membrane should eliminate surfactant-induced changes to the FAME profile.
To improve understanding of interactions among surfactants, biomass, and solvents, we selected the surfactants for subsequent evaluations that showed the best FAME recovery from each surface-charge category: zwitterionic – 3_DAPS; cationic – MTAB; and anionic-SDS.
TEM ultra-structure/SYTOX assay
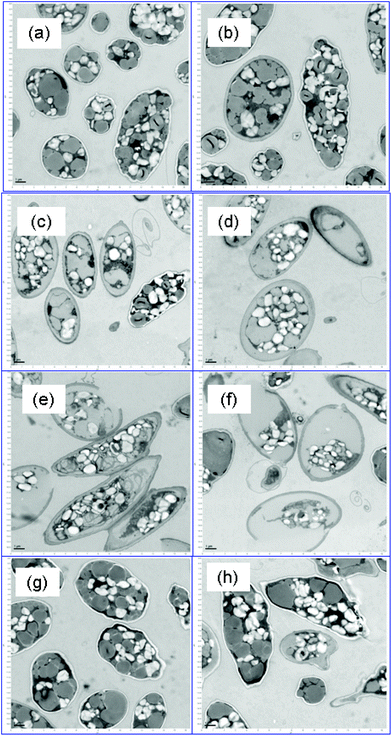
Fig. 2 shows representative TEM images of control and surfactant-treated biomass (lipid-rich condition) after a 48 h incubation. The control biomass (Fig. 2a) had morphology representative of nutrient-depleted Scenedesmus cells: a high proportion of lipid inclusions (gray color) and intact cell walls.41 Surfactant treatment significantly altered the cell morphology in the case of MTAB and 3_DAPS, and damaged cell walls are particularly evident for MTAB-treated cells (Fig. 2e and f). In particular, the cytoplasm and cell wall cannot be clearly distinguished for both types of surfactant treatments, while the control cells (Fig. 2a) show a clear white separation due to the presence of intact membrane-bound carbohydrates and proteins that were extracted by the TEM fixation reagents. The disruptions of the cell membrane resemble those from treatment of Scenedesmus with pulsed-electric fields.14The intact lipid inclusions of Fig. 2a clearly are disrupted in Fig. 2c and e, supporting one means by which the surfactants enhanced FAME extraction. These effects were not isolated, but existed throughout the sample space (Fig. 2c and e). SDS (Fig. 2g and h) did not affect cell disruption significantly.
FC with the SYTOX stain (Fig. S5†) gave another, more quantitative gauge of the efficiency of cell lysis. The green fluorescence intensity (M2 section for inactivated cells) increased by orders of magnitude and followed the order of 3_DAPS ≥ MTAB > SDS > control, which corresponded well to extractable FAME efficiency as discussed for Fig. 1, 2 and 4. The FC assay was consistent with the TEM images: i.e., low FC signal for SDS, indicating mild cell disruption, versus a strong FC signal for MTAB, which caused significant cell disruption.
Surfactant treatment also disrupted other organelles for protein-rich and intermediate-lipid conditions (shown in Fig. S3 and S4†), but the degree of cell disruption was milder than for the high-lipid biomass. SDS showed the opposite trend from the other two surfactants: greater cell disruption and cell wall damage for the protein-rich biomass, compared to the lipid-rich biomass (in Fig. 2vs. Fig. S3†). The disruption trends agree with the lipid recovery trends for SDS, which is discussed in the following section.
MTAB showed significant cell disruption for intermediate-lipid biomass (in Fig. S4e and f†), with clear separation between the cytoplasm and the cell wall. In contrast, 3_DAPS (in Fig S4c and d†) did not show a significant impact on the cell walls, which retained the white color for the membranes of the control biomass.
Wet biomass extraction and its kinetics of FAME recovery
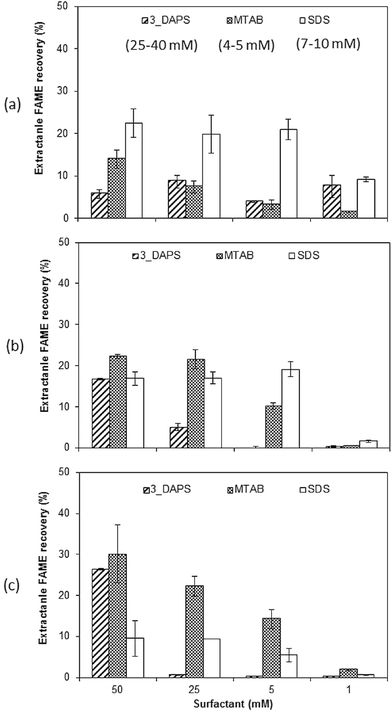
Effective wet-biomass extraction of lipid is highly desirable for eliminating the intense energy requirements for dewatering and drying.11,16,22–24Fig. 3 shows that the efficiency of wet-biomass extraction depended on the type of surfactant and the growth phase of the Scenedesmus biomass. Amongst all growth conditions, the best FAME recovery was 32% of total FAME for MTAB and 3_DAPS with 72 h incubation and for the high-lipid biomass; this was as much as 160-fold higher than control without surfactant treatment. In most cases, FAME recovery reached its maximum within 24 hours.The different biomass growth stages affected surfactant performance dramatically. For the protein-rich biomass, FAME recovery followed the order SDS > MTAB > 3_DAPS, but the high-lipid biomass gave lipid recovery in the order MTAB = 3_DAPS > SDS. The efficacy of SDS for FAME recovery was the highest with low lipid content, while MTAB and 3_DAPS showed the opposite trend. The chemical composition of the cell membrane changes with growth phase for microalgae species, and this leads to differences in charge-specific interactions that could explain the variation of surfactant performance.36,37
The trend with SDS probably is related to its affinity for proteins, as SDS is universally used in polyacrylamide gel electrophoresis (PAGE) for determining the protein molecular weight.42 It is likely that polypeptides that make up glycoproteins, a major component of the cell wall of Scenedesmus biomass, had a major impact on the binding capacity of SDS. The high-lipid biomass had fewer glycoproteins located in the cell membrane, leading to a lower binding by SDS and hence lower cell disruption and lower FAME recovery. Up to now, information describing how the composition of the cell wall of Scenedesmus varies with growth stage is missing, although such information is well documented for several other microalgal species.36,37
Impact of surfactant concentration on FAME recovery
The correlation of surfactant concentration to FAME recovery and Scenedesmus growth stages is shown in Fig. 4. Consistent with Table 1, the most effective extraction of lipids assayed by FAME required a surfactant concentration greater than its CMC. As shown in Fig. 4(a) and (b), SDS, which has a relatively low CMC value (7–10 mM), was not sensitive to decreased surfactant concentration. MTAB, with an even lower CMC (4–5 mM), maintained its ability to enhance extraction down to a concentration of 5 mM (Fig. 4(b) and (c)). 3_DAPS, which has the highest CMC value (25–40 mM), was the most sensitive to decreased surfactant concentration. It lost its capability to enhance FAME extraction when its concentration was close to its CMC (25 mM), as shown in Fig. 4(b) and (c).The growth stage of the biomass had a more defining impact on FAME recovery than CMC, i.e., 3_DAPS and MTAB had lower extraction efficiency even at higher surfactant concentrations for protein-rich biomass, while SDS had lower extraction efficiency at all surfactant concentration for the high-lipid biomass.
With the minimum surfactant dosage we used (5 mM), the maximum extractable FAME was achieved with SDS (20% of total FAME from protein-rich and intermediate-lipid biomass). For the high-lipid biomass, at least 25 mM MTAB was required to obtain the same extraction efficiency. Thus, achieving a high efficiency of lipid recovery with the least surfactant amendment demands knowledge of the interaction among the microalgae growth stage, surfactant type, and its concentration.
FAME profiles
The surfactant amendment improved FAME recovery substantially for all growth conditions (shown in Fig. 3 and 4). From the standpoint of biofuel production, addition of surfactant to enhance lipid extraction is a highly desired result, as long as the FAME profile is not changed in an adverse way.Fig. 5 shows that the different surfactant treatments had significant impacts on the extracted-FAME distribution for the protein-rich biomass, although it did not show changes for the intermediate- and high-lipid biomasses. The changes for the protein-rich biomass may be explained by some form of stress during surfactant treatment. Possible stress factors are nutrient depletion during incubation and stress caused by the surfactant itself. Although we cannot determine which of these factors accounted for the profile shift, the large differences shown in Fig. 5 may suggest that the effect more likely was nutrient depletion, since the intermediate- and high-lipid biomass already had been subjected to stress due to nutrient depletion before surfactant amendment.
The positive impacts of surfactant-assisted microalgae lipid extraction will be sustainable only if the downstream negative impacts of the surfactants added are minimal and the solvent mixture can be effectively reused. To mitigate the hazard risk from hexane, a solvent recycle line can be incorporated into the downstream refinery to recycle solvent use and minimize its discharge.11,15 Hexane accumulated in the residual (non-lipid) biomass should be decomposed during anaerobic digestion,43–45 and surfactant amendments used to improve the hydrolysis of waste activated sludge have been documented.46–48
Conclusions
Surfactant treatment of Scenedesmus biomass disrupted the cells in ways that made solvent extraction more efficient. Surfactant treatment increased the FAME recovery efficiency from dried biomass by 16-fold using isopropanol extraction, and 3_DAPS achieved nearly 100% FAME recovery without any Folch-solvent addition. Wet-biomass extraction with hexane + isopropanol was achieved after surfactant treatment, and it increased the extractable FAMEs by several orders of magnitude compared to wet extraction of control biomass. The CMC values of surfactants had an important influence on the FAME-extraction efficiency, but the biomass growth stage played an even greater role in determining the effectiveness of surfactant treatment.This work shows that surfactant treatment offers a novel strategy for more efficient, sustainable, and economical extraction of fuel feedstock from microalgae. Given the important interactions among surfactant, growth stage, and solvent, future research should emphasize improved understanding of these interactions with the goal of optimizing surfactant/solvent selection.
Acknowledgements
The project was supported by LightWorks, Arizona State University. We thank Dr John McGowen and the Arizona Center for Algal Technology and Innovation (AzCATi) for generously supplying algal biomass. We thank Mr David Lowry at the Electron microscopy facility at the School of Life Sciences (SoLS) at Arizona State University with his expertise in sample preparation and use of the TEM.References
- Y. Chisti, Biotechnol. Adv., 2007, 25, 294–306 CrossRef CAS PubMed.
- B. E. Rittmann, Biotechnol. Bioeng., 2008, 100, 203–212 CrossRef CAS PubMed.
- A. Singh, P. S. Nigam and J. D. Murphy, Bioresour. Technol., 2010, 102, 26–34 CrossRef PubMed.
- A. Parmar, N. K. Singh, A. Pandey, E. Gnansounou and D. Madamwar, Bioresour. Technol., 2011, 102, 10163–10172 CrossRef CAS PubMed.
- Y. Liang, N. Sarkany and Y. Cui, Biotechnol. Lett., 2009, 31, 1043–1049 CrossRef CAS PubMed.
- L. Rodolfi, G. Chini Zittelli, N. Bassi, G. Padovani, N. Biondi, G. Bonini and M. R. Tredici, Biotechnol. Bioeng., 2009, 102, 100–112 CrossRef CAS PubMed.
- L. Xin, H. Y. Hu, G. Ke and Y. X. Sun, Bioresour. Technol., 2010, 101, 5494–5500 CrossRef CAS PubMed.
- P. Bondioli, L. Della Bella, G. Rivolta, G. Chini Zittelli, N. Bassi, L. Rodolfi, D. Casini, M. Prussi, D. Chiaramonti and M. R. Tredici, Bioresour. Technol., 2012, 114, 567–572 CrossRef CAS PubMed.
- Q. Hu, M. Sommerfeld, E. Jarvis, M. Ghirardi, M. Posewitz, M. Seibert and A. Darzins, Plant J., 2008, 54, 621–639 CrossRef CAS PubMed.
- K. Liang, Q. Zhang and W. Cong, J. Agric. Food Chem., 2012, 60, 11771–11776 CrossRef CAS PubMed.
- R. Halim, M. K. Danquah and P. A. Webley, Biotechnol. Adv., 2012, 30, 709 CrossRef CAS PubMed.
- M. D. Zbinden, B. S. Sturm, R. D. Nord, W. J. Carey, D. Moore, H. Shinogle and S. M. Stagg-Williams, Biotechnol. Bioeng., 2013, 110, 1605–1615 CrossRef PubMed.
- J. Sheng, R. Vannela and B. E. Rittmann, Environ. Sci. Technol., 2011, 45, 3795–3802 CrossRef CAS PubMed.
- Y. J. S. Lai, P. Parameswaran, A. Li, M. Baez and B. E. Rittmann, Bioresour. Technol., 2014, 173, 457–461 CrossRef CAS PubMed.
- M. Mubarak, A. Shaija and T. V. Suchithra, Algal Res., 2015, 7, 117–123 CrossRef.
- K.-Y. Show, D.-J. Lee, J.-H. Tay, T.-M. Lee and J.-S. Chang, Bioresour. Technol., 2015, 184, 258–266 CrossRef CAS PubMed.
- W. W. Christie, Preparation of clean lipid extracts from tissues, in Advances in Lipid Methodology—Two, The Oily Press Ltd, Dundee, 1993, pp. 195–213 Search PubMed.
- J. Folch, M. Lees and G. H. S. Stanley, J. Biol. Chem., 1957, 226, 497–509 CAS.
- E. G. Bligh and W. J. Dyer, Can. J. Biochem. Physiol., 1959, 37, 911–917 CrossRef CAS PubMed.
- C. Dejoye, M. A. Vian, G. Lumia, C. Bouscarle, F. Charton and F. Chemat, Int. J. Mol. Sci., 2011, 12, 9332–9341 CrossRef CAS PubMed.
- L. Lardon, A. Hélias, B. Sialve, J.-P. Steyer and O. Bernard, Environ. Sci. Technol., 2009, 43, 6475–6481 CrossRef CAS PubMed.
- L. M. L. Laurens, N. Nagle, R. Davis, N. Sweeney, S. Van Wychen, A. Lowell and P. T. Pienkos, Green Chem., 2015, 17, 1145–1158 RSC.
- D. L. Sills, V. Paramita, M. J. Franke, M. C. Johnson, T. M. Akabas, C. H. Greene and J. W. Tester, Environ. Sci. Technol., 2013, 47, 687–694 CrossRef CAS PubMed.
- G. Yoo, Y. Yoo, J.-H. Kwon, C. Darpito, S. K. Mishra, K. Pak, M. S. Park, S. G. Im and J.-W. Yang, Green Chem., 2014, 16, 312–319 RSC.
- H. Kim, S.-Y. Kim, N. Han and B. Tao, Biotechnol. Bioprocess Eng., 2007, 12, 542–547 CrossRef CAS.
- M. le Maire, P. Champeil and J. V. Møller, Biochim. Biophys. Acta, Biomembr., 2000, 1508, 86–111 Search PubMed.
- B. T. Arachea, Z. Sun, N. Potente, R. Malik, D. Isailovic and R. E. Viola, Protein Expression Purif., 2012, 86, 12–20 CrossRef CAS PubMed.
- N. Nasirpour, S. M. Mousavi and S. A. Shojaosadati, Bioresour. Technol., 2014, 169, 33–37 CrossRef CAS PubMed.
- J. Cheng, Y. Yu and M. Zhu, Green Chem., 2014, 16, 2689–2695 RSC.
- W.-C. Huang and J.-D. Kim, Bioresour. Technol., 2013, 149, 579–581 CrossRef CAS PubMed.
- G. Ulloa, C. Coutens, M. Sanchez, J. Sineiro, J. Fabregas, F. J. Deive, A. Rodriguez and M. J. Nunez, Green Chem., 2012, 14, 1044–1051 RSC.
- G. Zeng, H. Fu, H. Zhong, X. Yuan, M. Fu, W. Wang and G. Huang, Biodegradation, 2007, 18, 303–310 CrossRef CAS PubMed.
- M. Bergero and G. Lucchesi, Biodegradation, 2013, 24, 353–364 CrossRef CAS PubMed.
- W. C. N. Mulligan, R. N. Yong and B. F. Gibbs, Eng. Geol., 2001, 60, 371–380 CrossRef.
- W. Zhou and L. Zhu, Water Res., 2008, 42, 101–108 CrossRef CAS PubMed.
- H. G. Gerken, B. Donohoe and E. P. Knoshaug, Planta, 2013, 237, 239–253 CrossRef CAS PubMed.
- A. J. Simpson, X. Zang, R. Kramer and P. G. Hatcher, Phytochemistry, 2003, 62, 783–796 CrossRef CAS PubMed.
- G. Corre, J. Templier, C. Largeau, B. Rousseau and C. Berkaloff, J. Phycol., 1996, 32, 584–590 CAS.
- E. W. Rice, R. B. Rodger, A. D. Eaton and L. S. Clesceri, Standard Methods for the Examination of Water and Wastewater, American Public Health Association, Washington, DC, 22nd edn, 2012 Search PubMed.
- J. Sheng, R. Vannela and B. E. Rittmann, Bioresour. Technol., 2011, 102, 1697–1703 CrossRef CAS PubMed.
- L. Wang, Y. Li, M. Sommerfeld and Q. Hu, Bioresour. Technol., 2013, 129, 289–295 CrossRef CAS PubMed.
- A. Rath, M. Glibowicka, V. G. Nadeau, G. Chen and C. M. Deber, Proc. Natl. Acad. Sci. U. S. A., 2009, 106, 1760–1765 CrossRef CAS PubMed.
- H. Wilkes, R. Rabus, T. Fischer, A. Armstroff, A. Behrends and F. Widdel, Arch. Microbiol., 2002, 177, 235–243 CrossRef CAS PubMed.
- E.-H. Lee, J. Kim, K.-S. Cho, Y. Ahn and G.-S. Hwang, Environ. Sci. Pollut. Res., 2010, 17, 64–77 CrossRef CAS PubMed.
- S. N. Singh, S. N. Singh, B. Kumari and S. Mishra, in Microbial Degradation of Xenobiotics, Springer, Berlin, Heidelberg, 2012, pp. 439–469 Search PubMed.
- S. Jiang, Y. Chen and Q. Zhou, Chem. Eng. J., 2007, 132, 311–317 CrossRef CAS.
- A. Zhou, C. Yang, Z. Guo, Y. Hou, W. Liu and A. Wang, Biochem. Eng. J., 2013, 77, 240–245 CrossRef CAS.
- A. Zhou, W. Liu, C. Varrone, Y. Wang, A. Wang and X. Yue, Bioresour. Technol., 2015, 192, 835–840 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c5gc02159f |
| This journal is © The Royal Society of Chemistry 2016 |