Open Access Article
Open Access ArticleDeterministic modeling of the exposure of individual participants in the National Health and Nutrition Examination Survey (NHANES) to polychlorinated biphenyls†
Stephen A.
Wood
ab,
James M.
Armitage
a,
Matthew J.
Binnington
a and
Frank
Wania
*ab
aDepartment of Physical and Environmental Sciences, University of Toronto Scarborough, 1265 Military Trail, Toronto, ON M1C 1A4, Canada. E-mail: frank.wania@utoronto.ca; Tel: +1-416-287-7225
bDepartment of Chemistry, University of Toronto Scarborough, 1265 Military Trail, Toronto, ON M1C 1A4, Canada
First published on 15th August 2016
Abstract
A population's exposure to persistent organic pollutants, e.g., polychlorinated biphenyls (PCBs), is typically assessed through national biomonitoring programs, such as the United States National Health and Nutrition Examination Survey (NHANES). To complement statistical methods, we use a deterministic modeling approach to establish mechanistic links between human contaminant concentrations and factors (e.g. age, diet, lipid mass) deemed responsible for the often considerable variability in these concentrations. Lifetime exposures to four PCB congeners in 6128 participants from NHANES 1999–2004 are simulated using the ACC-Human model supplied with individualized input parameters obtained from NHANES questionnaires (e.g., birth year, sex, body mass index, dietary composition, reproductive behavior). Modeled and measured geometric mean PCB-153 concentrations in NHANES participants of 13.3 and 22.0 ng g−1 lipid, respectively, agree remarkably well, although lower model-measurement agreement for air, water, and food suggests that this is partially due to fortuitous error cancellation. The model also reproduces trends in the measured data with key factors such as age, parity and sex. On an individual level, 62% of all modeled concentrations are within a factor of three of their corresponding measured values (Spearman rs = 0.44). However, the model attributes more of the inter-individual variability to differences in dietary lipid intake than is indicated by the measured data. While the model succeeds in predicting levels and trends on the population level, the accuracy of individual-specific predictions would need to be improved for refined exposure characterization in epidemiological studies.
Environmental impactThis study describes a model evaluation that tries to use to the fullest a measured dataset of very large scope and quality to gain an appreciation of how well human PCB exposure can presently be predicted from emissions. While the model approach succeeds in reproducing population average exposure and relationships with age and sex, less success at the level of the individual is attributed to the difficulty in establishing reliably what individuals habitually eat. While this challenge may be well recognized in the field of nutrition,1 the literature reports many statistical associations between dietary composition (e.g. based on 24 hour recall) and measured concentrations of POPs in humans. How valid are such associations, if it is questionable that food recall data are representative of an individual's diet over time periods long enough to affect POP exposure? |
1. Introduction
Polychlorinated biphenyls (PCBs) are a class of Persistent Organic Pollutants (POPs). They have a variety of industrial uses, e.g., as coolants in electrical transformers, sealants, and insulating fluids. In the United States, PCBs were first produced in 1929, and due to their potentially harmful effects on wildlife and humans, their production (and import) was prohibited in 1979.2 The predominant source of exposure for the general population is the ingestion of PCB contaminated food stuffs, particularly fish and livestock.3,4 Adverse health outcomes stemming from PCB exposure are of particular concern; for example, some evidence has been found of associations between PCB serum levels and diabetes,5–7 hypertension,8,9 and endocrine disruption.10,11Human biomonitoring (HBM) studies involve the collection of blood, urine, or other tissues for analysis of environmental contaminants. These studies aim to identify which environmental contaminants a population is exposed to, and quantify the level of these exposures.12 With repeated sampling over time, HBM studies can also provide insight on how a population's exposure to a contaminant is changing. One prominent example is the United States National Health and Nutrition Examination Survey (NHANES). NHANES represents a stratified multistage probability sample of the civilian non-institutionalized population of the United States. It is conducted on a continuous basis and includes an extensive questionnaire with information on diet, health, and demographics.13 Blood and urine samples are collected and analyzed for a suite of organic chemicals. The biomonitoring results in NHANES includes data on many POPs such as PCBs, polybrominated diphenyl ethers (PBDEs), and pesticides, e.g., hexachlorobenzene (HCB) and dichlorodiphenyltrichloroethane (DDT).14,15
Measured PCB concentrations from HBM studies often vary widely; for example, 2003–2004 NHANES PCB-153 levels range from 1.05 ng g−1 lipid to 986 ng g−1 lipid across the analyzed serum samples (although a majority of levels are in a much more confined range, e.g., first quartile = 10.4 ng g−1 lipid and third quartile = 47.4 ng g−1 lipid). Thus, it is of interest to ascertain the extent to which various factors – age, dietary composition, sex, body mass index (BMI), etc. – contribute to differences in levels. Traditionally, this has been accomplished by identifying statistical associations between measured contaminant body burdens and these factors. In non-occupationally exposed populations age, sex, and BMI have been shown to significantly associate with PCB body burdens.16–22 Age is frequently positively associated with PCB body burden.14,15,20 Generally, the male sex is associated with higher PCB levels,18 while associations between BMI and PCB body burden are inconsistent.23–25
Statistical associations do not necessarily imply causal relationships and thus provide limited mechanistic insight into the sources of variability in contaminant levels. They are also generally not suited for making predictions. Toxicokinetic models of varying complexity that mechanistically estimate time-variant POP concentrations in humans26–29 constitute a complementary approach to statistical methods. In these models, POP concentrations in humans are calculated using information on intake rates, partitioning properties, and elimination kinetics (e.g., biotransformation half-life). These calculations generally require time-variant POP intakes as an input parameter. More ambitious approaches seek to also predict intakes by calculating the transfer of POPs through the food chains leading to humans (e.g., ACC-Human).30 If such human food chain models are further combined with mechanistic models of chemical fate in the physical environment (e.g., CoZMo-POP 2 (ref. 31)), integrated models can mechanistically describe the journey of POPs from their initial release into the environment to their accumulation in humans; CoZMoMAN is one model example,32 which has been used previously to simulate human exposure to PCBs for different purposes.33–37 A particularly important feature of such an approach is its dynamic nature, which allows for simulations of time-variant chemical emissions covering multiple decades. Because such emission estimates are available for PCBs,38 the application of integrated models has focused on this chemical group.
In the present study, we combine a global-scale fate model with a human food chain bioaccumulation model to predict the exposure of Americans to PCBs. Specifically, we use time-variant global emissions of PCBs to mechanistically quantify their global fate and transport and transfer through aquatic and agricultural food chains, in order to predict the concentration of four PCB congeners during the entire life of 1999–2004 NHANES participants. By comparing individual predicted PCB concentrations (at time of sampling) to measured PCB levels, this approach utilizes a large, high quality, and diverse empirical dataset (wide age range, both sexes) to perform a novel model evaluation. It also allows for an assessment of different aspects of model performance, including our ability to accurately predict (i) individual contaminant levels, (ii) the mean and range of total population contaminant levels, and (iii) the relationship between contaminant levels and certain demographic factors (age, diet, sex, BMI). The results of such an evaluation should then be able to inform what aspects of the model prediction need improvement, and how the model can be applied with confidence.
2. Methods
2.1 Overview and justification of modeling approach
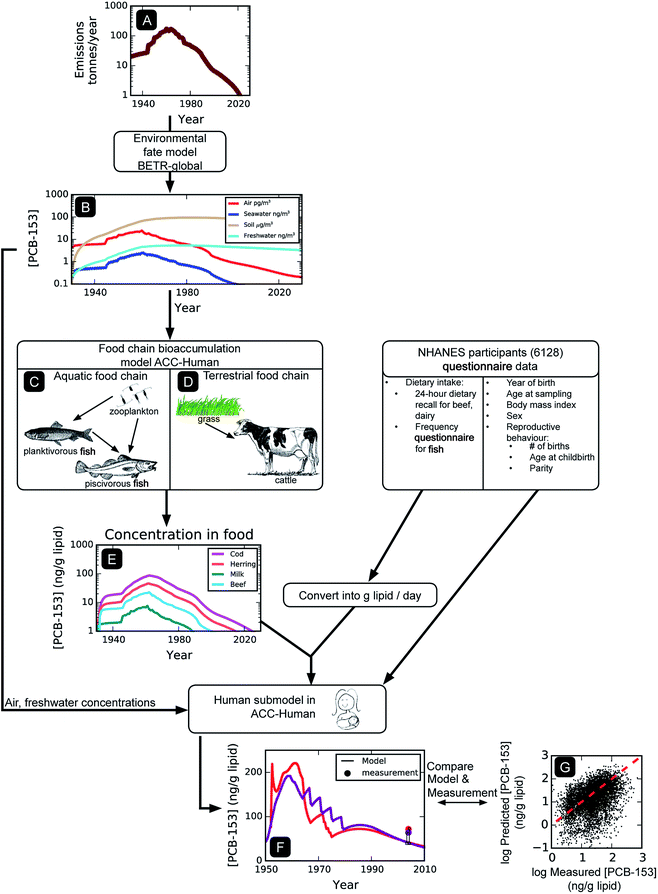
We aim to simulate human exposure to PCBs at both the population level (average) and on an individual basis. Fig. 1 illustrates the modeling approach used to simulate PCB exposures of individual NHANES participants. Since the diet represents the main source of general population PCB exposure, and the intrinsic elimination half-lives of some PCBs in humans exceed a decade, the calculation of historical PCB levels in foodstuffs is required. We focused on PCB congeners 118, 138, 153, and 180, because of their well-defined physical–chemical properties, historical emissions, and frequent detection within NHANES. Starting with historical emissions (Fig. 1A), the global-scale fate model is used to calculate the ambient concentrations of these four PCBs in the United States over time (Fig. 1B). These concentrations serve as input to the human food chain bioaccumulation model (Fig. 1C and D), which outputs concentration as a function of time for the various organisms in the model assumed to constitute the human diet (e.g., fish, beef, dairy, Fig. 1E). Air, freshwater, and food item PCB concentrations (Fig. 1B and E) are then combined with individualized human demographic input data to derive longitudinal time trends of PCB exposure for each individual (Fig. 1F). Finally, the concentration at the time the individual was sampled is compared with the measured concentration reported in NHANES (n = 6128, Fig. 1G). Population level PCB concentrations are obtained by averaging the results of modeled individuals.We recognize that the complexity inherent to the modeling approach described here may not be necessary for simulating exposure to PCBs. In principle, it would have been also possible to predict PCB concentrations in individuals and the population using empirical data on food contamination. While such an approach would undoubtedly be simpler and subject to fewer uncertainties, we prefer an approach that mechanistically calculates food contamination.
One reason is that empirical data are inevitably incomplete and require extrapolation. For chemicals other than PCBs the empirical database is often completely insufficient to reliably define the time course of concentrations in different food items. Even for PCBs, monitoring data are very limited before the 1980s (whereas the simulations cover the period 1930–2010). To address this data gap, contemporary data from the 1990s/2000s could be used to estimate an average intake rate for this period and then scale backwards in time using the temporal trend in emissions.28,47 This approach requires the assumption that primary emissions strongly dominate ambient environmental concentrations (vs. secondary emissions from reservoirs) over the entire simulation period. The validity of this assumption is becoming increasingly tenuous as primary emissions of PCBs continue to decline. In other words, this approach is best suited for retrospective analyses of PCB exposures whereas the holistic approach proposed here could also be used prospectively.
It is also inherently more advantageous to derive human exposure from a mechanistic understanding of the underlying processes rather than literature data. Establishing a quantitative link between emissions and human exposure concentrations is a prerequisite for establishing “safe” levels of emissions. And only such an approach is suited to explore the impact of various factors, such as chemical management strategies or global climate change on human exposure.39,40
2.2 Prediction of PCB concentrations in air, water and soil
Concentrations in the physical environment are calculated using BETR-Global, which is a dynamic, fugacity-based, global scale environmental fate and transport model.41 It separates the physical world into 288 cells based on a 15° longitude by 15° latitude grid. Each cell is composed of seven environmental compartments: upper air, lower air, vegetation, freshwater, soil, coastal water, and sediment. Contaminants are allowed to transfer between compartments and between neighbouring cells. The model requires information on a contaminant's physical–chemical properties, e.g., the octanol–water partition coefficient KOW and the air–water partition coefficient KAW, environmental degradation half-lives, and historical time-variant emissions (Fig. 1A); PCB emission data are from Breivik et al.38 A complete description of input properties can be found in the ESI, Table S1.† Because of the long residence time of PCBs in the environment, global PCB fate and transport is simulated from the beginning of PCB production in 1930 (ref. 2) until 2010.2.3 Prediction of PCB concentrations in food
The dynamic, fugacity-based, mechanistic bioaccumulation model ACC-Human30 is used to describe the uptake of POPs in the human food chain from concentrations in air, water, and soil. It includes an aquatic food chain (Fig. 1C, consisting of zooplankton, planktivorous fish, and piscivorous fish) and an agricultural food chain (Fig. 1D, consisting of grass, milk cows, and beef cattle). Dietary intake of PCBs is assumed to occur through the consumption of three dietary items only (beef, fish, and dairy products). Dietary intake of PCBs from other foodstuffs is either deemed negligible (e.g., plant based food42) or is represented by one of the three (e.g., fowl, pork is represented by beef, ESI, Table S3†). The latter assumption is based on lipid-adjusted PCB concentrations in different meats that are generally within a factor of 2 of each other (see ESI, Table S8†).The PCB concentration calculated for coastal water in cell 76 of BETR-Global, corresponding to the Pacific Ocean adjacent to the California coast, is the basis for the aquatic food chain calculations in ACC-Human, whereas the PCB levels in air, fresh water, and soil in cell 78, corresponding to the central United States, are inputs for the agricultural food chain calculation. There are several assumptions inherent to this approach. Due to the lack of geographical information (i.e., no information on the source of food or geographical location of NHANES participants), the contamination of all foodstuffs is based on the environmental contamination of the atmosphere over the central United States (for the agricultural food chain) and in the seawater off the west coast (for the aquatic food chain). In other words, there is no regional differentiation in the food supply, and the food sources are the same for Americans everywhere. Modelled air concentrations in other regions of the United States were generally within a factor of 2 of those in the central United States, suggesting that regional differences in food chain contamination may be relatively minor. Our approach also ignores changes in food production over the entire simulation time period (1930–2010), for example, the transition from locally produced food and livestock to a nationally integrated food industry, and the transition from grass-fed to corn-fed beef. Additional modeling indicated that PCB levels in grass and corn are similar (i.e., within a factor of 1.5, data not shown). Additionally, dietary transitions on the population level are also ignored (e.g., the shift to leaner meats).43
2.4 Prediction of PCB concentrations in individual humans
The human sub-model within ACC-Human30 is used to calculate the four PCB concentrations in each NHANES 1999–2004 participant (n = 6128) at their time of sampling. Because of the long residence time of PCBs in humans, this requires the calculation of each participant's lifetime exposure history. Input parameters that are adjusted for each study participant using information extracted from their NHANES questionnaire include year of birth, sex, BMI, the dietary intake of fish, beef lipids and dairy lipids, and – in the case of mothers – number of children, mother's age at each childbirth, and nursing duration(s) (for a summary of input parameters, see ESI, Table S2†). In brief, the ACC-Human model calculates PCB levels in humans by considering uptake from the diet, inhalation, and drinking water, and also the elimination of the contaminant by fecal egestion, biotransformation, exhalation, urinary excretion, and skin shedding, and in the case of mothers, childbirth and nursing. To arrive at a lipid-normalized concentration, the total mass of chemical in the human body is divided by the total lipid mass of the human, which is a function of age, sex, and BMI.Breastfeeding is parameterized as follows: mothers who responded “Yes”, “Don't know” or did not respond to the question “Breastfed any of your children?” are assumed to have breastfed all children for 6 months (as recommended by the American Academy of Pediatrics).44 Participants who responded “no” did not breastfeed any of their children. All participants are assumed to have been breastfed for 6 months during their own infancies. Exposure from breastfeeding is unlikely to have a significant impact on model concentrations, as the minimum participant age in the NHANES PCB biomonitoring data is 12 years.
Because PCBs are associated mostly with lipids within the body, differences in the lipid content of study participants could be a source of concentration variability.45 In order to account for the wide range in lipid contents, we modified the original ACC-Human model to allow the user to assign each study participant to one of 22 BMI classes (from BMI = 17 to 38 kg m−2, in increments of 1). For each BMI class, a growth curve is defined which determined the change in lipid content with age. Details on these lipid weight and body weight growth curves can be found in the ESI, Section S3.†
Each individual's intake of beef lipids and dairy lipids is estimated using responses from a 24 hour dietary recall interview on individual foods.13 Fish intake is estimated using a food frequency questionnaire (FFQ) on fish consumption.13 Each individual is assumed to eat the same diet throughout his or her entire lifetime, with a consistent composition based on their dietary recall. On average, the dietary intake values derived from the NHANES data are approximately 1.5 times lower than average American food consumption reported in the USDA Agriculture Fact Book.43 Because of the potential for dietary intake underreporting46 to influence estimated PCB exposures, a scaling factor is implemented for each food item to bring the NHANES average diet in line with the USDA average. The scaling factors applied to all individuals are approximately 1.5 for beef and dairy lipids, and approximately 2 for fish. The application of scaling factors requires the assumption that the PCB biomonitoring subsample of the NHANES population is identical to the US population, i.e., we did not take into account the sampling weights assigned to each NHANES participant. A full description of the process used to convert the dietary questionnaire information into daily intake rates of fish, beef, and dairy can be found in the ESI, Section S4.† The possibility to specify an individualized BMI/growth curve and individualized diet represents a significant expansion in the capabilities of the ACC-Human model, which previously allowed for only a single growth curve and diet for each gender.
Humans additionally take up PCBs by inhaling air and drinking water. These exposures are calculated using the concentrations in lower air and fresh water, respectively, from cell 78 of BETR-Global, i.e., all Americans are assumed to inhale air and drink water from the central USA. Although environmental contamination in air and freshwater may be greater in other parts of the US (e.g., the East coast and urban locations),47 exposure of the four PCB congeners from inhalation and drinking water is insignificant compared to exposure from dietary intake. For the same reason, the inhalation rate (15 m3 d−1) and water consumption rate (3 L d−1) are not individualized. Other, non-individualized model input parameters include the human biotransformation half-life (HLb) for PCBs and the body lipid excretion rate (i.e., skin shedding, 0.8 g lipid per day).
Our approach assumes that exposure only occurs through far-field sources, i.e., from general environmental contamination and not from occupational or indoor exposure. Since dietary lipid intake2,48 is the main source of PCB exposure to the general American population, which is targeted by NHANES sampling, this assumption is appropriate.
While the geographic location of NHANES participants is confidential, reported regional differences in human PCB concentrations are minor: the geometric mean concentration was only 1.4 times greater for people living in the Northeastern US than elsewhere in the nation.49 While this justifies our approach of using the output of a single BETR-Global cell as the input for the food chain calculations, we also used our model to explore the extent of regional concentration differences that could be expected if the American diet were sourced regionally. By varying the BETR-Global cells chosen to drive contamination in the food chain, we predict regional geographic differences in human PCB-153 concentrations of at most a factor of two compared to our reference calculations (data not shown). This is larger than the differences reported by Wattigney et al.,49 which can be explained by the fact that the diet of most Americans will include items sourced from outside their region of residence.
2.5 Constraining emissions timing and human biotransformation half-life
The emission history, in particular the time of peak emissions (E(t)), and the human biotransformation half-life (HLb) are uncertain model input parameters, yet can have a strong impact on the relationship between PCB body burden and age for a population cross-section.29,35 Furthermore, Breivik et al.32 has previously stated that the emissions of PCBs prior to 1980 are potentially underestimated. However, it is difficult to assess the potential bias in emission estimates for this period because monitoring data for PCBs in abiotic and biotic samples collected prior to 1970 are sparse.32As a preliminary exploration, we developed an algorithm to optimize the selection of E(t) and HLb values for the PCBs simulated here. Specifically, three different emissions scenarios [(i) the default emission inventory from Breivik et al. (emissions peak in 1970),38 or peak in emissions occurring (ii) 5 or (iii) 10 years earlier] were combined with HLb values ranging from 1 to 300 years (every 1 year between 1 and 30, and every 10 thereafter) to yield the smallest sum of squared residuals (SSR) between the modeled concentrations of PCB congeners 118, 138, 153, and 180, and the measured concentrations reported for NHANES participants. Data from 6 years of NHANES (2003–04, 2001–02, and 1999–2000) were considered. The use of multiple NHANES years, combined with data on four congeners, provided us confidence in the validity of this fitting procedure. The implication here is that the model is being refined to generate data that best fit the measured data. However, as mentioned previously, these major input parameters are uncertain and we confirmed that the values obtained for E(t) and HLb are within the plausible range. See details in the ESI, Section S5.† We note that this optimization procedure is similar to the use of HBM data in the derivation of intrinsic human elimination half-lives as described by Ritter et al.50 However, here only the biotransformation half-life is optimized. The overall intrinsic elimination half-life is still influenced by other depuration processes (e.g., fecal egestion, breastfeeding) and hence varies according to the related input parameters (e.g., BMI/lipid content, reproductive history). The fact that the model calculates an overall intrinsic elimination half-life for each individual over time in a consistent manner is another advantage of the adopted modeling approach.
2.6 Data visualization, statistical methods, and other software
All statistical and data analysis were performed using R (version 3.1.3) and Python (version 2.7.7)51 with additional libraries NumPy (ver. 1.8.1),52 SciPy (ver. 0.14.0), and pandas (ver. 0.14.0).53 Additionally, all graphical representations of data were generated using matplotlib (ver. 1.3.1).54 Associations between PCB-153 exposure (modeled and measured values) and model input parameters were assessed using linear regression.3. Results
3.1 Optimized emission history and human biotransformation half-life
For all congeners (118, 138, 153, and 180), assuming that the peak in emissions occurred 10 years earlier than reported in Breivik et al.38 results in the lowest SSR, i.e. the best fit between model and measurement requires a shift in peak emissions from 1970 to 1960. While this shift is not derived mechanistically, it is not unreasonable. Whereas production of PCBs peaked in the 1970s, it is possible that emissions of PCBs were higher in the previous decade (e.g., due to industrial emission not accounted for in Breivik et al.38 or emission factors that were higher before the problematic nature of PCBs became obvious).32 We consider this emission time shift only a preliminary hypothesis and suggest that the historical emission history of PCBs should be revisited to find a rigorous mechanistic explanation for higher emissions in the past. However, such an effort is considered outside the scope of the current study.The optimized HLb values for PCB congeners 118, 138, 153, and 180 are 8, 25, 35, and 300 years, respectively. In order to compare these results with those reported in the literature, the modeled total intrinsic elimination half-lives over time were extracted from a model simulation and are as follows: 2 to 7 years for PCB-118, 2 to 18 years for PCB-138, 3 to 24 years for PCB-153, and 3 years to 50 years for PCB-180 (values range depending on sex, age, and BMI, for details see ESI, Section S8 and Fig. S11†). Our estimates compare favorably with those calculated by Ritter et al.:50 9.3, 10.8, 14.4 and 11.5 years for PCB-118, 138, 153, and 180, respectively, and Aylward et al.:55 5, 11, 14.4, and >20 years for PCB-118, 138, 153, and 180, respectively. Considering the large uncertainty of HLb, the optimized values obtained here do not deviate unreasonably from earlier estimates.35 In particular, the differences in the HLb values between the four congeners conform to expectations. Moreover, the modeled total intrinsic elimination half-lives are reasonable. All results presented below are based on these optimized emission history and biotransformation half-lives.
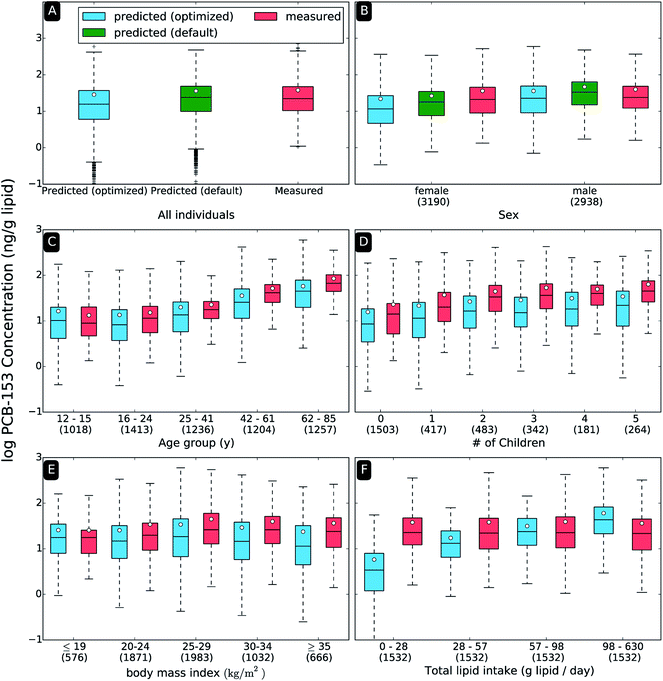
3.2 Comparison of model predictions and measured data at the population level
Fig. 2A compares predicted and measured PCB-153 concentrations at the population level for the original and revised emission scenario (i.e., E(t) = 1970 and E(t) = 1960). In general, for the optimized scenario, the model slightly underestimates PCB-153 concentrations when compared to the measured data. The geometric mean modeled concentration of 13.3 ng g−1 lipid is close to the geometric mean measured concentration of 22.0 ng g−1 lipid. Similar agreement is found when comparing median concentrations, where the modeled value of 15.7 ng g−1 lipid is only marginally lower than the median measured concentration of 22.2 ng g−1 lipid. Similar results are observed for the other PCB congeners, see ESI, Table S4, and Fig. S5–S7.† This level of agreement is quite remarkable, considering the complexity of the model approach, the number of required assumptions, and the uncertainty of many input parameters. The default emission scenario (E(t) = 1970; default 15 years biotransformation half-life for PCB-153) also performs well: the geometric mean modeled concentration of 19.2 ng g−1 lipid is very similar to the geometric mean measured concentration of 22.0 ng g−1 lipid. However, model results based on the default/original scenario fail to reproduce trends with age (R2 = 0.04), a very important predictor of PCB level. Predicted concentrations in younger individuals (age classes 12–15 years, 16–24 years) tend to overestimate the empirical data whereas predicted concentrations in older individuals (age classes 42–61 years, 61–85 years) tend to underestimate the empirical data. See ESI, Fig. S4† for a comparison of the default/original model results and measured levels.The range of predicted concentrations (6![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) log10
log10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) units, 0.001 to 598 ng g−1 lipid) is much greater than that of the measured concentrations (3
units, 0.001 to 598 ng g−1 lipid) is much greater than that of the measured concentrations (3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) log10
log10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) units, 1.05 to 986 ng g−1 lipid). However, the similar variance for the measured and modeled datasets in Fig. 2A (i.e. box size and whisker length), suggests that outliers drive the overall range difference. Particularly, this discrepancy is due to the lower bound of the model output, whereas the upper bound is in good agreement with the empirical data. In other words, for a relatively small number of individuals (n = 338) the model predicts concentrations that are much lower than measurements. A majority of these individuals (n = 260) reported no consumption of meat, fish or dairy products (<1.0 g lipid per day) on their 24 hour dietary recall surveys. Because the model assumes that only these three food categories contribute to a person's dietary PCB intake, modeled body burdens of these individuals are unreasonably low (<1 ng g−1 lipid) due to exposure from inhalation and drinking water only.
units, 1.05 to 986 ng g−1 lipid). However, the similar variance for the measured and modeled datasets in Fig. 2A (i.e. box size and whisker length), suggests that outliers drive the overall range difference. Particularly, this discrepancy is due to the lower bound of the model output, whereas the upper bound is in good agreement with the empirical data. In other words, for a relatively small number of individuals (n = 338) the model predicts concentrations that are much lower than measurements. A majority of these individuals (n = 260) reported no consumption of meat, fish or dairy products (<1.0 g lipid per day) on their 24 hour dietary recall surveys. Because the model assumes that only these three food categories contribute to a person's dietary PCB intake, modeled body burdens of these individuals are unreasonably low (<1 ng g−1 lipid) due to exposure from inhalation and drinking water only.
It is likely that some of these 260 individuals actually eat meat or dairy products on a regular basis, but did not during the 24 hours to which the dietary recall survey applied. It is also likely that some individuals in NHANES ate a vegetarian diet. While the assumption that plant-based food contributes negligibly to PCB intake among those who also eat meat, fish and/or dairy is suitable, it is clearly inappropriate for vegetarians/vegans.4 Unlike more recent NHANES, there was no survey question that explicitly asked the participant if they consider themselves to be a vegetarian. The failure of the model to correctly predict the lower end of PCB exposure among NHANES participants could be addressed by not relying exclusively on 24 hour recall data for estimating dietary intake and by including foods such as grains, vegetables, and fruits in the model calculations.56
3.3 Comparison of model predictions and measured data when stratified by sex, age, parity, BMI, and dietary lipid intake
Fig. 2 also includes panels where measured and modeled PCB-153 concentrations are compared when the data are stratified by sex (Fig. 2B), age (Fig. 2C), number of children (Fig. 2D), BMI (Fig. 2E) and dietary lipid intake (Fig. 2F). Only model output for the revised emission scenario (i.e., E(t) = 1960) are presented here. Similar figures for PCB-118, -138, and -180 are shown in the ESI, Fig. S5–S7.† The model correctly reproduces associations between PCB exposure and certain variables (sex, age, number of children, BMI), but fails for others (dietary lipid intake).Both the model estimates and measured data indicate that on average males have a slightly higher PCB-153 body burden than females (Fig. 2B), a sex-mediated effect supported by previous HBM studies.18,57 There are several factors contributing to higher predicted levels in males: first, they generally have a higher total dietary lipid intake (82 g lipid per day) than females (60 g lipid per day). Secondly, the model assumes that females have higher lipid contents than males for a given BMI, which could contribute to the lower levels observed in females (“solvent dilution”). Lastly, reproduction affords females two additional PCB loss processes: childbirth and breastfeeding.
When stratifying the data by age (Fig. 2C), the model reproduces the trend of rising PCB-153 body burdens with increasing age in the population cross-section. Such agreement is not surprising, because the agreement between modeled and measured trends with age served as a criterion during the optimization of E(t) and HLb. The model results for the non-optimized HLb and E(t) over-predict exposures for individuals older than 70 (see ESI, Fig. S3†). Concentrations of PCB-153 that increase monotonically with age in a cross sectional HBM study conducted during time periods of declining emissions have previously been explained by the older participants' bodies retaining a “memory” of past exposures.29,35 Younger individuals, born after the peak in emissions, do not experience comparable PCB exposures.
In both the measured and modeled data, PCB-153 body burden increases with number of children (females only, Fig. 2D). This result may at first seem counterintuitive, since childbirth and nursing can significantly reduce PCB body burdens.33 However, this may simply be because NHANES participants with more children are generally older. For example, the average birth year of mothers with 5 children is 1939, which resulted in higher PCB-153 body burdens because their lifetimes directly overlapped with the peak emission period. Similarly, mothers with 0 children include younger NHANES participants (i.e., <25 years of age), who have the lowest PCB-153 body burdens.
The relationship between PCB-153 body burden and BMI on the population level is subtle, but there is a trend of increasing model and measured PCB-153 concentration with increasing BMI, with the highest concentrations in those with BMIs in the 24.5–35 range (Fig. 2E). The literature is largely inconsistent in regards to associations between PCB body burden and BMI.23–25 The role of BMI in influencing PCB levels, and how it is related to birth cohort, age, and time of sampling is investigated in detail elsewhere.58 We note that BMI was not correlated with our derived daily food intake levels, i.e., higher levels of PCBs in more obese people appear to not be a result of higher rates of lipid intake.58
Unlike other examined variables, the model identifies total dietary lipid intake as a significant contributor to PCB-153 concentrations, while the measured data indicate no differences in PCB-153 concentrations between the four lipid intake quartiles (Fig. 2F). The model suggests that PCB concentrations should increase with increasing lipid intake, a trend that will be discussed further in the next section describing individual model results.
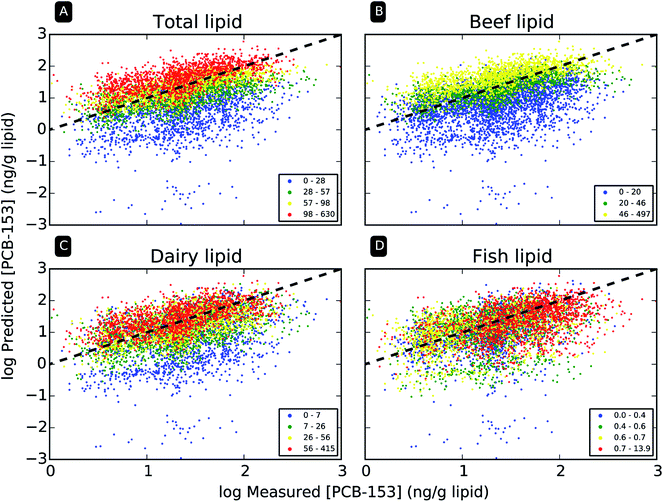
3.4 Comparison of model predictions and measured data for individuals
Fig. 3 directly compares modeled and measured PCB-153 concentrations for each individual NHANES participant (see ESI, Fig. S8–S10,† for the other congeners). Again, only model output for the revised emission scenario (i.e., E(t) = 1960) are presented here. Model performance for the individual predictions is modestly successful. For example, 62% of all predicted data are within a factor of 3 of corresponding measured values, while 89% fall within one order of magnitude of measurements. Rank correlation between modeled and measured PCB-153 concentration was highly significant, with a Spearman rs = 0.44. Considering the scope (i.e., source to receptor) of, and uncertainties inherent to, the model calculations, such model performance is encouraging. However, the model fails to capture all sources of variability.Discrepancies between modeled and measured PCB-153 levels are mainly due to the divergent impact of dietary lipid intake, as mentioned for population level predictions above (Fig. 2F). In each subplot of Fig. 3, the data are colored according to the estimated individual intake of total lipids (panel A), beef lipids (B), dairy lipids (C), and fish lipids (D). Total dietary lipid intake (Fig. 3A) has a significant impact on the modeled concentrations (R2 = 0.44), but virtually no impact on measured concentrations (R2 = 0.00). This is readily observed in Fig. 3A, where dietary intake quartile coloring stratifies along the predicted concentration axis (y axis), but not the measured concentration axis (x axis). The same is observed when only beef lipid intake (Fig. 3B, modeled data: R2 = 0.49, measured R2 = 0.00) and dairy lipid (Fig. 3C, modeled data: R2 = 0.11, measured R2 = 0.00) intake is considered. For fish lipid intake (Fig. 3D), there seems to be little association with either modeled (R2 = 0.02) or measured concentrations (R2 = 0.04). This is due to the fact that a majority of participants (n = 3162, or 52%) are assigned the default US average fish consumption (18.9 g ww per day before age adjustment), as they could not recall (“don't know”) fish intake in their FFQ, or the data were missing.
4. Discussion
4.1 Comparison with previous studies predicting PCB exposure in individuals
This work complements two earlier studies that sought to mechanistically predict PCB exposure in individual humans. Nøst et al.59 predict PCB concentrations in 554 Norwegian women who were either pregnant or postmenopausal; Binnington et al.60 predicted PCB concentrations in 298 Arctic aboriginal mothers. Both studies used the PCB emissions by Breivik et al.;38 although without the shift of peak PCB emissions back 10 years that we employed.Like the present study, Nøst et al. and Binnington et al. successfully reproduced mean population PCB exposures. For example, concentrations of PCB-153 predicted for Norwegian women were within one order of magnitude of measured values.59 However, on an individual level the predictions were less effective, with the models again attributing a larger share of the exposure variability to dietary differences than was observed in the measured data. Our rank correlation coefficient between measured and modeled data (rs = 0.44) is similar to those observed by Binnington et al. (rs > 0.40 for each study group), and lower than the value calculated by Nøst et al. (rs = 0.67).
4.2 Reconciling differences between average and individual prediction success
Because the model is only moderately successful in predicting individuals' PCB exposure using 24 hour dietary recall and FFQ data, it may at first be surprising that the population level predictions are so close to the average measured levels, considering that they are based on the average of those individual predictions. In parameterizing dietary intakes, our model approach requires a major assumption: the dietary recall from NHANES (24 hour period) is extrapolated to an individual's entire lifetime, i.e., there are no changes in dietary composition with age or season. It is quite likely that an individual's dietary intake within one particular 24 hour period is not entirely representative of their diet over an entire year, let alone a lifetime. On the other hand, the average of several thousand individuals' dietary intakes from one particular 24 hour period may in fact be a reasonable estimate of the average of those individuals' lifetime dietary intake. It is thus possible that personal dietary reports may poorly describe actual individual intakes, but when sampled together in sufficiently large numbers may give an accurate representation of actual mean population consumption. In other words, a cancellation of errors in the individual NHANES estimated dietary intakes may be partly responsible for satisfactorily approximating dietary intake at the population level.In addition, the unreliability of dietary recall data likely also contributed to the model's ineffectiveness in accurately predicting individual exposures. In particular, the 24 hour recall and FFQ reports (which are used to estimate beef, dairy and fish consumption, respectively) are prone to recall bias, which leads to uncertain food consumption estimates.61–63 It has been reported previously that such methods tend to appreciably underestimate actual individual consumption rates.46,64 For example, NHANES energy intakes may be underreported by upwards of 800 kcal per day.46 Although we scaled up modeled dietary intakes to agree with the national US average diet, this does not address other known shortcomings of dietary intake data. For example, Freedman et al.65 observed that “across a diverse sample of Americans, subjective estimates of energy intake explained <10% of the variance of true intake”. Binnington et al.60 also identified the unreliability of dietary recall data as a key contributor to poor model performance for individuals. In particular, they noted that reported traditional food intake among Arctic populations had increased when compared to the previous decade, contradicting an expected decline in traditional food consumption.60 Furthermore, Shin et al.66 looked at estimating exposure to perfluorinated compounds, and also highlighted the need for better intake data to improve model estimates.
4.3 Evaluation of model predictions for environmental and food chain contamination
Considering the limitations of the dietary data, it is appropriate to also ask whether model-measurement agreement at the population level may be fortuitous. In order to explore this, we also compared the predicted PCB concentrations in air, sea water, fish, beef, and dairy lipids with measured values reported in the literature (ESI, Section S7†). Such a comparison is limited by the considerable variability of PCB concentrations in such samples and their potential to inadequately represent the average US food supply. Nevertheless, our modeled calculations are generally well within an order of magnitude of reported environment and food chain measurements. Often the agreement is much better, e.g. dairy concentrations (ESI, Table S9†). There is tendency for concentrations in fish to be underpredicted (ESI, Table S7†) and those in beef/meat to be overpredicted (ESI, Table S8†).Because the model-measurement agreement for the average levels in humans is better than model-measurement agreement for the food items, we suspect that the former is to some extent fortuitous, i.e., is a result of error cancellation. In particular, an overestimation of concentrations in meat could affect model results for humans, because NHANES participants on average consumed 47 times more meat lipids than fish lipids, and also because beef lipid intake correlates well with predicted PCB-153 exposure in our model calculations (R2 = 0.49, data not shown). Overall, it is thus likely that discrepancies in model to measurement agreement for individuals are not only due to shortcomings in the reported dietary intake data (and its extrapolation to lifetime consumption), but also to deviations between predicted and observed PCB levels in various food items.
Nevertheless, we note that a simpler modeling approach relying on empirical food contamination data would be similarly affected by the questionable reliability of dietary intake estimates at the individual level, i.e. it is not primarily the estimation of the food contamination that presently limits the accuracy of predicted individual exposures to PCBs, but the difficulty in reliably establishing what individuals eat.
4.4 Other limitations of the model approach
Individuals with unique exposure scenarios, such as those who lived in close proximity to a PCB manufacturing plant,67 are difficult to model with our approach. Additionally, individuals who consume locally produced livestock are difficult to describe because our model approach assumes that all individuals obtain their food from the same source, specifically the central US for beef and dairy, and the Pacific Ocean for fish. However, with geographical information and application of a regional-scale or ‘nested’ fate and transport model, parameterized for particular locations/food origin scenarios, prediction of PCB levels for both groups of individuals could be achieved.The low reliability of FFQs and 24 hour dietary recall data presently limits the feasibility of reconstructing individual exposure histories based on data typically collected in HBM studies. This implies that it may be difficult to improve exposure characterization in epidemiological studies of the health effect of contaminants through the use of such models,68–70 unless the quality of dietary intake information is improved. It also implies that the presence or absence of statistical associations between measured PCB levels and the reported intake of certain dietary items may be more uncertain than previously recognized. On the other hand, the averages of dietary intake data appear to allow for reasonably good predictions of both the mean and range of measured population PCB levels, especially if adjustment for the known bias towards underestimation of energy intake is performed.46,64 In particular, the ability to reproduce statistical associations (or the lack thereof) between PCB concentrations and non-dietary individual attributes such as age, sex, BMI or parity with the model approach is encouraging. It implies that our model can mechanistically explain such associations and also make predictions of such relationships. Lastly, the modelling approach described here can be adapted to national biomonitoring campaigns of other countries, for example, the Canadian Health Measures Survey (CHMS), or for other long-lived contaminants, provided the necessary information is available.
Acknowledgements
We gratefully acknowledge support from an Ontario Graduate Scholarship to SAW, and the Natural Sciences and Engineering Research Council of Canada.References
- J. E. Blundell, Am. J. Clin. Nutr., 2000, 71, 3–5 CAS.
- Agency for Toxic Substances and Disease Registry, Toxicological Profile for Polychlorinated Biphenyls (PCBs), Atlanta, GA, 2000 Search PubMed.
- G. A. Moser and M. S. McLachlan, Environ. Sci. Technol., 2002, 36, 3318–3325 CrossRef CAS PubMed.
- A. Schecter, P. Cramer, K. Boggess, J. Stanley, O. Päpke, J. Olson, A. Silver and M. Schmitz, J. Toxicol. Environ. Health, Part A, 2001, 63, 1–18 CrossRef CAS PubMed.
- N. Codru, M. J. Schymura, S. Negoita, R. Rej and D. O. Carpenter, Environ. Health Perspect., 2007, 115, 1442–1447 CAS.
- L. Rylander, A. Rignell-Hydbom and L. Hagmar, Environ. Health, 2005, 4, 28 CrossRef PubMed.
- O. Vasiliu, L. Cameron, J. Gardiner, P. Deguire and W. Karmaus, Epidemiology, 2006, 17, 352–359 CrossRef PubMed.
- C. J. Everett, A. G. Mainous III, I. L. Frithsen, M. S. Player and E. M. Matheson, Environ. Res., 2008, 108, 94–97 CrossRef CAS PubMed.
- M.-H. Ha, D.-H. Lee, H.-K. Son, S.-K. Park and D. R. Jacobs, J. Hum. Hypertens., 2008, 23, 274–286 Search PubMed.
- V. Persky, M. Turyk, H. A. Anderson, L. P. Hanrahan, C. Falk, D. N. Steenport, R. Chatterton, S. Freels and Great Lakes Consortium, Environ. Health Perspect., 2001, 109, 1275–1283 CrossRef CAS PubMed.
- M. E. Turyk, H. A. Anderson and V. W. Persky, Environ. Health Perspect., 2007, 115, 1197–1203 CrossRef CAS PubMed.
- J. Angerer, U. Ewers and M. Wilhelm, Int. J. Hyg. Environ. Health, 2007, 210, 201–228 CrossRef CAS PubMed.
- Centers for Disease Control (CDC), National Center for Health Statistics (NCHS), National Health and Nutrition Examination Survey Data, U.S. Department of Health and Human Services, Centers for Disease Control and Prevention, Hyattsville, MD, http://www.cdc.gov/nchs/nhanes.htm, accessed 1 May 2014.
- A. Sjödin, R. S. Jones, S. P. Caudill, L.-Y. Wong, W. E. Turner and A. M. Calafat, Environ. Sci. Technol., 2014, 48, 753–760 CrossRef PubMed.
- D. A. Axelrad, S. Goodman and T. J. Woodruff, Environ. Res., 2009, 109, 368–378 CrossRef CAS PubMed.
- F. Laden, L. M. Neas, D. Spiegelman, S. E. Hankinson, W. C. Willett, K. Ireland, M. S. Wolff and D. J. Hunter, Environ. Health Perspect., 1999, 107, 75–81 CrossRef CAS PubMed.
- A. L. Choi, J. I. Levy, D. W. Dockery and L. M. Ryan, Environ. Health Perspect., 2006, 114, 1092–1098 CAS.
- P. G. Tee, A. M. Sweeney, E. Symanski, J. C. Gardiner, D. M. Gasior and S. L. Schantz, Environ. Health Perspect., 2003, 111, 702–707 CrossRef CAS PubMed.
- A. Agudo, F. Goñi, A. Etxeandia, A. Vives, E. Millán, R. López, P. Amiano, E. Ardanaz, A. Barricarte, M. D. Chirlaque, M. Dorronsoro, P. Jakszyn, N. Larrañaga, C. Martínez, C. Navarro, L. Rodríguez, M. J. Sánchez, M. J. Tormo and C. A. González, Environ. Res., 2009, 109, 620–628 CrossRef CAS PubMed.
- D. H. Garabrant, A. Franzblau, J. Lepkowski, B. W. Gillespie, P. Adriaens, A. Demond, E. Hedgeman, K. Knutson, L. Zwica, K. Olson, T. Towey, Q. Chen, B. Hong, C.-W. Chang, S.-Y. Lee, B. Ward, K. Ladronka, W. Luksemburg and M. Maier, Environ. Health Perspect., 2009, 117, 818–824 CrossRef CAS PubMed.
- D. Bachelet, T. Truong, M.-A. Verner, P. Arveux, P. Kerbrat, C. Charlier, C. Guihenneuc-Jouyaux and P. Guénel, Environ. Res., 2011, 111, 861–870 CrossRef CAS PubMed.
- M. Pavuk, J. R. Olson, W. A. Wattigney, N. D. Dutton, A. Sjödin, C. Shelton, W. E. Turner and S. M. Bartell, Sci. Total Environ., 2014, 496, 624–634 CrossRef CAS PubMed.
- R. A. James, I. Hertz-Picciotto, E. Willman, J. A. Keller and M. J. Charles, Environ. Health Perspect., 2002, 110, 617–624 CrossRef CAS PubMed.
- S. D. Stellman, M. V. Djordjevic, J. A. Britton, J. E. Muscat, M. L. Citron, M. Kemeny, E. Busch and L. Gong, Cancer Epidemiol., Biomarkers Prev., 2000, 9, 1241–1249 CAS.
- M. S. Wolff, J. A. Britton, S. L. Teitelbaum, S. Eng, E. Deych, K. Ireland, Z. Liu, A. I. Neugut, R. M. Santella and M. D. Gammon, Cancer Epidemiol., Biomarkers Prev., 2005, 14, 2224–2236 CAS.
- M.-A. Verner, M. Charbonneau, L. López-Carrillo and S. Haddad, Environ. Health Perspect., 2008, 116, 886–892 CrossRef CAS PubMed.
- M. Lorber, Sci. Total Environ., 2002, 288, 81–95 CrossRef CAS PubMed.
- M.-A. Verner, D. Sonneborn, K. Lancz, G. Muckle, P. Ayotte, É. Dewailly, A. Kocan, L. Palkovicová, T. Trnovec, S. Haddad, I. Hertz-Picciotto and M. Eggesbø, Environ. Health Perspect., 2013, 121, 131–137 Search PubMed.
- R. Ritter, M. Scheringer, M. MacLeod, U. Schenker and K. Hungerbühler, Environ. Health Perspect., 2009, 117, 1280–1286 CAS.
- G. Czub and M. S. McLachlan, Environ. Toxicol. Chem., 2004, 23, 2356–2366 CrossRef CAS PubMed.
- F. Wania, K. Breivik, N. J. Persson and M. S. McLachlan, Environ. Model. Software, 2006, 21, 868–884 CrossRef.
- K. Breivik, G. Czub, M. S. McLachlan and F. Wania, Environ. Int., 2010, 36, 85–91 CrossRef CAS PubMed.
- C. L. Quinn, F. Wania, G. Czub and K. Breivik, Environ. Health Perspect., 2011, 119, 641–646 CrossRef CAS PubMed.
- E. Undeman, T. N. Brown, F. Wania and M. S. McLachlan, Environ. Sci. Technol., 2010, 44, 6249–6255 CrossRef CAS PubMed.
- C. L. Quinn and F. Wania, Environ. Health Perspect., 2012, 120, 554–559 CrossRef CAS PubMed.
- M. J. Binnington, C. L. Quinn, M. S. McLachlan and F. Wania, Environ. Health Perspect., 2014, 122, 178–186 Search PubMed.
- T. H. Nøst, K. Breivik, O.-M. Fuskevåg, E. Nieboer, J. Ø. Odland and T. M. Sandanger, Environ. Health Perspect., 2013, 121, 1292–1298 Search PubMed.
- K. Breivik, A. Sweetman, J. M. Pacyna and K. C. Jones, Sci. Total Environ., 2007, 377, 296–307 CrossRef CAS PubMed.
- J. M. Armitage and F. Wania, Environ. Sci.: Processes Impacts, 2013, 15, 2263–2272 CAS.
- K. Breivik, J. M. Armitage, F. Wania, A. J. Sweetman and K. C. Jones, Environ. Sci. Technol., 2016, 50, 798–805 CrossRef CAS PubMed.
- M. MacLeod, H. von Waldow, P. Tay, J. M. Armitage, H. Wöhrnschimmel, W. J. Riley, T. E. McKone and K. Hungerbühler, Environ. Pollut., 2011, 159, 1442–1445 CrossRef CAS PubMed.
- A. De Mul, M. I. Bakker, M. J. Zeilmaker, W. A. Traag, S. P. J. V. Leeuwen, R. L. A. P. Hoogenboom, P. E. Boon and J. D. V. Klaveren, Regul. Toxicol. Pharmacol., 2008, 51, 278–287 CrossRef CAS PubMed.
- United States Department of Agriculture (USDA), in Agriculture Fact Book, 2014, pp. 13–21 Search PubMed.
- A. I. Eidelman, R. J. Schanler et al., Pediatrics, 2012, vol. 129, pp. 827–841 Search PubMed.
- M. S. Wolff, H. A. Anderson, J. A. Britton and N. Rothman, Cancer Epidemiol., Biomarkers Prev., 2007, 16, 1925–1930 CAS.
- E. Archer, G. A. Hand and S. N. Blair, PLoS One, 2013, 8, e76632 CAS.
- K. Pozo, T. Harner, F. Wania, D. C. G. Muir, K. C. Jones and L. A. Barrie, Environ. Sci. Technol., 2006, 40, 4867–4873 CrossRef CAS PubMed.
- M. D. Ampleman, A. Martinez, J. DeWall, D. F. K. Rawn, K. C. Hornbuckle and P. S. Thorne, Environ. Sci. Technol., 2015, 49, 1156–1164 CrossRef CAS PubMed.
- W. A. Wattigney, E. Irvin-Barnwell, M. Pavuk and A. Ragin-Wilson, J. Environ. Public Health, 2015, 2015, 1–12 CrossRef PubMed.
- R. Ritter, M. Scheringer, M. MacLeod, C. Moeckel, K. C. Jones and K. Hungerbühler, Environ. Health Perspect., 2011, 119, 225–231 CrossRef CAS PubMed.
- T. E. Oliphant, Comput. Sci. Eng., 2007, 9, 10–20 CrossRef CAS.
- S. Van Der Walt and S. C. Colbert, Comput. Sci. Eng., 2011, 13, 22–30 CrossRef.
- W. McKinney, Python for Data Analysis, O'Reilly Media, Sebastol, 1st edn, 2013 Search PubMed.
- J. D. Hunter, Comput. Sci. Eng., 2007, 9, 90–95 CrossRef.
- L. L. Aylward, J. J. Collins, K. M. Bodner, M. Wilken and C. M. Bodnar, Chemosphere, 2014, 110, 48–52 CrossRef CAS PubMed.
- E. Undeman, G. Czub and M. S. McLachlan, Environ. Sci. Technol., 2009, 43, 3751–3756 CrossRef CAS PubMed.
- M. S. Wolff, A. Fischbein and I. J. Selikoff, Environ. Res., 1992, 59, 202–216 CrossRef CAS PubMed.
- S. A. Wood, F. Xu, J. M. Armitage and F. Wania, Unravelling the relationship between body mass index and polychlorinated biphenyl concentrations using a mechanistic model, Environ. Sci. Technol., 2016 Search PubMed , in press.
- T. H. Nøst, K. Breivik, F. Wania, C. Rylander, J. Ø. Odland and T. M. Sandanger, Environ. Health Perspect., 2016, 124, 299–305 Search PubMed.
- M. Binnington, M. S. Curren, C. L. Quinn, J. M. Armitage, J. A. Arnot, H. M. Chan and F. Wania, Environ. Int., 2016, 92–93, 256–258 CrossRef CAS PubMed.
- D. Bedard, B. Shatenstein and S. Nadon, Publ. Health Nutr., 2004, 7, 675–681 Search PubMed.
- V. Kipnis, D. Midthune, L. S. Freedman, S. Bingham, A. Schatzkin, A. Subar and R. J. Carroll, Am. J. Epidemiol., 2001, 153, 394–403 CrossRef CAS PubMed.
- V. Kipnis, D. Midthune, L. Freedman, S. Bingham, N. E. Day, E. Riboli, P. Ferrari and R. J. Carroll, Publ. Health Nutr., 2002, 5, 915–923 CrossRef PubMed.
- N. V. Dhurandhar, D. Schoeller, A. W. Brown, S. B. Heymsfield, D. Thomas, T. I. A. Sørensen, J. R. Speakman, M. Jeansonne, D. B. Allison and Energy Balance Measurement Working Group, Int. J. Obes., 2015, 39, 1109–1113 CrossRef CAS PubMed.
- L. S. Freedman, J. M. Commins, J. E. Moler, L. Arab, D. J. Baer, V. Kipnis, D. Midthune, A. J. Moshfegh, M. L. Neuhouser, R. L. Prentice, A. Schatzkin, D. Spiegelman, A. F. Subar, L. F. Tinker and W. Willett, Am. J. Epidemiol., 2014, 180, 116–188 CrossRef PubMed.
- H.-M. Shin, V. M. Vieira, P. B. Ryan, K. Steenland and S. M. Bartell, Environ. Health Perspect., 2011, 119, 1760–1765 CrossRef CAS PubMed.
- M. Pavuk, J. R. Olson, A. Sjödin, P. Wolff, W. E. Turner, C. Shelton, N. D. Dutton and S. Bartell, Sci. Total Environ., 2014, 473–474, 286–297 CrossRef CAS PubMed.
- M.-A. Verner, P. Ayotte, G. Muckle, M. Charbonneau and S. Haddad, Environ. Health Perspect., 2008, 117, 481–487 CrossRef PubMed.
- C. Rylander, T. M. Sandanger, T. H. Nøst, K. Breivik and E. Lund, Environ. Res., 2015, 142, 365–373 CrossRef CAS PubMed.
- M. A. Verner, P. Plusquellec, G. Muckle, P. Ayotte, É. Dewailly, S. W. Jacobson, J. L. Jacobson, M. Charbonneau and S. Haddad, Neurotoxicology, 2010, 31, 424–431 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available: Tables summarizing model input parameters for BETR-Global and ACC-Human; description of the BMI parameterization used in the model, conversion of dietary questionnaire to model input, and optimization of emissions timing and human biotransformation half-life; model results for the default/original results; additional model results for PCB-118, -138, and -180, including figures similar to Fig. 2 and 3; tables for comparison of model estimates for food chain contamination and measured levels, and for environmental contamination; lastly estimates of intrinsic elimination half-lives for the 4 PCB congeners. See DOI: 10.1039/c6em00424e |
| This journal is © The Royal Society of Chemistry 2016 |