A perspective on structural and computational work on collagen
Carmen
Domene
*ab,
Christian
Jorgensen
a and
Sumra Wajid
Abbasi†
a
aDepartment of Chemistry, King's College London, Britannia House, 7 Trinity Street, London SE1 1DB, UK. E-mail: carmen.domene@kcl.ac.uk; Tel: +44 (0)207848754
bChemistry Research Laboratory, University of Oxford, Mansfield Road, Oxford OX1 3TA, UK
First published on 17th August 2016
Abstract
Collagen is the single most abundant protein in the extracellular matrix in the animal kingdom, with remarkable structural and functional diversity and regarded one of the most useful biomaterials. Etymologically, the term collagen comes from Greek kola ‘glue’ and gen ‘giving birth to’. Thus, it is not surprising that the various collagens and the structures they form all serve the same purpose, to help tissues withstand stretching. Among the functions the various collagens are involved in are cell adhesion and migration, tissue repair, scaffolding and morphogenesis. Thus knowledge about the structure and properties of collagen, how they change depending on the nature of the local environment as well as the nature and specificity of collagen interactions with its partners is central to discerning the role of collagen in medical applications such as imaging, drug delivery and tissue engineering, and in the design and construction of synthetic collagen-like materials for tools in biomaterial science and nanotechnology. The main focus of this perspective is to review the molecular and packing structures of collagen and the computer simulations work performed up to now to further highlight the significance of collagen.
Introduction
Collagen is a key structural protein responsible for the constitutional integrity of vertebrates.1–5 It is a representative of insoluble crystalline proteins and a model for its structure was first proposed by Wyckoff, Corey and Biscoe in 1935 when they could not associate categorically the diffraction pattern with a chain-like structure or an ordinary molecular crystal built up by the regular arrangement of very large molecules.6 Subsequently, Astbury & Bell7 proposed that the collagen molecule consisted of a single extended polypeptide chain in the cis conformation. They suggested that the data presented earlier by Wyckoff and co-workers was better fitted using a gliadin class molecule. The first suggestion that the collagen structure might be helical was proposed by Pauling & Corey in 1951.8 The model was a three helical polypeptide chain with a one third content of proline or hydroxyproline. In their manuscript, they affirmed that ‘the structure of the molecule provided an immediate explanation of the principal mechanical property of collagen’. Independently, Cohen & Bear and Cowan, North & Randall in 1953 suggested the correct non-integer screw axis.9,10 One of Ramachandran's major contributions to structural biology was the proposal, made along with Kartha, of a triple-helical structure for the fibrous protein collagen published in Nature in 1954.11,12 At a time when the structure of only one globular protein had been determined, the model they proposed solved the controversy regarding the so-called short inter-atomic distances in previous collagen models, and re-examination of them led to the famous ‘Ramachandran plot’ which graphically illustrates the restrictions on polypeptide conformation.13 In 1955, this structure was refined by Rich & Crick14 and by North and coworkers15 to the triple-helical structure accepted today, with modifications focused on the arrangement of the hydrogen bonds as Ramachandra's models presented some stereochemical limitations.The molecular organization in the collagen fiber was also later determined and the axial repeat usually designated as the D period was established. D-periodic fibrils contain intermolecular covalent cross-links that lead to their high tensile strength and mechanical stability.16
In 1994, Berman and co-workers reported the first high-resolution crystal structure of triple-helical collagen-related peptides17–20 and subsequently, several other high resolution crystal structures of oligopeptides related to collagen as well as synthetic mimics were determined.
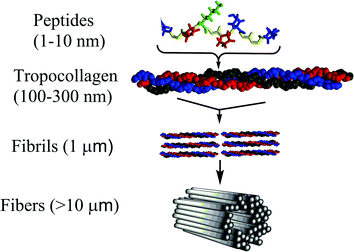
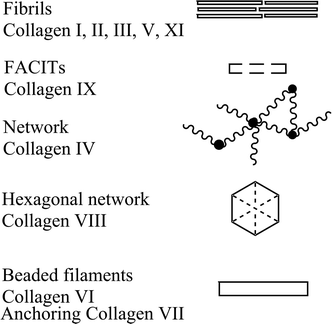
The umbrella term ‘collagen’ covers all proteins that form a right-handed three polypeptide three-dimensional complex (Fig. 1). This denomination is used for all the members belonging to the collagen family characterised by varying tissue distribution, size and function.1,21–25 Collagen is conserved in the animal kingdom, and it is a key component of the majority of tissues accounting for about 20–30% of total body proteins.26,27 Collagen is a primary component of bones, muscles, skin and tendons of vertebrates supporting delicate organs.28–34 Collagen types are classified in several sub-families according to sequence homologies and to similarities in their structural organization and supramolecular arrangement such as fibrils, networks and filaments (Scheme 1). To date, at least 29 different types of collagen have been identified with the designation I–XIX,35 along with additional proteins that have collagen-like domains including adiponectin, C1q macrophage receptor, acetyl cholinesterase, conglutinin, ectodysplasin collectin or ficolin among many others (Fig. 2).1 Crucially, insight into collagen–protein interactions will facilitate the progress in new approaches in drug discovery, targeting and delivery.36 Collagen types I, II and III account for the majority of collagen in the human body, and about one half of the total body collagen is in the skin, and about 70% of the material other than water present in dermis of skin and tendon is collagen.37 In contrast, invertebrate collagen genes encode only for fibrillar and basement membrane collagen.38 The presence of over a hundred bacterial protein sequences containing the collagen characteristic domain in genomic databases has been reported, which suggests a whole new family of collagen-like proteins.39
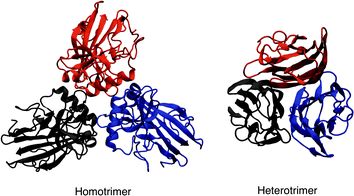 | ||
| Fig. 2 Example of homotrimeric (PDB id 4AE2) and heterotrimeric (PDB id 1PK6) collagen-like proteins. Each monomer is coloured differently. | ||
Therefore, understanding how the relatively simple molecular building blocks of collagen self-assemble is imperative for artificial tissue development, growth, regeneration, and disease, cosmetics formulation, pharmacology or in its application in plastic surgery and medicine in general.37
Structural characteristics of collagen
The molecular structure of collagen was assigned based on the evidence from studies of chemical composition, physicochemical analysis of solutions, X-ray crystallography and electron microscopy, and a variety of spectroscopy techniques.40,41 Under physiological conditions, the basic collagen molecule is rod-shaped with a length and a width of about 3000 and 15 Å, respectively.42 It consists of three polypeptide chains twined around one another with individual twists in the opposite directions and flanked at both termini by short non-helical telopeptides also known as amino-terminal collagen crosslinks. The three chains are staggered by one residue which creates non-identical environments for the three chains. Generally, the triple helix is composed by two identical chains and a third chain that differs in its chemical composition. The main constituents of collagen are glycine, which is found at almost every third residue, proline that makes up ∼28% of collagen, and two uncommon derivative amino acids, hydroxyproline and hydroxilysine both formed by enzymatic post-translational modification. Stereoelectronic effects and preorganization play a key role in collagen stability.4 Each chain contains a repeating (Gly-Pro-Y) or (Gly-X-Hyp) triad, consisting of a glycine repeat motif followed by proline or 4-hydroxyproline and various other residues in the X and Y positions, respectively (Fig. 1). The most common triplet in collagen is Pro-Hyp-Gly accounting for about 10% of the total sequence, and the peptide (Pro-Hyp-Gly)10 forms a very stable triple helix. A high content of glycine and amino acid residues dictates the principal features of collagen.43 The strands are held together primarily by inter-chain hydrogen bonds formed between individual chains with the involvement of hydroxyproline residues and facilitated by the high content of Gly residues.44 A single collagen molecule is referred as tropocollagen. Collagen structures are either homotrimers or heterotrimers with either two or three dissimilar chains.45 A group of tropocollagen molecules self-assemble to form fibrils having a distinct periodicity, and they also differ in their ability to form fibres and to organise the fibres into networks. Some covalent crosslinking within the triple helices can be identified as well as a variable amount of covalent crosslinking between tropocollagen molecules forming well organized aggregates.3 A group of tropocollagen may be broadly divided into fibrillar types and non-fibrillar collagens. The fibrillar collagen chains all have a perfect (Gly-X-Y)n repeating sequence. In contrast, all non-fibrillar collagens have sites where the repeating tripeptide pattern is interrupted.46Collagen I, IV, V, VI, IX, XI are heterotrimers, and the remaining are homotrimers. Among the 29 different types of collagens characterised up to now, type I, II, III, V, and XI are fibril-forming collagens Collagen fibrils are characterised by remarkable strength and stability conferred by its self-aggregation and cross-linking; for instance collagen type I fibrils are stronger than steel. These fibrils assemble into well-structured supra-molecular linear aggregates of length >1 μm with a characteristic supra-structure. Fibrillar collagens contain a relatively high content of charged residues (∼15–20%) and a small percentage of hydrophobic residues (∼6%).36 Fibers can form hydrogels, films or sponges. Several models have been also proposed for the arrangement of the tropocollagen within collagen fibrils. One of the initial ones was by Hodge and Petruska.47 In this model, five tropocollagen molecules are staggered side-by-side with an offset of 67 nm between two neighbors as revealed by transmission electron microscopy. Subsequently, Schmitt et al. proposed that 234 amino acid residues is the period of the helix. In other words, the length of the tropocollagen molecule is about 4.4 times that of the native collagen period.48 Not all collagens occur as periodic-structured fibrils though. However, the model cannot describe the spatial extension of the quarter staggered in two or three dimensions. This model is known as the Quarter staggered stacking model. Afterwards,48 the Smith model was proposed where five tropocollagen molecules are arranged concentrically into a hollow filament, known as the microfibril.49 Its limitation is that it cannot predict the organisation of fibrils with a diameter greater than 3.5 nm. Next, Hulmes and Miller proposed the Quasihexagonal packing model where periodic tropocollagen molecules were assigned the character of a molecular crystal and without microfibrillar sub-structures.50 The compressed microfibril model followed.51 In this model, five-stranded microfibrils are compressed to place the molecules in fibril cross-section on a pseudo-hexagonal lattice, and in the longitudinal direction the molecules are supercoiled with a left handed twist. Using X-ray diffraction, Orgel et al. presented the microfibrillar model which supported a microfibril structure composed by five staggered tropocollagen molecules arranged with a right-handed tilt, rather than just axially staggered.33 This model seems to be the one that best fits the native X-ray diffraction data and other experimental observations on the organization of the molecular segments in the overlap region of the fibril.
Type I collagen of fibril-forming collagens makes up over than 90% of organic mass of bones, as well as being the main collagen in a number connective tissues, including skin, tendons, cornea, ligaments, as well as vitreous body, brain, cartilage and hyaline tissues.2 Type II collagen makes up over 50% of all protein in cartilage and 85–90% of collagen of articular cartilage.2 Type III collagen is essential for type I fibrillogenesis. Normal type I collagen is a heterotrimer triple-helical linear molecule consisting of two α1 chains and one α2 chain.52 In contrast, type II and III form homotrimers, and can assemble into globular homotrimeric domains.
The fibril-associated collagens with interrupted triple helices (FACIT) subclass, comprise collagen types IX, XII, XIV, XVI, XIX, and XX; these structure are short non-helical domains.2 Type VI is a heterotrimeric collagen with small helical domains and stretched globular termini.53,54 Stretching the α3-chain of type VI, of double length when compared to the rest of chains, is due to the larger globular domain at the N- and C-termini. These extended collagen domains are subject to extensive intracellular and extracellular post-translational modifications.55,56
Collagen types VII and X are formed by short chains, and type X, is homotrimeric exhibiting long C-terminal and short N-terminal domains. In vivo studies revealed it assembles into hexagonal networks.57 Type VIII collagen is similar in structure to type X collagen, although it possesses a different spatial distribution which confers distinctive function.58
Collagen type IV is predominant in membranes, assimilating nitrogen atoms, the laminin proteins of the extracellular matrix, and other elements into a 2-dimensional supra-molecular aggregate. Type IV consists of a conformational flexible triple-helical structure with three domains. Until now, there are six recognised subunit chains (α1 (IV)–α6 (IV)). Among these chains, α1 (IV) 2α2 (IV) heterotrimers have been reported to be vital in forming important network in most embryonic as well as adult basement membranes.
Numerous experimental studies link the misregulation of collagen to a broad range of diseases. Collagen serves as binding sites for cytokines and multiple growth factor proteins. These cytokines and growth factors in turn regulate vital cell functions including survival, differentiation, motility and polarity.5,59 One of the therapeutic potential for collagen is in the field of drug delivery, where the binding ability of collagen makes it a promising agent in the delivery of drugs, while the anchoring and network-generation ability of some collagen types has potential in tissue regeneration and repair.60–62 Furthermore, experimental studies have widely pointed to deficiencies in type III collagen, as well as elastin, as linked to cardiac aneurysm formation.63,64 Finally, collagen bio prostheses have been studied for roles in surgery.65
Mutations in different regions can have different effects and defects at the molecular structure or collagen organization into mature fibers result in different diseases associated with connective tissues and even some types of osteoporosis and arthritis (1, 2). A number of excellent books and reviews on collagen describe the biochemical and biomedical aspects in detail,16,66 and some others focus on describing how understanding the key biochemical and physical properties of collagen lead to strategies to create, control and modify the structure and function of collagen-based biomaterials.36,67
Collagen-like peptide models and collagen–protein systems
The inability to crystallise collagen and its unsuitability for NMR characterisation rendered collagen model peptides an invaluable alternative to study native collagen structural features using biophysical techniques. Advances in peptide chemistry, in the 60's, set the stage for the synthesis and characterisation of poly-tripeptides to be used as collagen models.46 In these systems, the packing is reminiscent of collagen and usually, there is a high proportion of Pro-Hyp-Gly triplets. These studies have explored the effect of amino and imino acid content on the triple-helix structure, they have unravelled the basic principles of collagen self-assembly and into supramolecular structures and they have aided in clarifying the sequence dependent nature of biologically important features of native collagen and their alterations in diseased states as well as. Collagen mimetic peptide assemblies and functionalised collagen mimetic peptides have been also synthesised, and these have been reviewed elsewhere.68Bella and co-workers19 designed and crystallised at 1.9 Å resolution, a peptide to model the effect of interrupting the repeating (Gly-X-Y) motive with a single Gly substitution to Ala at the center of a 30-amino acid peptide as these substitution had been identified in several diseases. The crystal structure published in 1994 provided structural information on the effect of a glycine substitution in a triple helix, an alteration which usually leads to pathological states in fibrillar collagens.
A first structural analysis of the polypeptide (Pro-Gly-Pro)n was reported by Yonath and Traub in 196969 followed by work on fiber diffraction by Scheraga70–73 and Blout's groups.74–78 In 1981, Okuyama et al.18 presented crystallographic studies on the polypeptide model (Pro-Pro-Gly)10 and reported 72 symmetry as opposed to the 103 triple helical symmetry in earlier natural collagen studies which animated a debate around the actual symmetry of natural collagen. In the 90's, new polypeptide structural models were synthesised and characterised, and continued to illustrate key features of collagen related systems. More recently, the first report of the full-length structure of the collagen-like polypeptide [(Pro-Pro-Gly)10]3 at 1.3 Å was given by Berisio and co-workers in 2001. Model peptides have also been used to define the basic principles of collagen self-association to the supramolecular structures found in tissues.46,79
The interactions between the triple helical structure of collagen and proteins play important roles in collagen binding and degradation and for example in healing and repair of the body's tissues. Crystallography has yielded atomistic structures of a variety of collagen types, which has allowed for detailed studies of collagen complexes. Over the past decades, much information has been also gained about the interactions of collagen with cell surface receptors, extracellular matrix components and enzymes such as matrix metalloproteinases (MMPs). Currently, around over 300 different crystal structures of collagen in complex with other proteins have been reported, and some of these selected examples will be described next to illustrate relevant pivotal interactions and their relation to function. In addition, the triple helix structural motif is found in a few non-collagenous proteins.
The binding of a monoclonal antibody (MAb) to the triple-helical region of type III collagen was one of the first examples where a region of collagen which binds to another molecule was studied in some detail to clarify specific recognition and binding properties.80 The molecular features involved in triple-helix interactions with another macromolecule were characterised, and the observation of unstable Gly-Gly-Y triplets adjacent to the recognition region was made, suggesting involvement of some flexibility or instability near the actual binding site.
Human cysteine cathepsin is a protein crucial in pathophysiological and physiological cellular mechanisms, and is a key therapeutic agent for a range of diseases as it hydrolyses various extracellular matrix components among which are some types of collagens. Sage et al. have described an inhibition mechanism of this protein mediated by glycosaminoglycan that involved in vivo modulation of its collagenase activity.81
Another process of vital importance where protein–collagen interactions are crucial is the degradation of collagen to maintain the correct collagen homeostasis in tissues. In the collagenases, hemopexin C domain exosites bind native collagen, which is required for triple helicase activity during collagen cleavage. The active site of collagenolytic matrix metalloproteinases can only accommodate a single-chain of collagen. Thus the collagen helix must be initially unwound by a triple helicase in order to expose the scissile bonds, and then cleavage of the chains occurs sequentially. Models were proposed for the regulation of type I collagen levels upon stimulation of the activity of several matrix metalloproteinases. The collagen binding properties and the role of the ectodomain and the hemopexin C domain of the collagenolytic membrane type-1 matrix metalloproteinase (MT1-MMP) in collagenolysis were detailed charaterised.82 It was reported that collagen is a unique substrate for the proteases responsible for its cleavage, and these interactions recruit and regulate collagenolytic and gelatinolytic activities in a homeostatic manner.82
Computer simulations and modelling of collagen systems
It has become possible to clarify at the atomic level the features and interactions of collagen in various forms and environments, primarily due to progress in the experimental determination of three-dimensional structures of collagen and collagen–protein complexes together with improved algorithms for computer simulations and the increasing speed and availability of supercomputers.Molecular dynamics (MD) is a powerful computational technique that provides accurate descriptions of the structure and dynamics of biological systems, contributing to their understanding at an atomic level. In MD simulations, the motion of interacting atoms is calculated by integrating Newton's equations of motion. The potential energy of the system and the forces, derived from the negative gradient of the potential with respect to displacements in a specified direction, are used to forecast the time evolution of the system in the form of a trajectory. Equilibrium quantities are then calculated using statistical mechanics by averaging over trajectories of sufficient length which would have sampled a representative ensemble of the state of the system. Specific MD procedures to study tightly packed collagen have been described in the literature.83 These protocols are notably different from conventional MD simulations of proteins, which generally only treat individual protein molecules or complexes fully solvated.83 These alternative approaches exploit ideas borrowed from modelling crystalline solids such as periodic boundaries to replicate the super molecular arrangement of collagen proteins within fibrils.83 Numerous MD simulation studies applied to collagen have provided useful information to relate the diverse structural characteristics with the function.84–86 A representative example of a simulation system of tropocollagen–peptide complex in solution is illustrated in Fig. 3.
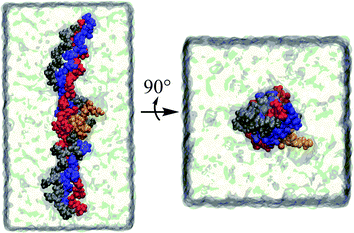 | ||
| Fig. 3 A tropocollagen–peptide complex in solution. Each of the four strands is coloured differently. | ||
Early computational studies in this area focused on microfibril and fibril packing issues such as longitudinal alignment and stabilization of fibrils.3,87–91 Subsequently, collagen-like peptides were used in combined experimental and theoretical calculations to get insight into the structural features of collagen.92–94 The structural importance of prolines and hydroxyproline for helix and fibril stability was established using computational models by Scheraga and coworkers.91 However, the first simulations in this area are the 0.5–1.0 ns molecular dynamics simulations of collagen-like peptides performed by Klein and Huang.95 This was followed by MD simulations studying the role hydroxylated prolines play in stabilising the collagen triple helix.96
Posterior work focused on the modelling of telopeptides, which are crucial for the formation of enzymatic covalent crosslinks in collagens near their N- and C-ends, as these crosslinks provide structural integrity, strength, and stiffness to collagenous tissues.97 One study reported conformational and packing studies of cross-linked structures of the fibril-forming type I collagen N-telopeptide heterotrimer.98 Due to the absence of high-resolution crystallographic structures of telopeptides, a triple-helical structure was built on the basis of crystallographic coordinates of a collagen-like sequence coordinates and then replaced with the actual bovine collagen residues. It was further found that if individual N-telopeptides were considered, their chain structures were essentially random, but when they were docked to their helix domain receptors, very ordered and specific conformations were created.98 A second study addressed type I collagen C-telopeptide conformations using all three chains of the heterotrimer before and after it was docked to its receptor domain.99 The computational models showed that the N- and C-telopeptide regions have different molecular packing and intrafibrillar crosslinking patterns that control the relative azimuthal orientations of molecules in the fibril.99 In a later study, the deformation mechanisms of N- and C-crosslinks and the functional roles for the N- and C-telopeptide conformations were investigated via MD simulations.97
Other computational work focused on mutagenic disruptions of collagen functioning, and their associated pathologies.100 In one study,101 collagen-like molecules designed to mimic the site of mutations in collagen type I are used in combination with MD simulations to contrast general structural properties of the peptides with and without the mutation to examine the effect of the single-point mutation on the surrounding residues.
On the development front, a novel set of molecular mechanics parameters for hydroxyproline by Park et al. allowed for the reproduction of the correct pucker preference of the collagen backbone motif, which were tested in a set of simulations of collagen-like peptides. The role of hydroxylation in the stability of the collagen triple helix by adjusting to the right pucker conformation was reproduced.102 Various other studies have been performed by several groups to investigate the relationship between interchain salt bridge formation and triple-helical stability using detailed molecular simulations with the aim to guide the design of collagen-like peptides that have specific interchain interactions.103 To further clarify the stereospecificity of ion pairs, MD simulations were computed for triple-helical peptides containing reversed sequences, comparing EGK with KGE, for example. In combination with experimental studies, the results indicated that the reversal of charges lowered thermal stability, highlighting the importance of cross-chain ionic interactions for the stability of the collagen triple helix in solution.104
MD simulations together with experiment have investigated the pathways and molecular mechanisms for peptide assembly into triple-helical protomers, as well as their subsequent organisation into structurally defined, linear assemblies.105 These studies showed that collagen-mimetic fibrils and microfibers, which are very similar to those formed in vivo, could be obtained through the linear assembly of a small collagen-mimetic peptide driven through electrostatic interactions with precisely defined periodic features with potential applications in material design. Experimental studies showed the positional preference of different amino acids to form a stable triple helical collagen motif, the structural basis for the variations in the sequence. The positional propensity was systematically investigated with computational techniques.106 Specifically, MD simulations were carried out on 39 collagen-like peptides showing that the propensity of the different amino acids to adopt collagen-like conformations depends primarily on their φ and ψ angle preferences.106
Several experimental and modelling studies have been carried out to understand mechanical properties of bone, a biological nanocomposite that exhibits a highly optimised and complex multi-level hierarchical structure composed primarily of type I collagen and hydroxyapatite.107 Among the computational studies, molecular dynamics and steered molecular dynamics were employed to characterise directional dependence of deformation response of collagen with respect to the hydroxyapatite surface107 and collagen interactions with rutile surfaces without hydroxylation.108 The early process in the nucleation of hydroxyapatite at a collagen template was studied by immersing a triple helical collagen molecule in a stoichiometric solution of Ca2+, PO43− and OH− ions and compared with simulations of collagen interacting with surfaces of hydroxyapatite from the crystal.109 In the context of drug delivery, medical diagnosis and molecular engineering, the interactions of collagen-like peptides with carbon nanotubes (CNTs) were also investigated with MD simulations.110,111 A collagen-like peptide with a hydrophobic center and hydrophilic surfaces could be inserted spontaneously but slowly and the mechanism of the encapsulation process was characterised. Two studies related to esthetic dentistry112,113 focused on dentin collagen fibrils which are formed during development. Dentin is one of the four major components of teeth. It is composed by 45% of hydroxylapatite and 33% of organic materials out of which 90% is collagen type 1, and the remaining dentine-specific proteins among which are proteases. These proteins add water across specific peptide bonds to solubilise ‘insoluble’ collagen. After development, apatite crystallites replace some of the water molecules in collagen but it is not clear the mechanism by which this occurs. Both studies focused on characterising how collagen interacts with adhesive monomers and whether these could displace all or just some proportion of water molecules from collagen intermolecular spaces using experimental or computational approaches. Other examples of computational studies of the interaction of collagen with materials are those with gold nanoparticles,114,115 graphene nanoribbons116 as well as those to get insight into collagen self-assembly on substrates.117
There are numerous computational studies illustrating collagen–protein interactions, and a few selected representative ones are highlighted below. To start, two related studies,118,119 were reported employing MD simulation to analyse (i) the structural effect on heterotrimeric models of triple helical peptides with interruptions in the Gly-X-Y repeats and (ii) the interactions of collagen with gelatinase-A, a matrix metalloproteinase, and the role of each domain of the protein in hydrolysing collagens with and without interruptions. Matrix metalloproteinases are members of the endogenous proteases mentioned earlier that hydrolyse collagen. The process of hydrolysis is relevant in a variety of physiological and pathological conditions and it involves breaking down the molecular bonds between individual collagen strands and peptides. Results from the first study118 showed the formation of a kink in the interrupted region of the triple-helical peptides and significantly differences in the hydrogen-bonding pattern due to singularities in the staggering of chains. In the second study,119 the authors proposed that the collagen binding domain binds to the C-terminal of collagen like peptides with interruption, helping in unwinding the loosely packed interrupted region. They speculated that the role of the hemopexin domain of the metalloproteinase is to prevent further unwinding of collagen by binding to the other end of the collagen like peptide. It was also postulated that subsequently, the catalytic domain would orient to interact with the partially unwound triple helix structure of the peptide to carry out hydrolysis. Next, extended MD simulations were reported to determine the most likely rearrangements of the domains of metalloproteinase-2 in response to the presence of the collagen triple helix.120 The authors pointed out that in spite of its physiological and pathological relevance, detailed structural information about the enzyme–substrate interactions during collagen hydrolysis catalysed by MMPs is not available. Different models for the interaction of the full-length MMP-2 enzyme and the synthetic collagen-like fTHP-5 were considered concluding that the full multidomain structure of MMP-2 is required for the studies of the interactions with collagen owing to its characteristic flexibility.120 The most significant MMP-2/fTHP-5 interactions at the catalytic and non-catalytic domains were also detailed gathering some clues about the role of the different domains during collagenolysis.
The binding of a C-terminal fragment of collagen XVIII, endostatin, to heparin and heparan sulfate was studied experimentally and further characterised by docking and molecular dynamics simulations.121 Endostatin interacts with the heparan sulfate chains of the cell surface contributing to its biological activities. The aim of this study was to determine the affinity of these interactions, identify the structural features of heparin/heparan sulfate–endostatin complexes, as well as to investigate the effect of divalent cations on the interaction.121
Collagen type II is a specific target in the collagen-induced arthritis model. In a study using a homology model of an antigen–antibody complex, using 200 ns MD trajectory, the critical amino acids conferring Collagen type II epitope specificity to a variety of autoantibodies were investigated. The presence of a few anchoring residues in the antibody regions was shown to be probably sufficient to confer a moderately high affinity key for the recognition.122
A combined computational and experimental study illustrated the nature of the ligand–receptor interactions between single and triple-helical strands of collagen and integrin. Integrins are the main receptor proteins that cells use to both bind to and respond to the extracellular matrix, with these interactions regulating many different cell functions, and thus a detailed understanding of the fine-tuning of collagen binding to integrin is essential as it might be a potential tool for therapeutic purposes.123 Combined NMR and MD simulation methods addressed the question of why single-stranded collagen fragments are unable to establish a stable specific binding interaction with the integrin receptor, finding only weak complexes in solution. In another study,124 a biomimetic design strategy of platelet adhesion inhibitors was proposed to develop potent inhibitors for the integrin α2β1-collagen binding, using a combination of molecular docking, structure similarity analysis, MD simulations and experimental validation.
All these studies highlight the potential for combined in silico and in vitro studies for extending our understanding of collagen–protein interactions.
Conclusions
Collagen is an example of a naturally occurring structural material composed by relatively simple chemical building blocks, the amino acids. The numerous types of collagen described in the literature have been reviewed in terms of their structure and interactions with other proteins and peptides. In addition, an overview of the computational studies carried out up-to-date has been presented, emphasising the potential of MD simulations to enhance our understanding of collagen. The importance of collagen has been highlighted through various of its applications, including the design of modern engineered biomaterials with precise formulated properties to serve a unique purpose, as well as constituting novel potent targets for the pharmaceutical industry for de novo drug design against a number of diseases as well as a drug delivery system owing to its excellent biocompatibility and safety. More than a thousand mutations in collagen are known to result in various diseases, including osteoporosis, arterial aneurysms, osteoarthrosis, as well as rare diseases.1 Tropoelastin has emerged as key biomarker to identify patients that are at risk of a heart attack, and binding imaging agents are under development to target tropoelastin and to provide an imagible signal that will allow clinicians to identify the location and extend of vulnerable plaques.125 The biomarker consists of a small peptide biding sequence coupled to a radioactive element that is detectable by MRI.125A wide variety of computational methods are currently used in the field of computational chemistry. Despite the universal availability of MD algorithms and forcefields applicable to macromolecules, the size of model systems and the computing resources simulations require pose inherent limitations. Recent expansion in computer hardware and high-performance computing facilities means MD simulations on a nanosecond timescale are now standard with microsecond simulations attainable in recent years. Using reduced representations, what is known as coarse-grained (CG) models, is one such approach, as this reduces the number of degrees of freedom in a simulation system by treating a group of atoms as a single entity, significantly curtailing the computational expense. Several algorithms also exist to accelerate sampling along a pre-defined set of reaction coordinates and estimate the potential of mean force providing a wealth of information about the simulation system at a fraction of the expense of traditional all-atom MD. Such methodologies are relevant to study and some have already been used to study some phenomena in which collagen is involved, for example, complex associations and conformational changes that are generally unattainable by atomistic equilibrium MD simulations. Computational simulations are steadily guiding the development of promising novel imaging agents for clinical use to facilitate personalised medicine by optimising the selection and dosing of disease therapies, and by improving the understanding of the underlying biology of a disease. By gleaning new insights into collagen interactions in bulk materials and in protein environments, computer simulations may accelerate the ability to understand the potential role of collagen in the design of tools for medical applications and broadly speaking in biomaterial science and nanotechnology. For example, the holy grail in cardiovascular prevention is to identify individuals at risk for myocardial infarction or stroke,126 and this is becoming possible by using non-invasive plaque detection where understanding of collagen–protein interactions at atomistic level is fundamental. Progress in these areas may allow earlier detection, may facilitate monitoring the response of the treatment, and overall, the provision of a more effective treatment.37,127 The increasing availability of high-resolution structural information, growth in computer capabilities and the development of state-of-the art algorithms and accompanying force fields will markedly amplify the use of computational simulations for the study of collagen–protein interactions in the coming years.
Acknowledgements
C. Domene thanks the Biotechnology and Biological Sciences Research Council and the Engineering and Physical Sciences Research Council for financial support. C. Jorgensen thanks King's College London for a Graduate Teaching Assistant studentship. S. W. Abbasi thanks the High Commission of Pakistan for a travel fellowship award.Notes and references
- J. Myllyharju and K. I. Kivirikko, Ann. Med., 2001, 33, 7–21 CrossRef CAS PubMed.
- E. P. K. Gelsea and T. Aigner, Adv. Drug Delivery Rev., 2003, 55, 1531–1546 CrossRef.
- S. Perumal, O. Antipova and J. P. R. O. Orgel, Proc. Natl. Acad. Sci. U. S. A., 2008, 105, 2824–2829 CrossRef CAS PubMed.
- M. D. Shoulders and R. T. Raines, Annu. Rev. Biochem., 2009, vol. 78, pp. 929–958 Search PubMed.
- J. C. Brown and R. Timpl, Int. Arch. Allergy Immunol., 1995, 107, 484–490 CrossRef CAS PubMed.
- R. W. Wyckoff, R. B. Corey and J. Biscoe, Science, 1935, 82, 175–176 CAS.
- W. Astbury and F. Bell, Nature, 1940, 145, 421–422 CrossRef CAS.
- L. Pauling and R. B. Corey, Proc. Natl. Acad. Sci. U. S. A., 1951, 37, 272–281 CrossRef CAS.
- C. Cohen and R. S. Bear, J. Am. Chem. Soc., 1953, 75, 2783–2784 CrossRef CAS.
- P. M. Cowan, A. T. C. North and J. T. Randall, in The Nature and Structure of Collagen, ed. J. T. Randall, Butterworths, London, 1953, p. 241 Search PubMed.
- G. N. Ramachandran and G. Kartha, Nature, 1954, 174, 269–270 CrossRef CAS PubMed.
- G. N. Ramachandran and G. Kartha, Nature, 1955, 176, 593–595 CrossRef CAS PubMed.
- A. Bhattacharjee and M. Bansal, IUBMB Life, 2005, 57, 161–172 CrossRef CAS PubMed.
- A. Rich and F. H. C. Crick, J. Mol. Biol., 1961, 3, 483–710 CrossRef CAS PubMed.
- P. M. Cowan, S. McGavin and A. C. T. North, Nature, 1955, 176, 1062–1064 CrossRef CAS PubMed.
- Collagens and Gelatins, ed. B. B. Brodsky, J. Werkmeister and J. Ramshaw, Wiley-VCH, ch. 6, 2005, vol. 108, pp. 119–128 Search PubMed.
- R. Z. Kramer, J. Bella, B. Brodsky and H. M. Berman, J. Mol. Biol., 2001, 311, 131–147 CrossRef CAS PubMed.
- K. Okuyama, K. Okuyama, S. Arnott, M. Takayanagi and M. Kakudo, J. Mol. Biol., 1981, 152, 427–443 CrossRef CAS PubMed.
- J. Bella, M. Eaton, B. Brodsky and H. M. Berman, Science, 1994, 266, 75–81 CAS.
- R. Berisio, L. Vitagliano, L. Mazzarella and A. Zagari, Protein Sci., 2002, 11, 262–270 CrossRef CAS PubMed.
- K. V. D. Mark, in Dynamics of Bone and Cartilage Metabolism: Principles and Clinical Applications, ed. S. R. Markus Seibel and J. Bilezikian, Academic Press, 2006, ch. 1, pp. 3–26 Search PubMed.
- K. Kühn, Rheumatology, 1986, 10, 29–69 Search PubMed.
- R. Mayne and R. G. Brewton, Curr. Opin. Cell Biol., 1993, 5, 883–890 CrossRef CAS PubMed.
- M. van der Rest and R. Garrone, FASEB J., 1991, 5, 2814–2823 CAS.
- K. Sato, K. Yomogida, T. Wada, T. Yorihuzi, Y. Nishimune, N. Hosokawa and K. Nagata, J. Biol. Chem., 2002, 277, 37678–37684 CrossRef CAS PubMed.
- K. Kadler, Protein Profile, 1995, 2, 491–619 CAS.
- R. D. Harkness, Biol. Rev. Cambridge Philos. Soc., 1961, 36, 399–463 CrossRef CAS PubMed.
- J. F. V. Vincent, Structural biomaterials, Princeton University Press, Princeton, NJ, Rev. edn, 1990 Search PubMed.
- R. W. Peter Fratzl, Prog. Mater. Sci., 2007, 52, 1263–1334 CrossRef.
- R. K. Nalla, J. J. Kruzic, J. H. Kinney and R. O. Ritchie, Biomaterials, 2005, 26, 217–231 CrossRef CAS PubMed.
- R. O. Ritchie, J. J. Kruzic, C. L. Muhlstein, R. K. Nalla and E. A. Stach, Int. J. Fract., 2004, 128, 1–15 CrossRef CAS.
- D. J. S. Hulmes, T. J. Wess, D. J. Prockop and P. Fratzl, Biophys. J., 1995, 68, 1661–1670 CrossRef CAS PubMed.
- J. P. R. O. Orgel, T. C. Irving, A. Miller and T. J. Wess, Proc. Natl. Acad. Sci. U. S. A., 2006, 103, 9001–9005 CrossRef CAS PubMed.
- J. W. Freeman and F. H. Silver, J. Theor. Biol., 2004, 229, 371–381 CrossRef CAS PubMed.
- S. Ricard-Blum, Cold Spring Harbor Perspect. Biol., 2010, 3, a004978 Search PubMed.
- B. An, Y.-S. Lin and B. Brodsky, Adv. Drug Delivery Rev., 2016, 97, 69–84 CrossRef CAS PubMed.
- C. H. Lee, A. Singla and Y. Lee, Int. J. Pharm., 2001, 221, 1–22 CrossRef CAS PubMed.
- J. Huxley-Jones, D. L. Robertson and R. P. Boot-Handford, Matrix Biol., 2007, 26, 2–11 CrossRef CAS PubMed.
- C. Xu, Z. Yu, M. Inouye, B. Brodsky and O. Mirochnitchenko, Biomacromolecules, 2010, 11, 348 CrossRef CAS PubMed.
- J. Gross, Biochim. Biophys. Acta, 1963, 71, 250–252 CrossRef CAS.
- G. N. Ramachan and V. Sasisekh, Biochim. Biophys. Acta, 1965, 109, 314–316 CrossRef.
- W. Traub and K. A. Piez, Adv. Protein Chem., 1971, 25, 243–352 CrossRef CAS PubMed.
- K. A. Piez, in Extracellular Matrix Biochemistry, ed. A. H. R. E. K. A. Piez, Elsevier, New York, 1984, pp. 1–40 Search PubMed.
- B. Brodsky and A. V. Persikov, Adv. Protein Chem., 2005, 70, 301–339 CrossRef CAS PubMed.
- G. N. Ramachandran, Nature, 1956, 177, 710–711 CrossRef CAS PubMed.
- B. Brodsky, G. Thiagarajan, B. Madhan and K. Kar, Biopolymers, 2008, 89, 345–353 CrossRef CAS PubMed.
- J. A. Petruska and A. J. Hodge, Proc. Natl. Acad. Sci. U. S. A., 1964, 51, 871–876 CrossRef CAS.
- C. Schmitt, H. CE and M. Jakns, J. Cell. Comp. Physiol., 1942, 11–33 CrossRef.
- J. W. Smith, Nature, 1968, 219, 157–158 CrossRef CAS PubMed.
- D. J. S. Hulmes and A. Miller, Nature, 1979, 282, 878–880 CrossRef CAS PubMed.
- K. A. Piez and B. J. Trus, Biosci. Rep., 1981, 1, 801–810 CrossRef CAS PubMed.
- S.-W. Chang, S. J. Shefelbine and M. J. Buehler, Biophys. J., 2012, 102, 640–648 CrossRef CAS PubMed.
- D. Weil, M. G. Mattei, E. Passage, V. C. N'Guyen, D. Pribula-Conway, K. Mann, R. Deutzmann, R. Timpl and M. L. Chu, Am. J. Hum. Genet., 1988, 42, 435–445 CAS.
- M.-L. Chu, K. Mann, R. Deutzmann, D. Pribula-Conway, C.-C. Hsu-Chen, M. P. Bernard and R. Timpl, Eur. J. Biochem., 1987, 168, 309–317 CrossRef CAS PubMed.
- T. Aigner, L. Hambach, S. Soder, U. Schlotzer-Schrehardt and E. Poschl, Biochem. Biophys. Res. Commun., 2002, 290, 743–748 CrossRef CAS PubMed.
- M.-L. C. Rupert Timpl, in Extracellular Matrix Assembly and Structure, ed. R. P. M. a. D. E. B. P. D. Yurchenco, Academic Press, Orlando, 1994, pp. 207–242 Search PubMed.
- T. F. L. T. M. Schmidin Structure and Function of Collagen Types, ed. R. E. B. E. R. Mayne, Academic Press, Orlando, 1987, pp. 195–222 Search PubMed.
- N. Yamaguchi, R. Mayne and Y. Ninomiya, J. Biol. Chem., 1991, 266, 4508–4513 CAS.
- B. D. a. M. v. d. Ricard-Blum, Unconventional Collagens Types VI, VII, VIII, IX, X, XIV, XVI and XIX, Oxford University Press, 2000 Search PubMed.
- D. Schuppan, M. Schmid, R. Somasundaram, R. Ackermann, M. Ruehl, T. Nakamura and E. O. Riecken, Gastroenterology, 1998, 114, 139–152 CrossRef CAS.
- S. R. Frenkel, B. Toolan, D. Menche, M. I. Pitman and J. M. Pachence, J. Bone Jt. Surg., Br. Vol., 1997, 79B, 831–836 CrossRef.
- S. Wakitani, T. Kimura, A. Hirooka, T. Ochi, M. Yoneda, N. Yasui, H. Owaki and K. Ono, J. Bone Jt. Surg., Br. Vol., 1989, 71, 74–80 CAS.
- R. J. Rizzo, W. J. McCarthy, S. N. Dixit, M. P. Lilly, V. P. Shively, W. R. Flinn and J. S. Yao, J. Vasc. Surg., 1989, 10, 365–373 CrossRef CAS PubMed.
- P. Dobrin and R. Mrkvicka, Vascular, 1994, 2, 484–488 CAS.
- V. Samouillan, A. Lamure, E. Maurel, J. Dandurand, C. Lacabanne, F. Ballarin and M. Spina, Med. Biol. Eng. Comput., 2000, 38, 226–231 CAS.
- E. A. Abou Neel, L. Bozec, J. C. Knowles, O. Syed, V. Mudera, R. Day and J. K. Hyun, Adv. Drug Delivery Rev., 2013, 65, 429–456 CrossRef CAS PubMed.
- B. D. Walters and J. P. Stegemann, Acta Biomater., 2014, 10, 1488–1501 CrossRef CAS PubMed.
- Y. Li and S. M. Yu, Curr. Opin. Chem. Biol., 2013, 17, 968–975 CrossRef CAS PubMed.
- A. Yonath and W. Traub, J. Mol. Biol., 1969, 43, 461–477 CrossRef CAS PubMed.
- M. H. Miller, G. Nemethy and H. A. Scheraga, Macromolecules, 1980, 13, 470–478 CrossRef CAS.
- M. H. Miller, G. Nemethy and H. A. Scheraga, Macromolecules, 1980, 13, 910–913 CrossRef CAS.
- M. H. Miller and H. A. Scheraga, J. Polym. Sci., Part C: Polym. Symp., 1976, 171–200 CAS.
- G. Nemethy, M. H. Miller and H. A. Scheraga, Macromolecules, 1980, 13, 914–919 CrossRef CAS.
- F. R. Brown, E. R. Blout and A. J. Hopfinge, J. Mol. Biol., 1972, 63, 101–115 CrossRef CAS PubMed.
- B. B. Doyle, W. Traub, G. P. Lorenzi and E. R. Blout, Biochemistry, 1971, 10, 3052–3060 CrossRef CAS PubMed.
- B. B. Doyle, E. G. Bendit and E. R. Blout, Biopolymers, 1975, 14, 937–957 CrossRef CAS PubMed.
- B. B. Doyle, W. Traub, G. P. Lorenzi, F. R. Brown and E. R. Blout, J. Mol. Biol., 1970, 51, 47–59 CrossRef CAS PubMed.
- F. R. Brown, E. R. Blout, A. D. Corato and G. P. Lorenzi, J. Mol. Biol., 1972, 63, 85–89 CrossRef CAS PubMed.
- K. V. Simon-Lukasik, A. V. Persikov, B. Brodsky, J. A. M. Ramshaw, W. R. Laws, J. B. A. Ross and R. D. Ludescher, Biophys. J., 2003, 84, 501–508 CrossRef CAS PubMed.
- N. K. Shah, M. Sharma, A. Kirkpatrick, J. A. M. Ramshaw and B. Brodsky, Biochemistry, 1997, 36, 5878–5883 CrossRef CAS PubMed.
- J. Sage, F. Mallevre, F. Barbarin-Costes, S. A. Samsonov, J. P. Gehrcke, M. T. Pisabarro, E. Perrier, S. Schnebert, A. Roget, T. Livache, C. Nizard, G. Lalmanach and F. Lecaille, Biochemistry, 2013, 52, 6487–6498 CrossRef CAS PubMed.
- E. M. Tam, Y. I. Wu, G. S. Butler, M. S. Stack and C. M. Overall, J. Biol. Chem., 2002, 277, 39005–39014 CrossRef CAS PubMed.
- I. Streeter and N. H. de Leeuw, J. Phys. Chem. B, 2010, 114, 13263–13270 CrossRef CAS PubMed.
- P. H. M. Torres, G. L. S. C. Sousa and P. G. Pascutti, Proteins, 2011, 79, 2684–2692 CrossRef CAS PubMed.
- I. E. Andersson, C. D. Andersson, T. Batsalova, B. Dzhambazov, R. Holmdahl, J. Kihlberg and A. Linusson, PLoS One, 2011, 6, e17881 CAS.
- S. H. Prior, T. S. Byrne, D. Tokmina-Roszyk, G. B. Fields and S. R. Van Doren, J. Biol. Chem., 2016, 291, 7888–7901 CrossRef CAS PubMed.
- L. W. Cunningham, H. A. Davies and R. G. Hammonds, Biopolymers, 1976, 15, 483–502 CrossRef CAS PubMed.
- E. Y. Jones and A. Miller, J. Mol. Biol., 1991, 218, 209–219 CrossRef CAS PubMed.
- R. D. B. Fraser, T. P. Macrae, A. Miller and E. Suzuki, J. Mol. Biol., 1983, 167, 497–521 CrossRef CAS PubMed.
- E. Lee, G. Nemethy, H. A. Scheraga and V. S. Ananthanarayanan, Biopolymers, 1984, 23, 1193–1206 CrossRef CAS PubMed.
- G. Nemethy and H. A. Scheraga, Biochemistry, 1986, 25, 3184–3188 CrossRef CAS PubMed.
- B. Brodsky, M. H. Li, C. G. Long, J. Apigo and J. Baum, Biopolymers, 1992, 32, 447–451 CrossRef CAS PubMed.
- C. G. Long, E. Braswell, D. Zhu, J. Apigo, J. Baum and B. Brodsky, Biochemistry, 1993, 32, 11688–11695 CrossRef CAS PubMed.
- M. G. Venugopal, J. A. M. Ramshaw, E. Braswell, D. Zhu and B. Brodsky, Biochemistry, 1994, 33, 7948–7956 CrossRef CAS PubMed.
- T. E. Klein and C. C. Huang, Biopolymers, 1999, 49, 167–183 CrossRef CAS PubMed.
- S. D. Mooney, P. A. Kollman and T. E. Klein, Biopolymers, 2002, 64, 63–71 CrossRef CAS PubMed.
- A. L. Kwansa, R. De Vita and J. W. Freeman, Matrix Biol., 2014, 34, 161–169 CrossRef CAS PubMed.
- J. P. Malone, A. George and A. Veis, Proteins: Struct., Funct., Genet., 2004, 54, 206–215 CrossRef CAS PubMed.
- J. P. Malone and A. Veis, Biochemistry, 2004, 43, 15358–15366 CrossRef CAS PubMed.
- K. H. Lee and M. M. B. Holl, Biopolymers, 2011, 95, 401–409 CrossRef CAS PubMed.
- R. J. Radmer and T. E. Klein, Biochemistry, 2004, 43, 5314–5323 CrossRef CAS PubMed.
- S. Park, R. J. Radmer, T. E. Klein and V. S. Pande, J. Comput. Chem., 2005, 26, 1612–1616 CrossRef CAS PubMed.
- T. Gurry, P. S. Nerenberg and C. M. Stultz, Biophys. J., 2010, 98, 2634–2643 CrossRef CAS PubMed.
- N. Keshwani, S. Banerjee, B. Brodsky and G. I. Makhatadze, Biophys. J., 2013, 105, 1681–1688 CrossRef CAS PubMed.
- S. Rele, Y. Song, R. P. Apkarian, Z. Qu, V. P. Conticello and E. L. Chaikof, J. Am. Chem. Soc., 2007, 129, 14780–14787 CrossRef CAS PubMed.
- S. S. Raman, R. Gopalakrishnan, R. C. Wade and V. Subramanian, J. Phys. Chem. B, 2011, 115, 2593–2607 CrossRef PubMed.
- D. R. Katti, S. M. Pradhan and K. S. Katti, J. Biomech., 2010, 43, 1723–1730 CrossRef PubMed.
- M. Chen, T. Zheng, C. Wu and C. Xing, Colloids Surf., B, 2014, 121, 150–157 CrossRef CAS PubMed.
- N. Almora-Barrios and N. H. De Leeuw, Cryst. Growth Des., 2012, 12, 756–763 CAS.
- Y. Kang, Q. Wang, Y. C. Liu, T. Wu, Q. Chen and W. J. Guan, J. Phys. Chem. B, 2008, 112, 4801–4807 CrossRef CAS PubMed.
- R. Gopalakrishnan, K. Balamurugan, E. R. A. Singam, S. Sundaraman and V. Subramanian, Phys. Chem. Chem. Phys., 2011, 13, 13046–13057 RSC.
- M. Takahashi, M. Nakajima, J. Tagami, D. L. S. Scheffel, R. M. Carvalho, A. Mazzoni, M. Cadenaro, A. Tezvergil-Mutluay, L. Breschi, L. Tjäderhane, S. S. Jang, F. R. Tay, K. A. Agee and D. H. Pashley, Acta Biomater., 2013, 9, 9522–9528 CrossRef CAS PubMed.
- S. E. Jee, J. Zhou, J. Tan, L. Breschi, F. R. Tay, G. Grégoire, D. H. Pashley and S. S. Jang, Acta Biomater., 2016, 36, 175–185 CrossRef CAS PubMed.
- R. Gopalakrishnan, E. R. Azhagiya Singam, J. Vijaya Sundar and V. Subramanian, Phys. Chem. Chem. Phys., 2015, 17, 5172–5186 RSC.
- R. Gopalakrishnan, E. R. Azhagiya Singam, J. Vijaya Sundar and V. Subramanian, Phys. Chem. Chem. Phys., 2015, 17, 5172–5186 RSC.
- S. Ebrahimi, A. Montazeri and H. Rafii-Tabar, Comput. Mater. Sci., 2013, 69, 29–39 CrossRef CAS.
- B. Narayanan, G. H. Gilmer, J. Tao, J. J. De Yoreo and C. V. Ciobanu, Langmuir, 2014, 30, 1343–1350 CrossRef CAS PubMed.
- E. R. A. Singam, K. Balamurugan, R. Gopalakrishnan, S. R. Subramanian, V. Subramanian and T. Ramasami, Biopolymers, 2012, 97, 847–863 CrossRef CAS PubMed.
- E. R. Azhagiya Singam, V. Rajapandian and V. Subramanian, Biopolymers, 2014, 101, 779–794 CrossRef CAS PubMed.
- N. Díaz and D. Suárez, Biochemistry, 2015, 54, 1243–1258 CrossRef PubMed.
- S. Ricard-Blum, O. Feraud, H. Lortat-Jacob, A. Rencurosi, N. Fukai, F. Dkhissi, D. Vittet, A. Imberty, B. R. Olsen and M. van der Rest, J. Biol. Chem., 2004, 279, 2927–2936 CrossRef CAS PubMed.
- U. Boiers, H. Lanig, B. Sehnert, R. Holmdahl and H. Burkhardt, Eur. J. Immunol., 2008, 38, 2784–2795 CrossRef PubMed.
- M. B.-R. Hans-Christian Siebert, T. Eckert, S. Stötzel, U. Kirch, T. Diercks, M. J. Humphries, M. Frank, R. Wechselberger, E. Tajkhorshid and S. Oesser, Protein Cell, 2010, 1, 393–405 CrossRef PubMed.
- L. Zhang, C. Zhang and Y. Sun, Langmuir, 2014, 30, 4734–4742 CrossRef CAS PubMed.
- G. Passacquale, P. Di Giosia and A. Ferro, Cardiovasc. Res., 2016, 109, 9–23 CrossRef PubMed.
- F. A. Jaffer and J. W. Verjans, Heart, 2014, 100, 1469–1477 CrossRef PubMed.
- A. Phinikaridou, M. E. Andia, S. Lacerda, S. Lorrio, M. R. Makowski and R. M. Botnar, Molecules, 2013, 18, 14042–14069 CrossRef CAS PubMed.
Footnote |
| † Present address: Computational Biology Lab, National Center for Bioinformatics, Quaid-i-Azam University, Islamabad, Pakistan. |
| This journal is © the Owner Societies 2016 |