Peptide microarray-based fluorescence assay for simultaneously detecting matrix metalloproteinases†
Zhen
Lei
ab,
Jiaxue
Gao
ab,
Xia
Liu
ab,
Dianjun
Liu
a and
Zhenxin
Wang
*a
aState Key Laboratory of Electroanalytical Chemistry, Changchun Institute of Applied Chemistry, Chinese Academy of Sciences, Changchun, 130022, P. R. China. E-mail: wangzx@ciac.ac.cn
bUniversity of Chinese Academy of Sciences, No. 19A Yuquan Road, Beijing, 100049, P. R. China
First published on 8th September 2015
Abstract
Matrix metalloproteinases (MMPs) are believed to play an important role in tumor invasion. Herein, a peptide microarray-based fluorescence assay has been proposed for simultaneously determining the activities of MMP-2 and MMP-9 through the strong binding affinity of fluorescein isothiocyanate modified avidin (avidin-FITC) with the immobilized biotinylated peptide substrate on the microarray. In the presence of MMPs, the biotin moiety is released from the microarray by enzymatic cleavage of the peptide substrate, resulting in a significant decrease of the fluorescence signal. Under the optimal experimental conditions, the fluorescence intensity changes (ΔF%) are proportional to the concentrations of MMP-2 and MMP-9 within the ranges of 50 pg mL−1 to 50 ng mL−1 and 50 pg mL−1 to 100 ng mL−1 in the enzyme mixture, respectively. The detection limits are 45 pg mL−1 for MMP-2, and 60 pg mL−1 for MMP-9. In particular, the activities of extracellular MMP-2 and MMP-9 are determined by the peptide microarray-based fluorescence assay, and satisfactory results are obtained.
1 Introduction
Matrix metalloproteinases (MMPs) are a family of zinc-dependent endopeptidases that are able to digest various kinds of extracellular matrix (ECM) proteins and basement membrane components.1 MMPs have been found to play a critical role in several pathological and physiological processes, such as inflammation, arthritis and embryonic development.1–4 Particularly, the upregulation of MMPs is closely related to the metastasis of cancer,5 making MMPs important biomarkers for early cancer diagnosis and targets for therapeutic drug development.6 Among MMPs, MMP-2 (gelatinase A) and MMP-9 (gelatinase B) are considered as important enzymes in tumor invasion, because they are able to degrade type IV collagen which is the main structural protein component in the ECM and basement membrane.7 Hence, the sensitive and accurate analysis of MMP-2 and MMP-9 functionalities is of great importance for understanding the disease mechanism, and developing disease diagnostic and therapeutic methods.Until now, a number of bioanalytical approaches have been developed for detecting MMPs including enzyme-linked immunosorbent assay (ELISA),8,9 gelatinase zymography,10,11 fluorescence resonance energy transfer (FRET) assay,12,13 surface plasmon resonance (SPR)14,15 and electrochemical detection.16,17 The ELISA is the most widely used assay which can offer satisfactory specificity, sensitivity, and good selectivity of MMP determination. However, the ELISA requires high-quality anti-MMP antibodies, leading to a significant increase of testing cost. Although gelatinase zymography is an accurate and sensitive electrophoretic method to analyze the activities of MMPs, it is time-consuming and limited to assay of a small number of MMPs. Recently, peptide microarrays, as important tools for high-throughput screening and multiple analysis, have gained great interest in studying the functionalities and inhibition of enzymes including kinases, proteases and phosphatases.18–22 For instance, Yao and co-authors have developed a peptide aldehyde small-molecule microarray (SMM) for profiling the activity of cysteine proteases in native biological samples.18 Microarray-based assays enable simultaneous examination of enzyme activities across different cell states by taking advantage of the key characteristics of microarrays (miniaturization, high density and high throughput).
In the present work, we report a peptide microarray-based fluorescence assay for measuring the activities of MMPs through the high affinity of biotin with avidin. In the proof of principle experiment, we demonstrated that the assay can be employed to determine simultaneously MMP-2 and MMP-9 activities in both pure enzyme mixtures and cellular secretion.
2 Experimental section
2.1 Materials and reagents
The latent forms of matrix metalloproteinase-2 (proMMP-2) and matrix metalloproteinase-9 (proMMP-9) were purchased from Sino Biological Inc. (Beijing, China). 4-Aminophenyl mercuric acetate (APMA) and the MMP-2 and MMP-9 activity quantitative determination kits were offered by GenMed Medical Science and Technology Ltd (Shanghai, China). Peptide substrates (S0, CAEGFFSARGHRPLK (biotin); S1, CGGKGPRSLSGK (biotin); S2, CGGKGPLGLARK (biotin); and S3, CGGKGPLGVRGAK (biotin)) were synthesized by ChinaPeptides Ltd (Shanghai, China). Bovine serum albumin (BSA) was purchased from GEN-VIEW Scientific Inc. (USA), and avidin-FITC was acquired from Sigma-Aldrich Chemical Co. (USA). Phorbol-12-myristate-13-acetate (PMA) was purchased from Beyotime Institute of Biotechnology Ltd (Jiangsu, China). The human breast cancer cell line (MDA-MB-231) was obtained from the Chinese Academy of Sciences Cell Bank (Shanghai, China). Leibovitz's L-15 Medium was purchased from Sigma-Aldrich Chemical Co. (USA), and fetal bovine serum (FBS) was obtained from HyClone Co. (USA). Aldehyde 3-D glass slides and polytetrafluoroethylene (PTFE) masks were purchased from CapitalBio Ltd (Beijing, China). All other chemicals were of analytical grade. Milli-Q water (18.2 MΩ cm) was used in all experiments.2.2 Fabrication of peptide microarrays and MMP cleavage
Peptide microarrays were manufactured by the standard procedure using a SmartArrayer 136 system as our previously reported method with a slight modification (see the ESI† for details).23 Various concentrations of proMMPs in TCNB buffer were incubated with 1 mM APMA at 37 °C for various times. After being activated with APMA, 27 μL solutions of pure MMP-2, MMP-9 or the MMP mixture (MMP-2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) MMP-9 = 1
MMP-9 = 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 (mass ratio)) were applied to each subarray and incubated at 37 °C for desired times, respectively. The subarray incubated with blank TCNB buffer was employed as a control experiment. Then, the PTFE masks were removed and the microarrays were subjected to a series of washing steps: (1) 30 mL of washing buffer for 5 min (3 times), (2) 30 mL of washing buffer without Triton X-100 for 5 min (3 times), and (3) 30 mL Milli-Q water for 3 min (3 times), respectively. After being dried by centrifugation (300 g), the subarrays were incubated with 27 μL avidin-FITC (3 μM) in probe buffer (pH 7.5, 50 mM PB, 0.15 M NaCl, supplemented with 0.1% Tween-20 (v/v) and 1% BSA (w/v)) at 37 °C for 1 h. Subsequently, the microarrays were washed and dried as previously described.
1 (mass ratio)) were applied to each subarray and incubated at 37 °C for desired times, respectively. The subarray incubated with blank TCNB buffer was employed as a control experiment. Then, the PTFE masks were removed and the microarrays were subjected to a series of washing steps: (1) 30 mL of washing buffer for 5 min (3 times), (2) 30 mL of washing buffer without Triton X-100 for 5 min (3 times), and (3) 30 mL Milli-Q water for 3 min (3 times), respectively. After being dried by centrifugation (300 g), the subarrays were incubated with 27 μL avidin-FITC (3 μM) in probe buffer (pH 7.5, 50 mM PB, 0.15 M NaCl, supplemented with 0.1% Tween-20 (v/v) and 1% BSA (w/v)) at 37 °C for 1 h. Subsequently, the microarrays were washed and dried as previously described.
2.3 Data processing
The microarrays were scanned by using a LuxScan-10K fluorescence microarray scanner (CapitalBio, Beijing). The background signal originating from the slide was recorded and subtracted prior to evaluation. The mean values and standard deviations of the signals were determined from 6 spot replicates per sample, respectively. The change of fluorescence intensity (ΔF%) was calculated by the following equation: ΔF% = (F0 − F)/F0 × 100%; where F0 and F are the mean fluorescence signal intensity of 6 spots on the same microarray without and with treatment of MMPs, respectively.2.4 Detecting the activities of cell secreted MMP-2 and MMP-9
MDA-MB-231 cells were plated in a 48-well culture plate at various densities, and cultured in fresh L-15 medium supplemented with 10% FBS and 100 U mL−1 antibiotics (penicillin and streptomycin) in humidified air without CO2 at 37 °C for 24 h. Then, the cells were washed with PBS, and starved in serum-free medium for 12 h. The starved cells were stimulated by using PMA of different concentrations in serum-free L-15 medium for another 24 h. The culture media were collected and centrifuged (3000 rpm) at 4 °C for 10 min. Finally, the as-obtained supernatants were applied to subarrays (27 μL per subarray) and treated as previously described for evaluating the activities of cell secreted MMP-2 and MMP-9. For comparison, the activities of cell secreted MMP-2 and MMP-9 were also determined by using commercial determination kits as per the manufacturer's instructions.3 Results and discussion
3.1 Principle of experiment
Scheme 1 outlines the principle of the peptide microarray-based fluorescence assay for determining the activities of MMPs. Briefly, the biotinylated peptide substrates were spotted on the commercial aldehyde 3-D glass slides using a standard robotic procedure and immobilized through the formation of a Schiff base linkage between the aldehyde group on the slide surface and the N-terminal of the peptide.23 The biotin group of the peptide substrate can react with avidin-FITC, which generates a strong fluorescence signal. In the presence of MMPs, the peptide fragments carrying biotin moieties are released from the glass slide surface by enzymatic cleavage of peptide substrates. As a result, the fluorescence signal is decreased and the change of fluorescence intensity is dependent on the activity of MMPs. Due to the high throughput nature of the peptide microarray, the assay is capable of multiplex analysis by immobilizing different MMP substrates simultaneously. In this case, MMP-2 and MMP-9 were used as model MMPs to address the multiplexed detection ability of the assay because they belong to the same subfamily and have a striking resemblance in structure.24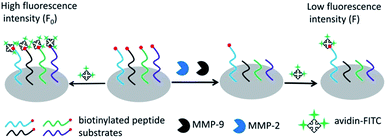 | ||
| Scheme 1 The schematic representation of the peptide microarray-based fluorescence assay for simultaneously determining MMPs. | ||
3.2 Specificity of peptide substrates
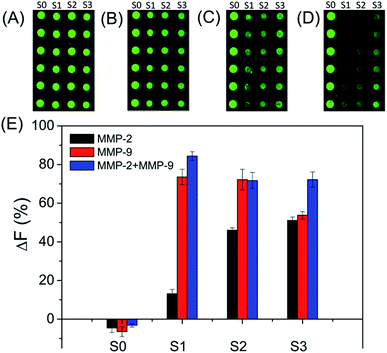
The specificities of four biotinylated peptides (S0 to S3) were firstly examined, since the non-specific reaction of the substrate with MMP can lead to false results (as shown in Fig. 1). As expected, S0 is not cleaved by both MMP-2 and MMP-9. S1 can be efficiently cleaved by MMP-9, suggesting that S1 has excellent specificity for MMP-9. The result is consistent with previously reported results.25–27 S2 and S3 are cleaved by both MMP-2 and MMP-9. The MMP-2 cleavage efficiency of S3 is better than that of S2 because S3 has a MMP-2 sensitive fragment ‘PLGVR’.28 In addition, the MMP-9 cleavage efficiency of S3 is lower than that of S2. Therefore, S1 and S3 were used for evaluating the amounts of MMP-9 and MMP-2, respectively.3.3 Optimization of experimental conditions
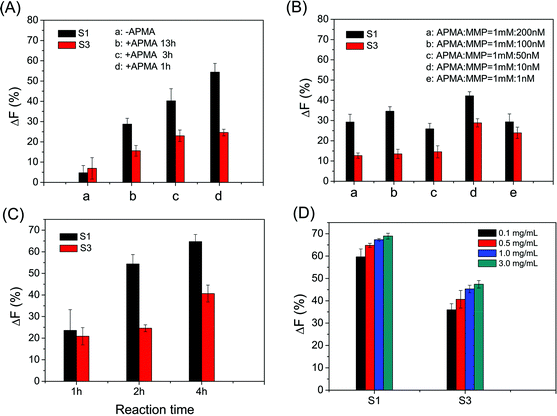
The cleavage efficiency of MMPs on the microarray could be affected by several conditions, such as the activation of MMPs, the peptide density on the microarray and the reaction time of activated MMPs with immobilized peptide substrates. The optimal activation of MMPs is achieved by the incubation of APMA and MMPs under the molar ratio of 100![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 for 1 h (as shown in Fig. 2A and B). Satisfactory ΔF% is obtained from the reaction of activated MMP with the immobilized peptide substrate for 4 h (as shown in Fig. 2C). Considering both ΔF% and assay cost, 0.5 mg mL−1 S1 and 1.0 mg mL−1 S3 in spotting solutions were used for determining MMP-9 and MMP-2 activities, respectively (as shown in Fig. 2D). In the following study, all results shown were obtained under the optimum conditions: MMP was activated by 100 times (molar ratio) APMA for 1 h, the activated MMP reacted with the immobilized peptide substrate for 4 h, and 0.5 mg mL−1 S1 and 1.0 mg mL−1 S3 in spotting solutions.
1 for 1 h (as shown in Fig. 2A and B). Satisfactory ΔF% is obtained from the reaction of activated MMP with the immobilized peptide substrate for 4 h (as shown in Fig. 2C). Considering both ΔF% and assay cost, 0.5 mg mL−1 S1 and 1.0 mg mL−1 S3 in spotting solutions were used for determining MMP-9 and MMP-2 activities, respectively (as shown in Fig. 2D). In the following study, all results shown were obtained under the optimum conditions: MMP was activated by 100 times (molar ratio) APMA for 1 h, the activated MMP reacted with the immobilized peptide substrate for 4 h, and 0.5 mg mL−1 S1 and 1.0 mg mL−1 S3 in spotting solutions.
3.4 Performance of peptide microarray-based fluorescence assay
In order to detect the amount of MMP-2 and MMP-9, a series of standard activated MMP solutions with various concentrations were prepared and applied to different subarrays. As shown in Fig. 3, the ΔF% increased linearly with the logarithm of MMP concentration. The ΔF% shows a linear correlation with the logarithm of the MMP concentration within the ranges from 100 pg mL−1 to 50 ng mL−1 for MMP-2, and 50 pg mL−1 to 50 ng mL−1 for MMP-9. The detection limits (3 times the standard deviation of ΔF% of control samples) are 84 pg mL−1 for MMP-2 and 10 pg mL−1 for MMP-9, respectively. The assay performance (dynamic range and detection limit) is much better than those of reported methods (e.g., electrophoresis-, SPR- and FRET-based assays).13–15,29–31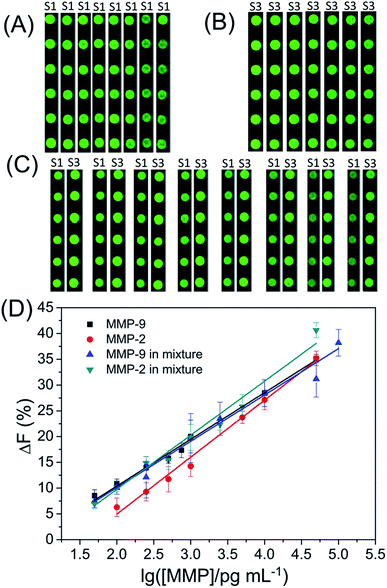 | ||
| Fig. 3 Fluorescence images of subarrays (A to C) and (D) the fluorescence intensity change (ΔF%) of the substrate S1 or S3 as a function of the logarithm of MMP concentration. | ||
As shown in Fig. S1,† the ΔF% of S1 shows a negligible change with increasing concentration of MMP-2 in the MMP mixture while the ΔF% of S1 shows a linear correlation with the logarithm of the MMP-9 concentration in the MMP mixture within the range from 50 pg mL−1 to 100 ng mL−1 (Fig. 3). The detection limit is 60 pg mL−1 for MMP-9 in the MMP mixture. The experimental result demonstrates that S1 can be used to detect MMP-9 in the MMP mixture. Most of the assays for the simultaneous determination of multiple protease activities are based on the assumption that the total observed cleavage for a particular substrate in the enzyme mixture is the addition of cleavages from each individual protease in the mixture.32,33 Although S3 is cleaved by both MMP-2 and MMP-9, the observed ΔF% of S3 for the MMP mixture is not equal to the sum of ΔF% of S3 for MMP-2 and ΔF% of S3 for MMP-9 in two separate experiments. After carefully comparing the ΔF% of S3 for MMP-2, MMP-9 or MMP mixture, we found that the ΔF% of S3 for the MMP mixture = 0.626 × ΔF% of S3 for MMP-2 + 0.488 × ΔF% of S3 for MMP-9 (as shown in Fig. S2 and S3†). Because the concentration of MMP-9 in the MMP mixture is easily defined by the ΔF% of S1, the ΔF% of S3 for MMP-2 in the MMP mixture can be calculated by the previous equation. Finally, the linear regression equation: ΔF% of S3 for the MMP mixture = −13.89 + 10.75 × log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) CMMP-2 (R2 = 0.98832) is obtained for evaluating the actual concentration of MMP-2 in the MMP mixture. Furthermore, the detection limit is 45 pg mL−1 for MMP-2 in the mixture. For comparison, the linear regression equations and detection limits are summarized in Table 1. The results demonstrate that our assay can be employed to determine multiple MMPs simultaneously.
CMMP-2 (R2 = 0.98832) is obtained for evaluating the actual concentration of MMP-2 in the MMP mixture. Furthermore, the detection limit is 45 pg mL−1 for MMP-2 in the mixture. For comparison, the linear regression equations and detection limits are summarized in Table 1. The results demonstrate that our assay can be employed to determine multiple MMPs simultaneously.
| Linear regression equations | Linear ranges | Detection limits | |
|---|---|---|---|
| MMP-2 | ΔF% = −17.15 + 11.05 × log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) CMMP-2 (R2 = 0.99275) CMMP-2 (R2 = 0.99275) |
100 pg mL−1 to 50 ng mL−1 | 84 pg mL−1 |
| MMP-9 | ΔF% = −7.71 + 9.06 × log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) CMMP-9 (R2 = 0.99589) CMMP-9 (R2 = 0.99589) |
50 pg mL−1 to 50 ng mL−1 | 10 pg mL−1 |
| MMP-2 in MMP mixture | ΔF% = −13.89 + 10.75 × log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) CMMP-2 (R2 = 0.98832) CMMP-2 (R2 = 0.98832) |
50 pg mL−1 to 50 ng mL−1 | 25 pg mL−1 |
| MMP-9 in MMP mixture | ΔF% = −7.87 + 8.98 × log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) CMMP-9 (R2 = 0.99115) CMMP-9 (R2 = 0.99115) |
50 pg mL−1 to 100 ng mL−1 | 60 pg mL−1 |
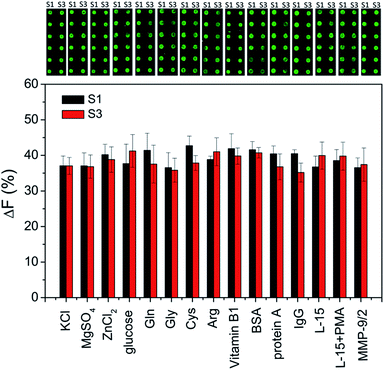
For addressing the practical applicability of the assay, the effects of potential interfering substances that may exist in biological samples (such as inorganic ions, sugars, amino acids, vitamins and other proteins) on the enzymatic cleavage are investigated. As shown in Fig. 4, a negligible effect on ΔF% is observed, indicating that the peptide microarray-based fluorescence assay may be used for detecting MMPs in complex biological samples.
3.5 Determination of cell secreted MMP-2 and MMP-9
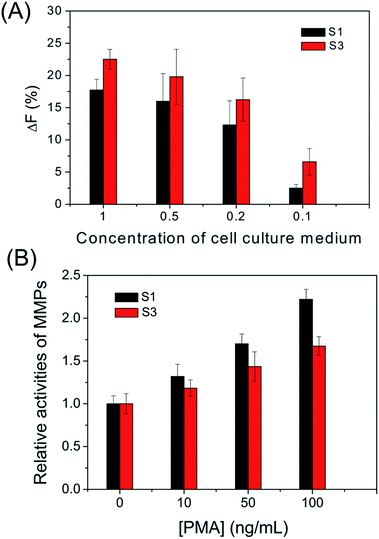
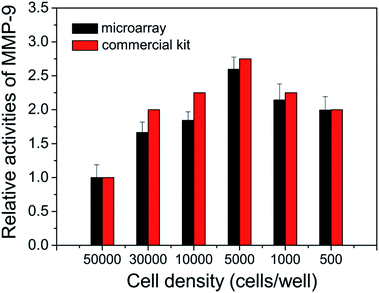
To further investigate its capability, the cell secreted MMPs are determined by the peptide microarray-based fluorescence assay. In this case, the well-known breast carcinoma cell line, MDA-MB-231, is selected as a model cell line.34,35 After being incubated with MDA-MB-231 cells, the cell culture medium was pretreated and applied to microarrays. As expected, the ΔF% of S1 and S3 increased with increasing concentration of cell culture medium. The experimental result indicates that MDA-MB-231 cells secrete MMP-2 and MMP-9 (as shown in Fig. 5A and S4†). The MMPs of PMA-stimulated MDA-MB-231 cells have also been measured, since the cellular expression of MMP-9 is highly regulated by cytokines and signal transduction pathways, and PMA is generally applied as a specific activator of the ERK pathway.36 As shown in Fig. 5B, the ΔF% of S1 for 100 ng mL−1 PMA-stimulated MDA-MB-231 cells is about 2.2 fold greater than that of the untreated MDA-MB-231 cells, indicating that the cell secretion level of MMP-9 is clearly activated by PMA. The result is consistent with the literature.36 In addition, the effect of cell density on MMP-9 secretion was studied, since high cell density can inhibit the expression level of MMPs of breast carcinoma cell lines.37 As shown in Fig. 6, the cell secretion level of MMP-9 increased with decreasing cell density. The result further demonstrates that the peptide microarray-based fluorescence assay can be used to screen the activities of the cell secreted MMPs.The activities of the cell secreted MMP-2 and MMP-9 are also tested by using commercially available MMP-2 and MMP-9 activity quantitative determination kits (as shown in Fig. S4, S5† and 6). The results of the peptide microarray-based fluorescence assay are comparable with the results of the commercial kits.
4 Conclusions
In summary, a peptide microarray-based fluorescence assay has been proposed for detecting MMP-2 and MMP-9 in both MMP mixtures and extracellular secretion with relative high sensitivity and a wide dynamic range. We have demonstrated the utility of the assay in determining extracellular MMP-2 and MMP-9 activity levels of untreated- and PMA-stimulated MDA-MB-231 cells. Although only MMP-2 and MMP-9 are determined in the proof of principle experiment, our approach provides a potential tool with a high throughput format for detecting MMPs in extracellular secretion and screening substrates of MMPs.Acknowledgements
The authors would like to thank the National Natural Science Foundation of China (Grant no. 21475126 and 21127010) and Chinese Academy of Sciences (YZ201354 and YZ201355) for financial support.Notes and references
- H. Nagase and J. F. Woessner, J. Biol. Chem., 1999, 274, 21491–21494 CrossRef CAS PubMed.
- O. Zitka, J. Kukacka, S. Krizkov, D. Huska, V. Adam, M. Masarik, R. Prusa and R. Kizek, Curr. Med. Chem., 2010, 17, 3751–3768 CrossRef CAS PubMed.
- G. Murphy and H. Nagase, Mol. Aspects Med., 2008, 29, 290–308 CrossRef CAS PubMed.
- M. Ram, Y. Sherer and Y. Shoenfeld, J. Clin. Immunol., 2006, 26, 299–307 CrossRef CAS PubMed.
- E. I. Deryugina and J. P. Quigley, Cancer Metastasis Rev., 2006, 25, 9–34 CrossRef CAS PubMed.
- M. Egeblad and Z. Werb, Nat. Rev. Cancer, 2002, 2, 161–174 CrossRef CAS PubMed.
- C. E. Brinckerhoff and L. M. Matrisian, Nat. Rev. Mol. Cell Biol., 2002, 3, 207–214 CrossRef CAS PubMed.
- S. Patel, G. Sumitra, B. Koner and A. Saxena, Clin. Biochem., 2011, 44, 869–872 CrossRef CAS PubMed.
- U. Pieper-Fürst, U. Kleuser, W. F. Stöcklein, A. Warsinke and F. W. Scheller, Anal. Biochem., 2004, 332, 160–167 CrossRef PubMed.
- F. Barbosa, F. Gerlach and J. E. Tanus-Santos, Basic Clin. Pharmacol. Toxicol., 2006, 98, 559–564 CrossRef CAS PubMed.
- T. Yamane, M. Mitsumata, N. Yamaguchi, T. Nakazawa, K. Mochizuki, T. Kondo, T. Kawasaki, S.-i. Murata, Y. Yoshida and R. Katoh, Cell Tissue Res., 2010, 340, 471–479 CrossRef CAS PubMed.
- S. Lee, E. J. Cha, K. Park, S. Y. Lee, J. K. Hong, I. C. Sun, S. Y. Kim, K. Choi, I. C. Kwon and K. Kim, Angew. Chem., 2008, 120, 2846–2849 CrossRef.
- Y. Wang, P. Shen, C. Li, Y. Wang and Z. Liu, Anal. Chem., 2012, 84, 1466–1473 CrossRef CAS PubMed.
- S.-H. Jung, D.-H. Kong, J. H. Park, S.-T. Lee, J. Hyun, Y.-M. Kim and K.-S. Ha, Analyst, 2010, 135, 1050–1057 RSC.
- O. R. Bolduc, J. N. Pelletier and J.-F. o. Masson, Anal. Chem., 2010, 82, 3699–3706 CrossRef CAS PubMed.
- D.-S. Shin, Y. Liu, Y. Gao, T. Kwa, Z. Matharu and A. Revzin, Anal. Chem., 2012, 85, 220–227 CrossRef PubMed.
- G.-C. Fan, L. Han, H. Zhu, J.-R. Zhang and J.-J. Zhu, Anal. Chem., 2014, 86, 12398–12405 CrossRef CAS PubMed.
- H. Wu, J. Ge, P.-Y. Yang, J. Wang, M. Uttamchandani and S. Q. Yao, J. Am. Chem. Soc., 2011, 133, 1946–1954 CrossRef CAS PubMed.
- M. Köhn, M. Gutierrez-Rodriguez, P. Jonkheijm, S. Wetzel, R. Wacker, H. Schroeder, H. Prinz, C. M. Niemeyer, R. Breinbauer and S. E. Szedlacsek, Angew. Chem., Int. Ed., 2007, 46, 7700–7703 CrossRef PubMed.
- C. M. Salisbury, D. J. Maly and J. A. Ellman, J. Am. Chem. Soc., 2002, 124, 14868–14870 CrossRef CAS PubMed.
- M. Schutkowski, U. Reimer, S. Panse, L. Dong, J. M. Lizcano, D. R. Alessi and J. Schneider-Mergener, Angew. Chem., 2004, 116, 2725–2728 CrossRef.
- H. Sun, C. H. Lu, M. Uttamchandani, Y. Xia, Y. C. Liou and S. Q. Yao, Angew. Chem., Int. Ed., 2008, 47, 1698–1702 CrossRef CAS PubMed.
- T. Li, D. Liu and Z. Wang, Anal. Chem., 2010, 82, 3067–3072 CrossRef CAS PubMed.
- Z. Yue, P. Lv, H. Yue, Y. Gao, D. Ma, W. Wei and G. Ma, Chem. Commun., 2013, 49, 3902–3904 RSC.
- S. J. Kridel, E. Chen, L. P. Kotra, E. W. Howard, S. Mobashery and J. W. Smith, J. Biol. Chem., 2001, 276, 20572–20578 CrossRef CAS PubMed.
- R. Fudala, A. P. Ranjan, A. Mukerjee, J. K. Vishwanatha, Z. Gryczynski, J. Borejdo, P. Sarkar and I. Gryczynski, Curr. Pharm. Biotechnol., 2011, 12, 834–838 CAS.
- R. Fudala, R. Rich, A. Mukerjee, A. P Ranjan, J. K Vishwanatha, A. K Kurdowska, Z. Gryczynski, J. Borejdo and I. Gryczynski, Curr. Pharm. Biotechnol., 2013, 14, 1134–1138 CAS.
- E. Song, D. Cheng, Y. Song, M. Jiang, J. Yu and Y. Wang, Biosens. Bioelectron., 2013, 47, 445–450 CrossRef CAS PubMed.
- R. B. Lefkowitz, G. W. Schmid-Schönbein and M. J. Heller, Anal. Chem., 2010, 82, 8251–8258 CrossRef CAS PubMed.
- A. Shoji, M. Kabeya and M. Sugawara, Anal. Biochem., 2011, 419, 53–60 CrossRef CAS PubMed.
- Z. Wang, X. Li, D. Feng, L. Li, W. Shi and H. Ma, Anal. Chem., 2014, 86, 7719–7725 CrossRef CAS PubMed.
- M. A. Miller, L. Barkal, K. Jeng, A. Herrlich, M. Moss, L. G. Griffith and D. A. Lauffenburger, Integr. Biol., 2011, 3, 422–438 RSC.
- F. H. Rasmussen, N. Yeung, L. Kiefer, G. Murphy, C. Lopez-Otin, M. P. Vitek and M. L. Moss, Biochemistry, 2004, 43, 2987–2995 CrossRef CAS PubMed.
- M. Roomi, J. Monterrey, T. Kalinovsky, M. Rath and A. Niedzwiecki, Oncol. Rep., 2009, 21, 1323–1333 CAS.
- S. Moulik, S. Pal, J. Biswas and A. Chatterjee, J. Tumor, 2014, 2, 87–98 Search PubMed.
- H. Ling, H. Yang, S. H. Tan, W. K. Chui and E. H. Chew, Br. J. Pharmacol., 2010, 161, 1763–1777 CrossRef CAS PubMed.
- B. E. Bachmeier, A. G. Nerlich, R. Vené, C. M. Iancu, B. Mayer, D. Noonan, A. Albini, M. Jochum and U. Pfeffer, Cancer Res., 2005, 65, 761–769 Search PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c5ay02041g |
| This journal is © The Royal Society of Chemistry 2016 |