Open Access Article
Open Access ArticleAn electrochemical biosensor for rapid detection of E. coli O157:H7 with highly efficient bi-functional glucose oxidase-polydopamine nanocomposites and Prussian blue modified screen-printed interdigitated electrodes†
Meng
Xu
a,
Ronghui
Wang
a and
Yanbin
Li
*ab
aDepartment of Biological and Agricultural Engineering, University of Arkansas, Fayetteville, AR 72701, USA. E-mail: yanbinli@uark.edu; Tel: +1 (479) 575-2881
bCenter of Excellence for Poultry Science, University of Arkansas, Fayetteville, AR 72701, USA
First published on 15th June 2016
Abstract
The presence of pathogenic bacteria in foods has always been a great threat to the wellbeing of people and the revenue of food manufacturers. Therefore, the demand for advanced detection methods that can sensitively and rapidly detect these pathogens has been of great importance. This study reports an electrochemical biosensor for rapid detection of E. coli O157:H7 with the integration of bifunctional glucose oxidase (GOx)–polydopamine (PDA) based polymeric nanocomposites (PMNCs) and Prussian blue (PB) modified screen-printed interdigitated microelectrodes (SP-IDMEs). The core–shell magnetic beads (MBs)–GOx@PDA PMNCs were first synthesized by the self-polymerization of dopamine (DA). Gold nanoparticles (AuNPs) were dispersed on the surface of PMNCs through biochemical synthesis to achieve further highly efficient adsorption of antibodies (ABs) and GOx. The final product ABs/GOxext/AuNPs/MBs–GOx@PDA PMNCs served as the carrier to separate target bacteria from food matrices as well as the amplifier for electrochemical measurement. The unbound PMNCs were separated by a filtration step and transferred into glucose solution to allow the enzymatic reaction to occur. The change of the current response was measured with an electrochemical detector using PB-modified SP-IDMEs. The constructed biosensor has been proven to be able to detect E. coli O157:H7 with the detection limit of 102 cfu ml−1. The bifunctional PMNCs contain a high load of enzyme and can optimally utilize the binding sites on bacterial cells, which efficiently amplify the signals for measurement. The biosensor in this study exhibited good specificity, reproducibility, and stability and is expected to have a great impact on applications in the detection of foodborne pathogens.
Introduction
Escherichia coli O157:H7 is the most common Shiga toxin-producing strain of E. coli in North America and can cause illness with a very low dose (10 to 100 cells). Symptoms include severe stomach cramps, bloody diarrhea, vomiting, or even life-threatening hemolytic uremic syndrome (HUS).1–3 Over many decades, every outbreak related to the presence of E. coli O157:H7 in food products has been a serious event that caused severe problems related to human health as well as brand damage and economic loss to food manufacturers.4 Most recent cases of multistate outbreaks related to E. coli O157:H7 include one case associated with alfalfa sprouts that had caused nine illnesses and two hospitalizations, and the company, Jack & the Green Sprouts, Inc., had to recall all of their alfalfa and alfalfa onion sprout products.5 Another case involving contaminated Costco rotisserie chicken salad had caused nineteen illnesses, five hospitalizations, and two cases of HUS.6 Taylor Farms Pacific, Inc. provided the celery and onion diced blend which was suspected to be the contamination source for Costco, and had to voluntarily recall all of their products using these two ingredients. Therefore, the development of detection methods for the purpose of monitoring or screening this pathogenic bacterium in foods to prevent the catastrophic foodborne illnesses have become vitally important and caught much attention of researchers working in the area related to food safety.There have been numerous detection methods developed over the past several decades that can detect foodborne pathogens efficiently and effectively.7–9 Electrochemical biosensors, one branch that belongs to the biosensor category for rapid detection, present advantages like good sensitivity, miniaturization potential, and mass production,10 and have proven to be very promising in the biosensor field. There are three common electrochemical methods: voltammetry, amperometry, and electrochemical impedance spectroscopy. Even though they are based on the measurement of the changes in different parameters, the strategies these methods employ have certain similarities.11,12 These electrochemical methods employ either (1) indirect detection by forming a sandwich-like structure comprising of the target recognition element that binds to the biosensor surface and captures the target, the target bacterial cell, and the biochemical label (commonly enzyme) that triggers the reaction of the analyst in the media, or (2) direct detection by adsorbing the target bacterial cells onto the surface of the biosensor.7 Both methods yield detectable changes as electric signals at the interface of the electrode and the media. With the recent advancements in nanomaterials, such as nanoporous films,13,14 nanochains,15 and nanotubes,16 and also microfluidics14,17 and screen-printed electrodes,18,19 electrochemical biosensors have become more sensitive, smaller, and cheaper.
Despite all the advantages of using electrochemical biosensors for the detection of pathogenic bacteria, there are still limitations that require novel ideas to improve the performance of these sensors. Just like the two strategies mentioned previously, indirect methods require the binding of two ligands successively to the target bacteria so the detection time is prolonged, whereas when dealing with direct methods, although they have a shorter detection time due to the label-free nature, they show a relatively higher limit of detection (LOD) because they lack additional amplification.11,20–25 Moreover, when using a sandwich-like mechanism which applies two ligands competing with each other for the limited binding sites and space on the surface of the bacterial cell, or using the direct adsorption of bacterial cells with spatial dimensions onto a plain surface of the electrode, it is apparent that neither of these two strategies can use the binding sites on the cell surface to the maximum potential. If there is an element that processes both functions of target recognition and electrochemical amplification, it can not only reduce the detection time, but also utilize the limited binding sites of the bacteria reaching the optimal conditions to improve the LOD.
The construction of such an element for serving dual functions as the carrier and the signal amplifier requires a good supporting matrix that can integrate individual components without losing their bioactivities as well as having a high process ability that can efficiently be modified with abundant biomolecules or nanoparticles. Since introduced by Messersmith's group,26 the mussel-inspired polydopamine has gained much attention because of its excellent properties. It is well-established that, at low concentrations, initial pH values of 8 or above, and room temperature, PDA can be deposited onto various substrates as a controlled thin layer of film.27–30 The rich reactive groups like catechol moieties and amines in the PDA backbone allow PDA to bind strongly to metal ions as well as synthetic entities, like nanoparticles, polymers, or other biomolecules.27–29,31,32 All of these great qualities allow PDA to be widely applied in the fabrication of nanocomposites.
Therefore, in this study, we report a novel electrochemical biosensor for the detection of E. coli O157:H7 based on the synthesized bifunctional polydopamine-polymeric nanocomposites that are comprised of both antibodies and glucose oxidase. For the fabrication of the electrochemical biosensor, MBs were first bound to GOx through a streptavidin–biotin reaction. After that, a thin layer of PDA film was synthesized on the MB–GOx conjugates through controlled self-polymerization of DA under alkaline conditions. The good biocompatibility of PDA allowed the GOx inside to still maintain the enzymatic activity to catalyse glucose to produce H2O2 which could further reduce HAuCl4 to generate AuNPs that are attached to the surface of the MBs–GOx@PDA PMNCs. With successive adsorption of ABs and additional GOx, the final product ABs/GOxext/AuNPs/MBs–GOx@PDA PMNCs was used to capture the target bacterial cells. By using the filtration technique, the free PMNCs were filtered out and concentrated in the glucose solution for measurement. The filtration treatment helped in the isolation and concentration of the free PMNCs, meanwhile the removal of the bonded PMNCs also reduced the background noise during measurement so that the sensitivity of the developed biosensor could be correspondingly improved. The developed biosensor was also validated to detect E. coli O157:H7 in ground beef by using a handheld electrochemical detector. In this study, the bifunctional PMNCs have presented some valuable qualities like a short detection time due to combining the target capturing and labeling steps, effective amplification because the PMNCs contain abundant enzyme, and efficient conversion of the biological recognition to the electrochemical signal because every possible binding site on the bacteria has an active enzyme attached. This new concept has opened a new direction to construct rapid, sensitive and highly efficient electrochemical biosensors.
Experimental
Materials and apparatus
Phosphate buffered saline 1 (PBS1, 0.1 mol l−1, pH 7.4), dopamine (DA), gold(III) chloride hydrate, potassium ferricyanide(III), glucose, and glucose oxidase (128![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 200 U g−1 solid) were bought from Sigma-Aldrich (St Louis, MI). Ferric chloride hexahydrate was purchased from MP Biomedicals, LLC. (Solon, OH). PBS2 (10 mmol l−1, pH 7.4) solution was prepared by diluting PBS1 at a ratio of 1
200 U g−1 solid) were bought from Sigma-Aldrich (St Louis, MI). Ferric chloride hexahydrate was purchased from MP Biomedicals, LLC. (Solon, OH). PBS2 (10 mmol l−1, pH 7.4) solution was prepared by diluting PBS1 at a ratio of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10, and used throughout all the tests. Ultrapure deionized water (18.2 MΩ cm) was obtained from Milli-Q (EMD Millipore, Billerica, MA). GOx was biotinylated based on the protocol reported by Kanayeva et al. using sulfo-NHS-biotin,33 and excessive biotin was removed with a Slide-A-Lyzer dialysis kit from Pierce Protein Research Product (10 K MWCO, Rockford, IL). Streptavidin-coated magnetic beads (MBs) with a diameter of 150 nm were manufactured by Ocean NanoTech, LLC. (San Diego, CA). Based on the information provided by the company, the superparamagnetic MBs contain 1 mg ml−1 solid content (Fe) with 2.7 × 1011 particles per mg. The surface of MBs was covalently modified with streptavidin with a binding capacity for biotin–BSA or biotin–IgG of over 50 μg per mg or per ml. The overall structure and TEM images of the MBs were also provided by the company (Fig. S1†).
10, and used throughout all the tests. Ultrapure deionized water (18.2 MΩ cm) was obtained from Milli-Q (EMD Millipore, Billerica, MA). GOx was biotinylated based on the protocol reported by Kanayeva et al. using sulfo-NHS-biotin,33 and excessive biotin was removed with a Slide-A-Lyzer dialysis kit from Pierce Protein Research Product (10 K MWCO, Rockford, IL). Streptavidin-coated magnetic beads (MBs) with a diameter of 150 nm were manufactured by Ocean NanoTech, LLC. (San Diego, CA). Based on the information provided by the company, the superparamagnetic MBs contain 1 mg ml−1 solid content (Fe) with 2.7 × 1011 particles per mg. The surface of MBs was covalently modified with streptavidin with a binding capacity for biotin–BSA or biotin–IgG of over 50 μg per mg or per ml. The overall structure and TEM images of the MBs were also provided by the company (Fig. S1†).
Electrochemical analysis was conducted with a CHI750B electrochemical workstation manufactured by CH Instruments (Bee Cave, TX). A BDI handheld electrochemical detector (BioDetection Instruments Inc., Fayetteville, AR) was used for amperometric measurement to test the food sample. The bare SP-IDME, which was designed by our group and customized by DropSens (Llanera, Spain), was constructed with six pairs of interdigitated concentric circles of gold fingers. The width of the gold fingers and the spacing between each two fingers are both 200 μm. The active area of the gold fingers is about 12.38 mm2 (Fig. S2†). The MS0206 magnetic separator with a magnetic strength of approximately 1.0 Tesla (T) was purchased from Aibit LLC. (Jiangyin, China).
Rabbit anti-E. coli O + K polyclonal antibodies (4.0–5.0 mg ml−1) were purchased from Meridian Life Science Inc. (Memphis, TN). 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 5 dilutions of the antibodies (0.8–1.0 mg ml−1) were prepared with PBS and stored at 4 °C for further use, and the storage time was not more than one month. Stock bacterial cultures of E. coli O157:H7 (ATCC 43888), E. coli K12 (ATCC 29425), and S. typhimurium (ATCC 14028) were obtained from American Type Culture Collection (ATCC, Manassas, VA). The stock cultures were stored at −80 °C, and revived gently at room temperature when needed.
5 dilutions of the antibodies (0.8–1.0 mg ml−1) were prepared with PBS and stored at 4 °C for further use, and the storage time was not more than one month. Stock bacterial cultures of E. coli O157:H7 (ATCC 43888), E. coli K12 (ATCC 29425), and S. typhimurium (ATCC 14028) were obtained from American Type Culture Collection (ATCC, Manassas, VA). The stock cultures were stored at −80 °C, and revived gently at room temperature when needed.
Methods for culture preparation and media plating enumeration
The test cultures were prepared by growing the stock cultures in brain heart infusion (BHI) broth (Remel Microbiology Products, Lenexa, KS) at 37 °C for 18–20 h. A series of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10 dilutions for each bacterial culture were made with PBS2. To determine the viable cell numbers of the tested bacteria, 100 μl of each dilution were plated onto the surface of either non-selective Trypsin Soy Agar (TSA) when preparing pure laboratory culture, or selective Sorbitol MacConkey (SMAC) agar when testing the food sample. The number of bacterial colonies formed on the media after incubation at 37 °C for 18 to 24 h was counted to determine the concentration of viable bacterial cells in terms of colony forming units per milliliter (cfu ml−1). All the cultures were prepared on the test days.
10 dilutions for each bacterial culture were made with PBS2. To determine the viable cell numbers of the tested bacteria, 100 μl of each dilution were plated onto the surface of either non-selective Trypsin Soy Agar (TSA) when preparing pure laboratory culture, or selective Sorbitol MacConkey (SMAC) agar when testing the food sample. The number of bacterial colonies formed on the media after incubation at 37 °C for 18 to 24 h was counted to determine the concentration of viable bacterial cells in terms of colony forming units per milliliter (cfu ml−1). All the cultures were prepared on the test days.
Food sample preparation and inoculation
The detection of E. coli O157:H7 in ground beef (freshly purchased from a local grocery store and transported to the lab within 15 min) was examined. 25 g of ground beef was weighed and transferred into a filtering stomacher bag. After that, 225 ml of sterile PBS solution was added to the stomacher bag, and mixed using a stomacher machine (Stomacher 400, Seward, UK) at 200 rpm for two min. Nine ml of ground beef rinse water was transferred into new tubes. Then one ml of the bacterial dilution (PBS as negative control, NC) was used to spike the ground beef rinse water to achieve the desired concentration. The plate counting method was used to determine the concentration of the bacteria in ground beef rinse water. All the liquid samples right after preparation were directly used in the electrochemical analysis for the detection of E. coli O157:H7.Preparation of bifunctional PMNCs
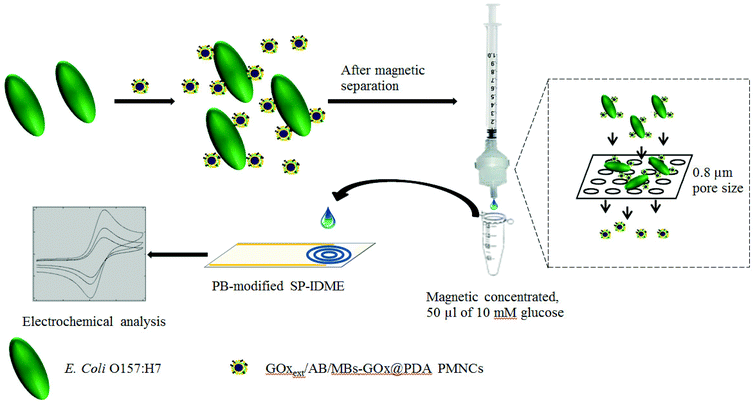
The schematic description of the preparation of the PMNCs is shown in Fig. 1. First, 20 μl of streptavidin-coated MBs were washed with 200 μl of PBS2 to remove the preservative content in 1.5 ml low protein binding tubes (SARSTEDT AG & Co., Germany) and magnetically separated with a magnetic separator for three min. The supernatant was removed with a pipette carefully (magnetic separation procedures are the same below unless specifically mentioned). Second, the remains were mixed in 180 μl of PBS2 and 20 μl of biotin-GOx (excessive) and rotated at 15 rpm and room temperature (RT) for one hour. After the rotation finished, the yielded mixture was magnetically separated and re-dispersed into 400 μl of 0.5 mg ml−1 DA (final concentration) in Tris buffer (pH 8.4, 10 mmol l−1). The mixture was rotated for another one hour to allow self-polymerization, yielding a suspension of the MBs–GOx@PDA biocomposites. After magnetic separation, the remaining biocomposites were rinsed with PBS2 three times and ultrasonically redispersed into 400 μl of PBS2. Third, the synthesis of AuNPs was based on a similar method used by Fu et al.34 Briefly, 0.20 mmol l−1 HAuCl4 (in PBS2, pH 7.4) and 5 mg ml−1 of glucose (both final concentration) were successively mixed in the yielded suspension for 5 h at 4 °C to allow biochemical synthesis of AuNPs on the surface of the MBs–GOx@PDA biocomposites. The dispersion of AuNPs on the surface of MBs–GOx@PDA PMNCs could help with adsorption of ABs and GOx more efficiently.34 Finally, the synthesized AuNPs/MBs–GOx@PDA biocomposites were suspended into the solution containing excessive anti-E. coli polyclonal ABs overnight at 4 °C. After magnetic separation, the ABs/AuNPs/MBs–GOx@PDA PMNCs were mixed with 5 mg ml−1 GOx for 1 h at room temperature to block the unspecific attachment and to allow an additional load of enzyme on the PMNCs. The prepared ABs/GOxext/AuNPs/MBs–GOx@PDA PMNCs were stored at 4 °C when not in use.Preparation of a Prussian blue modified SP-IDME
The bare SP-IDME was first thoroughly cleaned based on the reported protocol.35 The surface of the SP-IDME was carefully polished in alumina slurry with 0.02–0.05 μm particles. After rinsing with Milli-Q water thoroughly, the electrodes were ultrasonically treated for 2 min to remove the residual alumina particles. After that, the electrodes were rinsed with Milli-Q water again and dried in the oven at 80 °C for 1 h.The electrochemical deposition of Prussian blue on the surface of the SP-IDME was performed by using cyclic voltammetry (CV) based on the procedure described by Lin et al. with minor changes.36 First, the bare SP-IDME was voltammetrically pre-treated from −1.7 V to 1.7 V for 1 cycle. Second, 50 μl of aqueous solution containing 50 mmol l−1 of FeCl3, K4Fe(CN)6, 0.1 mol l−1 KCl, and 0.01 mol l−1 HCl (all final concentration) were dropped to cover the entire active surface of the SP-IDME. The deposition of the PB was accomplished by applying −0.5 V to 0.5 V at a rate of 0.05 V s−1 for 2 cycles. The excessive solution was carefully washed off by Milli-Q water and the electrode was dried with nitrogen gas. Finally, 50 μl of aqueous solution containing 0.1 mol l−1 KCl and 0.01 mol l−1 HCl was dropped onto the surface of the SP-IDME. The electrode was electrochemically cycled from −0.5 V to 0.5 V at a rate of 0.1 V s−1 for 15 cycles until a stable CV curve was obtained. When not in use, the modified SP-IDMEs were kept in the desiccator at room temperature.
Construction of the electrochemical biosensor for the detection of E. coli O157:H7
As illustrated in Fig. 2, the construction and mechanism of the electrochemical biosensor are described as follows. First, after blocking with GOx, the ABs/GOxext/AuNPs/MBs–GOx@PDA PMNCs were thoroughly washed with PBS2 three times and ultrasonically dispersed in PBS2 each time. The final ABs/GOxext/AuNPs/MBs–GOx@PDA PMNCs were mixed with 200 μl of E. coli O157:H7 dilutions with a controlled concentration for 45 min at room temperature to allow the capture of the target bacteria. Third, the PMNCs–cell conjugates were magnetically separated, washed with PBS2 three times, and suspended in 200 μl of PBS2. Then, the suspension was filtered through a filter paper (EMD Millipore) with a pore size of 0.8 μm using a syringe (BD Syringes, Franklin Lakes, NJ) and a filter holder (EMD Millipore) into a new tube (1.5 ml). 600 μl of PBS2 were used to wash the free PMNCs thoroughly off the filter paper. Finally, the solution containing filtered-out free PMNCs was magnetically separated for 5 min. Then 50 μl of 10 mmol l−1 glucose solution (in PBS2) were added to allow enzymatic reaction for 5 min. 50 μl of sample were dropped onto the PB-modified SP-IDME for electrochemical analysis. Both CV and amperometric detection were used to characterize and detect different concentrations of target bacteria.The specificity of the constructed biosensor was investigated by testing other non-target bacteria, such as E. coli K12 and S. typhimurium. The concentration of the target and the non-target bacteria tested was 104 cfu ml−1. The BDI handheld electrochemical detector was used to validate the concept for the detection of the target bacteria in the concentration range of 101 to 105 cfu ml−1 in ground beef.
The statistical analysis of data, such as the mean, standard deviation, and linear regression relationship, was performed using Excel 2010 software (Microsoft, Redmond, WA) with at least three replications for experiments.
Results and discussion
Characterization of the PB-modified SP-IDME
Since the concept of glucose biosensors was first introduced in 1962,37 there have been numerous electrochemical methods developed to monitor the concentration of glucose in solution.38–41 Currently, most commercial glucose biosensors do not operate very differently from the concept of the second-generation glucose biosensors which use soluble or immobilized mediators to help with the charge transfer between the enzyme and the electrode.41 Prussian blue, a mixture of ferric and ferrous cyanide, is one of the most common mediators. The mechanism of using PB to detect glucose is based on the following reactions (GOx as the catalytic enzyme).42,43| Glucose + GOx − FAD+ → Glucolactone + GOx − FADH2, | (1) |
| GOx − FADH2 + O2 → GOx − FAD + H2O2, | (2) |
| FeIII4[FeII(CN)6]3 + 4K+ + 4e− ↔ K4{FeII4[FeII(CN)6]3}, | (3) |
| K4{FeII4[FeII(CN)6]3} + 2H2O2 + 4H+ → FeIII4[FeII(CN)6]3 + 4K+ + 4H2O. | (4) |
Even now, PB mediated electrochemical biosensors are still under intensive study to construct sensitive, cheap, and reproducible electrodes or strips to monitor the concentration of glucose in different media.
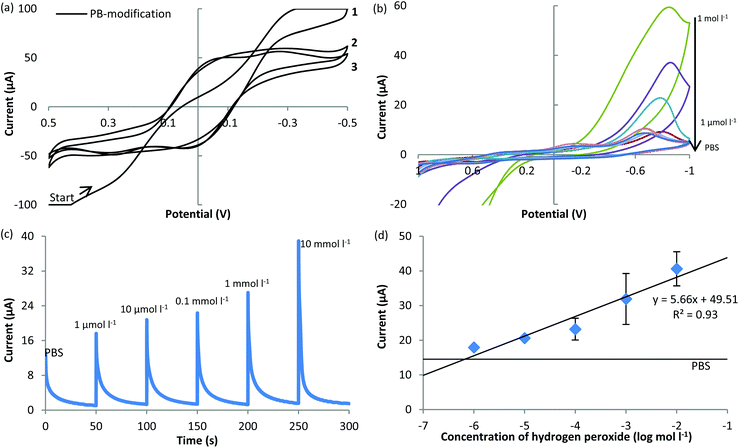
Therefore, PB was used as the mediator to modify the SP-IDME for the purpose of constructing a cheap and easily reproducible measurement method to monitor the enzymatic reaction of glucose in this study. In order to evaluate the performance of the constructed biosensor, the characteristics of the PB-modified SP-IDME must be clarified first (Fig. 3). During the electrodeposition of PB onto the surface of the SP-IDME (Fig. 3(a)), the CV curve of cycle 1 was different from those of cycles 2 and 3, indicating the formation of a PB film onto the electrode. The CV curves of cycles 2 and 3 show almost the same shape, indicating that there is no significant change of the PB film deposited onto the electrode surface after cycle 2, which was determined to be used as the duration of PB electrodeposition for the following electrode modification. Fig. 3(b) to (d) show the performance of the PB-modified SP-IDME when used for the electrochemical measurement of H2O2 at different concentrations. When H2O2was present in the solution, one redox peak was shown at −0.8 V, and a higher concentration of H2O2 caused a higher current response (Fig. 3(b)). The presence of 1 mol l−1 and 0.1 mol l−1 H2O2 gave an exceedingly large current response, and also damaged the PB film on the electrode surface based on the visual observation. Moreover, due to the electric connection of the SP-IDME, the working electrode and the reference electrode were connected to the same PB film, and the redox reaction of ferric ferrocyanide occurred simultaneously but individually on the two electrodes, doubling the redox potential reading (−0.75 V in Fig. 3(b) compared to −0.36 V vs. NHE). When the amperometric measurement at −0.75 V was used to detect different concentrations of H2O2 (Fig. 3(c)), the current decreased as the time increased, and the highest current response occurred at the beginning of the measurement. Considering that the sample stayed on the electrode surface during the measurement and no stirring was done to mix the sample, it appeared that the electrochemical reaction between PB and H2O2 at the interface of the electrode and the media was diffusion limited. The PB-modified SP-IDME showed a linear relationship between the current peaks and the log concentration of H2O2 in the range of 1 μmol l−1 to 10 mmol l−1 (Fig. 3(d)), which could satisfy the requirement for the following tests.
Characterization of the synthesized ABs/GOxext/AuNPs/MBs–GOx@PDA PMNCs
The synthesized ABs/GOxext/AuNPs/MBs–GOx@PDA PMNCs in this study had two responsibilities: (1) recognition of the target bacteria, which requires the PMNCs to have the acceptable capability to isolate and concentrate the target bacteria from the sample media. (2) Conversion of the biological recognition to the electrochemical signal through the enzymatic reaction. The PMNCs which contained a large amount of GOx were expected to have high catalytic activity to induce a significant concentration change of glucose which could efficiently amplify the electrochemical signal. Therefore, tests to examine the binding affinity between the synthesized PMNCs and the target bacterial cells to obtain the electrochemical characteristics of the PMNCs were conducted at first.The capture efficiency of using the synthesized PMNCs for E. coli O157:H7 at different concentrations is shown in Table 1. From the results, the PMNCs were able to capture approximately 89% or more of the target bacterial cells in the range of 102 to 105 cfu ml−1. The rest, about 10% of the bacterial cells, was lost during the operation. The significant lower capture efficiency of PMNCs at 101 cfu ml−1 was probably due to the fact that the total number of bacterial cells in the sample is too small (maybe only one or two). The loss of an individual cell in a small size group of cells was weighed much more than that in a large group of cells. Considering that there was an expected loss of bacterial cells due to the procedures like magnetic separation or pipetting, the capture efficiency of the PMNCs was acceptable, and the volume of PMNCs used in the test was followed throughout the study.
| # | Volume of PMNCs (μl) | Volume of sample (μl) | Captured bacteria (cfu ml−1) | Bacteria in waste (cfu ml−1) | Capture efficiency (%) |
|---|---|---|---|---|---|
| 1 | 2 × 101 | 1 × 101 | 66.7 | ||
| 2 | 0.80 ± 0.20 × 102 | 0.05 ± 0.05 × 102 | 94.1 | ||
| 3 | 20 | 200 | 1.04 ± 0.02 × 103 | 0.12 ± 0.00 × 103 | 89.6 |
| 4 | 1.01 ± 0.06 × 104 | 0.12 ± 0.00 × 104 | 89.4 | ||
| 5 | 0.90 ± 0.04 × 105 | 0.11 ± 0.01 × 105 | 89.1 |
To verify the enzymatic activity of the synthesized PMNCs, 20 μl of the ABs/GOxext/AuNPs/MBs–GOx@PDA PMNCs were mixed with 10 mmol l−1 glucose solution and measured by CV at different time points during the glucose catalysis (Fig. 4). As shown in Fig. 4(a), there was a clear redox peak at about −0.4 V and the current response at this redox peak potential increased when the reaction time was longer. The current responses at the redox peak were plotted against the enzymatic reaction time (Fig. 4(b)), showing that the enzyme activity reached the maximum rate at 5 min. This was used as the reaction time in the further test for the detection of E. coli O157:H7.
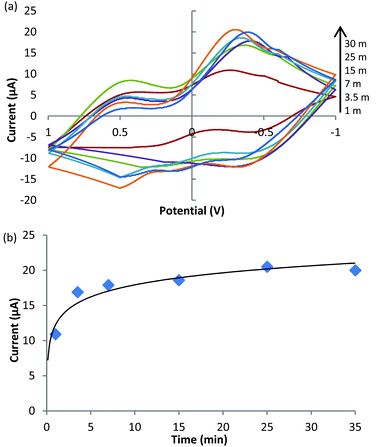 | ||
| Fig. 4 Enzymatic activity of the synthesized PMNCs. (a) Cyclic voltammetry at different times of enzymatic reaction. (b) Redox peaks at different time points of the enzymatic reaction. | ||
The constructed electrochemical biosensor for the detection of E. coli O157:H7 in pure culture and the specificity of the biosensor
The amperometric detection (at −0.4 V) of E. coli O157:H7 at different concentrations in the pure culture samples is shown in Fig. 5. From the figure, the current response is well-fitted into a simple linear regression against the concentration of E. coli O157:H7 within the range of 101 to 106 cfu ml−1 (R2 = 0.99). The obtained linear relationship between the current and the concentration of the bacteria has a negative slope, proving that more bacterial cells in the sample could retain more PMNCs at the filtration step and leave less PMNCs in the final glucose solution for the electrochemical measurement. The current response of negative control (NC) was 25.83 ± 2.34 μA. From the calibration curve, the limit of detection (LOD) of the constructed amperometric biosensor was 101.72 cfu ml−1 (or 52 cfu ml−1), which was determined by using the mean of the NC minus its standard deviation multiplied by three (S/N = 3). The large deviation shown at 102 cfu ml−1 was probably due to the small number of the bacterial cells present in the sample (approximately 20 cfu). At this concentration of the bacteria, a variation of several cells made a significant influence on the final results, where it is too weak to make a significant difference at lower concentrations (101 cfu ml−1, or 1 cfu per sample) and not strong enough to reveal the differences at higher concentrations (more than 103 cfu ml−1).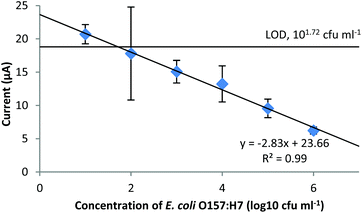 | ||
| Fig. 5 Amperometric detection of E. coli O157:H7 in pure culture samples. The potential applied was −0.4 V. | ||
The LOD of the constructed amperometric biosensor was competitive to that of other electrochemical methods reported in previous publications (Table 2). On comparing the detection time, LODs, and strategies, our biosensor possesses several merits such as it has a short detection time that is comparable to the electrochemical impedance sensors using a label-free strategy but yields an LOD that is similar to those obtained by the electrochemical sensors using labeling to amplify the signals. Based on the calculation of LOD × sample volume (200 μl), the obtained LOD equals 10 cells. This is the result without any pre-enrichment. The generation time of E. coli under normal laboratory conditions is 15 to 20 min. Therefore, if the developed electrochemical biosensor could be combined with a pre-enrichment procedure, it would only require less than 2 h for a single cell to multiply and reach the detection limit. The detection procedure in this study was simple, only requiring the capture of the bacteria and a filtration step, which is beneficial if this biosensor is applied to on-site or in-field detection of the target bacteria.
| Electrochemical method | Detection | Ref. | ||||
|---|---|---|---|---|---|---|
| Electrode | Strategy | Time | Range | |||
| Culture | Food | |||||
| a NA: not available. | ||||||
| Amperometry | 1st ABs/AuNPs–FeDC/SPCE | 2nd AB-HRP | ∼30 min | 5.75 × 101 to 5.75 × 107 cfu ml−1 with an LOD of 6 × 102 cfu ml−1 | NAa | 21 |
| Capture probe DNA/AuNPs/graphene oxide/GCE | Signal probe DNA/graphene oxide-thi-Au@SiO2/DNAzyme labeling | ∼1 h 50 min | 0.02 to 50.0 nmol l−1 with an LOD of 0.01 nmol l−1 | NA | 45 | |
| Electrochemical impedance spectroscopy | Platinum wire | Nanoporous alumina membrane–antibody | ∼2 h | 102 to 107 cfu ml−1 with an LOD of 102 cfu ml−1 | NA | 14 |
| Gold IDME | Magnetic NPs-AB-bacteria complex | ∼35 min | 7.4 × 104 to 7.4 × 107 cfu ml−1 with an LOD of 7.4 × 104 cfu ml−1 | 8.0 × 105 to 8.0 × 107 cfu ml−1 with an LOD of 8.0 × 105 cfu ml−1 | 17 | |
| SEMs of DTSP on SP-IDME | Wheat germ agglutinin labeling | ∼45 min without labeling | 102 to 106 cfu ml−1 with an LOD of 102 cfu ml−1 without WGA labeling | NA | 18 | |
| ∼90 min with labeling | ||||||
| Reduced graphene oxide paper electrode | ABs/AuNPs/rGOPE direct detection | ∼30 min | 1.5 × 102 to 1.5 × 107 cfu ml−1 with an LOD of 1.5 × 102 cfu ml−1 | 1.5 × 104 cfu ml−1 in ground beef, 1.5 × 103 cfu ml−1 in cucumber | 44 | |
| Cyclic voltammetry | Bio-AB/avidin/Au-SiO2/CHI-SH/Fc/C60/GCE | PtNCs–GOx–AB tag | ∼1 h | 3.2 × 101 to3.2 × 106 cfu ml−1 with an LOD of 15 cfu ml−1 | NA | 15 |
| Amperometry | PB-modified SP-IDME | Bifunctional ABs/GOxext/AuNPs/MBs–GOx@PDA PMNCs | ∼1 h | 102 to 106 cfu ml−1 with an LOD of 52 cfu ml−1 | 103 to 106 cfu g−1 with an LOD of 190 cfu g−1 | This work |
The specificity of the constructed biosensor for the detection of E. coli O157:H7 is shown in Fig. 6. At 104 cfu ml−1, the current response of the target bacteria had a difference of 12.60 μA to that of the NC (25.83 μA) which was significantly higher than that of E. coli K12 (3.77 μA) and S. typhimurium (3.02 μA). Moreover, neither the current responses of E. coli K12 nor S. typhimurium passed the LOD of current, which was 18.80 μA. All of the above results suggest that the constructed biosensor was very specific for the detection of E. coli O157:H7. The specificity of the developed method was primarily determined by the polyclonal anti-E. coli antibody used. Based on the information of this antibody provided by the company, it reacts with many E. coli serotypes containing “O” and ‘K’ antigens. The specificity of this method can be improved by using more specific antibodies if such a demand arises.
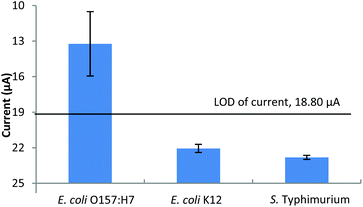 | ||
| Fig. 6 Specificity of the constructed electrochemical biosensor for the detection of E. coli O157:H7. | ||
The validation of the constructed biosensor for the detection of E. coli O157:H7 in ground beef by using a handheld electrochemical detector
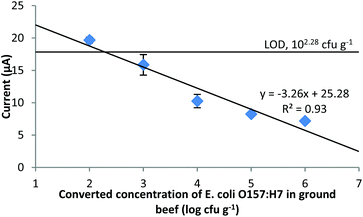
The validation for the detection of E. coli O157:H7 in ground beef using the established method and a handheld electrochemical detector to replace the benchtop electrochemical workstation was also conducted (Fig. 7). The current response of the NC was 20.64 ± 0.93 μA, which determined that the LOD of E. coli O157:H7 in ground beef was 102.28 cfu g−1 (1.90 × 102 cfu g−1) using similar calculations as in the section “The constructed electrochemical biosensor for the detection of E. coli O157:H7 in pure culture and the specificity of the biosensor”. This LOD is generally consistent with the result obtained from the pure culture if the ratio of the ground beef to the liquid sample (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10 w/v) is taken into consideration. Comparing the current responses of different concentrations of E. coli O157:H7, the results from the food samples were slightly lower than those from the pure culture samples. The reason for this is probably that the complex food matrix contains various ingredients (fat, proteins, etc.) which could obstruct the free PMNCs from being separated from the supernatant, or adsorbed onto the free PMNCs and hindered them from passing through the filter. However, considering the fact that the final LOD of E. coli O157:H7 in ground beef was comparable to that in pure culture and the filtration step also helped in retaining food residues, which could reduce the noise, the results using the proposed electrochemical method are acceptable.
10 w/v) is taken into consideration. Comparing the current responses of different concentrations of E. coli O157:H7, the results from the food samples were slightly lower than those from the pure culture samples. The reason for this is probably that the complex food matrix contains various ingredients (fat, proteins, etc.) which could obstruct the free PMNCs from being separated from the supernatant, or adsorbed onto the free PMNCs and hindered them from passing through the filter. However, considering the fact that the final LOD of E. coli O157:H7 in ground beef was comparable to that in pure culture and the filtration step also helped in retaining food residues, which could reduce the noise, the results using the proposed electrochemical method are acceptable.
Conclusion
In this study, we synthesized bifunctional ABs/GOxext/AuNPs/MBs–GOx@PDA PMNCs, and fabricated an electrochemical biosensor coupled with a PB-modified SP-IDME for the rapid and sensitive detection of E. coli O157:H7 in pure culture and in food. The developed biosensor showed a broad detection range from 102 to 106 cfu ml−1 in the pure culture within 1 h. The validation of the developed approach to detect the target bacteria in ground beef demonstrated that it could detect as low as 190 cfu g−1 without a pre-enrichment procedure, and the possibility of using a handheld device to achieve the electrochemical measurement which demonstrates the benefit for on-site or in-field applications by using the proposed method. To conclude, the developed electrochemical biosensor requires a short detection time similar to that of the sensors using a label-free strategy but also with efficient amplification to achieve low LOD which is comparable to others using various labeling strategies. The advantages of using bifunctional PMNCs in this study exhibited a new direction to construct electrochemical biosensors for sensitive and rapid detection of foodborne pathogens.Acknowledgements
This research was supported in part by ABI (project # 0380-43052-24-2333) and Aibit (project # 30-011905). The authors want to thank Dr David W. Paul and Zach Callaway for their help in revising the manuscript, and also thank Lisa Kelso for her help in microbial tests.References
- CDC (Centers for Disease Control and Prevention), E. coli (Escherichia coli). http://www.cdc.gov/ecoli/general/index.html#what-are-shiga-toxin, (last updated November 2015).
- P. M. Griffin and R. V. Tauxe, Epidemiol. Rev., 1991, 13, 60–98 CrossRef CAS PubMed.
- N. L. Padola, Front. Microbiol., 2014, 5, 1–2 CrossRef PubMed.
- S. Hoffmann and T. D. Anekwe, EIB-118, USDA, U.S. Department of Agriculture, Economic Research Service, 2013.
- CDC (Centers for Disease Control and Prevention), Multistate outbreak of Shiga toxin-producing Escherichia coli O157 infections linked to Costco rotisserie chicken salad. http://www.cdc.gov/ecoli/2015/o157h7-11-15/index.html (last updated 3.2.2016).
- CDC (Centers for Disease Control and Prevention), Multistate outbreak of Shiga toxin-producing Escherichia coli O157 infections linked to alfalfa sprouts produced by Jack & the Green Sprouts. http://www.cdc.gov/ecoli/2016/o157-02-16/index.html (last updated 12.22.2015).
- Z. Fu, S. Rogelj and T. L. Kieft, J. Food Microbiol., 2005, 99, 47–57 CrossRef CAS PubMed.
- H. P. Dwivedi and L.-A. Jaykus, Crit. Rev. Microbiol., 2011, 37, 40–63 CrossRef CAS PubMed.
- K. J. Yoshitomi, K. C. Jinneman, R. Zapata, S. D. Weagant and W. M. Fedio, J. Food Sci., 2012, 77, 481–489 CrossRef PubMed.
- I. Palchetti and M. Mascini, Anal. Bioanal. Chem., 2008, 391, 455–471 CrossRef CAS PubMed.
- J. Monzo, I. Insua, F. Fernandez-Trillo and P. Rodriguez, Analyst, 2015, 140, 7116–7128 RSC.
- A. J. Bard and L. R. Faulkner, Electrochemical methods. Fundamentals and Applications, John Wiley & Sons Inc., New York, 2nd edn, 2001 Search PubMed.
- C.-K. Joung, H.-N. Kim, M.-C. Lim, T.-J. Jeon, H.-Y. Kim and Y.-R. Kim, Biosens. Bioelectron., 2013, 44, 210–215 CrossRef CAS PubMed.
- F. Tan, P. H. M. Leung, Z.-B. Liu, Y. Zhang, L. Xiao, W. Ye, X. Zhang, L. Yi and M. Yang, Sens. Actuators, B, 2011, 159, 328–335 CrossRef CAS.
- Y. Li, L. Fang, P. Cheng, J. Deng, L. Jiang, H. Huang and J. Zheng, Biosens. Bioelectron., 2013, 49, 485–491 CrossRef CAS PubMed.
- E. I. Maurer, K. K. Comfort, S. M. Hussain, J. J. Schlager and S. M. Mukhopadhyay, Sens., 2012, 21, 8135–8144 CrossRef PubMed.
- M. Varshney, Y. Li, B. Srinivasan and S. Tung, Sens. Actuators, B, 2007, 128, 99–107 CrossRef CAS.
- Z. Li, Y. Fu, W. Fang and Y. Li, Sens., 2015, 15, 19212–19224 CrossRef CAS PubMed.
- M. Xu, R. Wang and Y. Li, Talanta, 2016, 148, 200–208 CrossRef CAS PubMed.
- E. B. Setterington and E. C. Alocilja, Biosensors, 2012, 2, 15–31 CrossRef CAS PubMed.
- Y.-H. Lin, S.-H. Chen, Y.-C. Chuang, Y.-C. Lu, T. Y. Shen, C. A. Chang and C.-S. Lin, Biosens. Bioelectron., 2008, 23, 1832–1837 CrossRef PubMed.
- S. Chemburu, E. Wilkins and I. Abdel-Hamid, Biosens. Bioelectron., 2005, 21, 491–499 CrossRef CAS PubMed.
- G. A. Zelada-Guillen, S. V. Bhosale, J. Riu and F. X. Rius, Anal. Chem., 2010, 82, 9254–9260 CrossRef PubMed.
- M. Varshney and Y. Li, Biosens. Bioelectron., 2007, 22, 2408–2414 CrossRef CAS PubMed.
- A. Shabani, M. Zourob, B. Allain, C. A. Marquette, M. F. Lawrence and R. Mandeville, Anal. Chem., 2008, 80, 9475–9482 CrossRef PubMed.
- H. Lee, S. M. Dellatore, W. M. Miller and P. B. Messersmith, Sci., 2007, 318, 426–430 CrossRef CAS PubMed.
- X. Liu, J. Cao, H. Li, J. Li, Q. Jin, K. Ren and J. Ji, ACS Nano, 2013, 7, 9384–9395 CrossRef CAS PubMed.
- Q. Wei, F. Zhang, J. Li, B. Li and C. Zhao, Polym. Chem., 2010, 1, 1430–1433 RSC.
- C.-C. Ho and S.-J. Ding, J. Biomed. Nanotechnol., 2014, 10, 3063–3084 CrossRef CAS PubMed.
- V. Ball, D. del Frari, V. Toniazzo and D. Ruch, J. Colloid Interface Sci., 2012, 386, 366–372 CrossRef CAS PubMed.
- X. Gu, Y. Zhang, H. Sun, X. Song, C. Fu and P. Dong, J. Nanomater., 2014, 2015, 1–12 Search PubMed.
- D. R. Dreyer, D. J. Miller, B. D. Freeman, D. R. Paul and C. W. Bielawski, Chem. Sci., 2013, 4, 3796–3802 RSC.
- D. A. Kanayeva, R. Wang, D. Rhoads, G. F. Erf, M. K. Slavik, S. Tung and Y. Li, J. Food Prot., 2012, 75, 1951–1959 CrossRef CAS PubMed.
- Y. Fu, P. Li, L. Bu, T. Wang, Q. Xie, X. Xu, L. Lei, C. Zou and S. Yao, J. Phys. Chem. C, 2010, 114, 1472–1480 CrossRef CAS.
- J. Zhang, S. Song, L. Wang, D. Pan and C. Fan, Nat. Protoc., 2007, 2, 2888–2897 CrossRef CAS PubMed.
- Y. Lin, L. Hu, L. Yin and L. Guo, Sens. Actuators, B, 2015, 210, 513–518 CrossRef CAS.
- L. C. Clark and C. Lyons, Ann. N. Y. Acad. Sci., 1962, 102, 29–45 CrossRef PubMed.
- A. A. Karyakin, O. V. Gitelmacher and E. E. Karyakina, Anal. Chem., 1995, 67, 2419–2423 CrossRef CAS.
- W.-Z. Jia, K. Wang and X.-H. Xia, Trends Anal. Chem., 2010, 29, 306–318 CrossRef CAS.
- M. M. Rahman, A. J. S. Ahammad, J.-H. Jin, S. J. Ahn and J.-J. Lee, Sens., 2010, 10, 4855–4886 CrossRef PubMed.
- E.-H. Yoo and S.-Y. Lee, Sens., 2010, 10, 4558–4576 CrossRef PubMed.
- A. L. Galant, R. C. Kaufman and J. D. Wilson, Food Chem., 2015, 188, 149–160 CrossRef PubMed.
- X. Ji, J. Ren, R. Ni and X. Liu, Analyst, 2010, 135, 2092–2098 RSC.
- Y. Wang, J. Ping, Z. Ye, J. Wu and Y. Ying, Biosens. Bioelectron., 2013, 49, 492–498 CrossRef CAS PubMed.
- Y. Li, J. Deng, L. Fang, K. Yu, H. Huang, L. Jiang, W. Liang and J. Zheng, Biosens. Bioelectron., 2015, 63, 1–6 CrossRef PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c6an00873a |
| This journal is © The Royal Society of Chemistry 2016 |