Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Whole-cell detection of live lactobacillus acidophilus on aptamer-decorated porous silicon biosensors†
K.
Urmann
ab,
S.
Arshavsky-Graham
b,
J. G.
Walter
a,
T.
Scheper
a and
E.
Segal
*b
aInstitute of Technical Chemistry, Leibniz University of Hannover, Callinstr. 5, 30167 Hannover, Germany
bDepartment of Biotechnology and Food Engineering, Technion Israel Institute of Technology, Technion City, 32000 Haifa, Israel. E-mail: esegal@tx.technion.ac.il; Tel: +972-4-8295071
First published on 28th June 2016
Abstract
This work describes the design of optical aptamer-based porous silicon (PSi) biosensors for the direct capture of Lactobacillus acidophilus. Aptamers are oligonucleotides (single-stranded DNA or RNA) that can bind their targets with high affinity and specificity, making them excellent recognition elements for biosensing applications. Herein, aptamer Hemag1P, which specifically targets the important probiotic L. acidophilus, was utilized for direct bacteria capture onto oxidized PSi Fabry–Pérot thin films. Monitoring changes in the reflectivity spectrum (using reflective interferometric Fourier transform spectroscopy) allows for bacteria detection in a label-free, simple and rapid manner. The performance of the biosensor was optimized by tuning the PSi nanostructure, its optical properties, as well as the immobilization density of the aptamer. We demonstrate the high selectivity and specificity of this simple “direct-capture” biosensing scheme and show its ability to distinguish between live and dead bacteria. The resulting biosensor presents a robust and rapid method for the specific detection of live L. acidophilus at concentrations relevant for probiotic products and as low as 106 cells per mL. Rapid monitoring of probiotic bacteria is crucial for quality, purity and safety control as the use of probiotics in functional foods and pharmaceuticals is becoming increasingly popular.
Introduction
Aptamers are short single-stranded oligonucleotides with specific target affinity which are yielded from an iterative selection process called SELEX (Systematic Evolution of Ligands by Exponential Enrichment), first established in 1990.1,2 Compared to antibodies, these oligonucleotides possess a number of advantages: aptamers can exhibit similarly high affinities while their production is fully performed in vitro, assuring fast, low-cost and reproducible synthesis.3,4 Aptamers can be designed to bind any desired target3 and their selection under conditions of the native target conformation and surrounding matrix fosters a highly selective and affine aptamer sequence as a result. When employed as capture probes in biosensors, the aptamers’ versatile chemical modification options facilitate their surface conjugation in a desired orientation and density5–8 and their small molecule size and high stability are highly advantageous.9–11When designing biosensors targeting whole cells, aptamer technology enables the development of highly affine capture probes even without exact knowledge of the molecular composition of the targeted structure. Whole-cell SELEX approaches utilize the cells in their native conformation and targeted structures do not need to be available in a purified form, as it would be necessary for antibody generation.3,4,12 Indeed, aptamer capture probes have been generated against a variety of bacteria (e.g. Escherichia coli,13Campylobacter jejuni,14Mycobacterium tuberculosis,15Staphylococcus aureus16 and Salmonella enterica17) and many aptamer-based capture assays have been reviewed recently.18 For Lactobacillus acidophilus (L. acidophilus), a common lactic acid bacteria, the aptamer Hemag1P was developed by Hamula et al.19L. acidophilus is the most commonly used probiotic bacteria with several health benefits, including stimulation of the immune system and better digestibility.20,21L. acidophilus is a Gram-positive, rod-shaped bacterium, widely found in fermented and dairy-containing food products.
The Hemag1P aptamer is a 78-nucleotide long sequence, selected against whole, live L. acidophilus cells by a SELEX approach. The aptamer is believed to target the abundantly present S-proteins on the outer bacteria membrane. This hypothesis was confirmed by flow cytometry studies of the aptamer-target interactions comparing Lactobacillus strains with and without S-layer proteins and its affinity was reported to be 13 ± 3 nM.19 Since its selection, Hemag1P has been employed twice as a capture probe. A preliminary work by Zhang et al.22 demonstrated the capture of fluorescently labelled L. acidophilus onto a Hemag1P-functionalized chip.22 Zuo et al.23 have developed a fluorescence-based biosensor in which fluorophore-labelled Hemag1P was adsorbed to a graphene oxide (GO). Bacteria detection was facilitated by monitoring changes in GO fluorescence quenching.23 Yet, these two studies required tedious labelling and employed a fluorescence microscope for signal readout. Thus, their potential application is confined to a laboratory setting.
Label-free optical biosensors can be designed using porous silicon-based systems. In the past few years, porous silicon (PSi) has received much attention as an optical transducer due to its facile fabrication, large surface-to-volume ratio and numerous available surface chemistries.24–37 Many PSi-based biosensors use reflective interferometric Fourier transform spectroscopy (RIFTS) to monitor target capture within the porous layer.32,38–40 However, for cellular targets (such as microorganisms), which are too large to penetrate into the pores, monitoring changes in the intensity of the reflectivity spectrum upon “direct cell capture” onto antibody-modified PSi allows their detection and quantification.41,42 This biosensing scheme has allowed for a sensitive detection of E. coli bacteria using a peptidomimetic antimicrobial compound as a recognition element.43 However, these biosensors are non-specific in their nature and require cell lysis to induce a response. Thus, in the present work, we design a highly specific biosensor for detection of L. acidophilus employing the Hemag1P aptamer as the capture probe. The biosensing concept relies on monitoring changes in the amplitude (intensity) of the FFT peak, which is obtained from the raw reflectivity spectra of the PSi, during exposure to bacteria suspensions. Bacteria capture onto the biosensor surface induce intensity changes. Recent work on aptamer-functionalized PSi has demonstrated their immense potential for designing highly stable and specific PSi biosensors for protein detection.44,45 Herein, we describe the design and characterization of a label-free optical PSi-based aptasensor, where an oxidized PSi (PSiO2) Fabry–Pérot thin film, used as the optical transducer, is conjugated with the L. acidophilus binding aptamer Hemag1P. We demonstrate the high selectivity and specificity of this simple “direct-capture” biosensing scheme and show its ability to distinguish between live and dead bacteria. The resulting biosensor presents a robust and rapid method for the specific detection of live L. acidophilus in concentrations as low as 106 cells per mL.
Experimental
Materials
Silicon wafers (0.0009 Ω cm resistivity, p-type, <100>-oriented, heavily boron-doped) were purchased from Siltronix Corp. Aqueous HF (48%) and absolute ethanol were supplied by Merck. (3-Mercaptopropyl)trimethoxysilane (MPTMS), maleimide, ethylenediaminetetraacetic acid (EDTA), 5,5′-dithiobis-(2-nitrobenzoic acid) (DTNB, Ellman reagent), culturing media components and all buffer salts were purchased from Sigma-Aldrich Chemicals. Buffers and media were all prepared with deionized water (18.2 MΩ cm) and filtered prior to use. Media were autoclaved prior to their use. Solvents (toluene, acetone) were purchased from Gadot Israel. The sequence of aptamer Hemag1P (5′-AGC AGC ACA GAG GTC AGA TGT AGC CCT TCA ACA TAG TAA TAT CTC TGC ATT CTG TGT GCC TAT GCG TGC TAC CGT GAA-3′) was published by Hamula et al.19 and purchased with a 5′-acrylamide phosphoramidite (Acrydite™ phosphoramidite; Mosaic Technologies) modification from Integrated DNA Technologies. Other aptamers included in this work as controls are listed in Table 1. Hemag1P was selected in 50 mM Tris-HCl (pH 7.4), 100 mM NaCl, 5 mM KCl, 1 mM MgCl2 (this buffer composition is subsequently abbreviated as SB). TE-buffer was composed of 10 mM Tris-HCl (pH 8.0) and 1 mM EDTA. PBS-buffer was composed of 137 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4 and 1.8 mM KH2PO4 (pH 7.0). Bacteria strain Lactobacillus acidophilus ATCC 4356 was obtained from Gamidor Diagnostics and Escherichia coli strain K12, as a negative control, was generously supplied by Prof. Sima Yaron (Technion – Israel Institute of Technology).| Sequence name | Sequence | Modification |
|---|---|---|
| Hemag1P | AGCAGCACAGAGGTCAGATGTAGCCCTTCAACATAGTAATATCTCTGCATTCTGTGTGCCTATGCGTGCTACCGTGAA | 5′-Acrydite |
| T10-Hemag1P | TTTTTTTTTTAGCAGCACAGAGGTCAGATGTAGCCCTTCAACATAGTAATATCTCTGCATTCTGTGTGCCTATGCGTGCTACCGTGAA | 5′-Acrydite |
| 6H746 | GCTATGGGTGGTCTGGTTGGGATTGGCCCCGGGAGCTGGC | 5′-Acrydite |
Bacteria culturing, sample preparation and cell count
L. acidophilus was grown in MRS medium (CM0359, OXOID) under 5% CO2 atmosphere at 37 °C. Bacterial growth was monitored by optical density (OD) measurements at 600 nm in order to determine the logarithmic phase of growth. To correlate OD600 values with bacteria cell concentration, a standard plate-counting technique47 was performed. Briefly, bacteria were grown to the mid-logarithmic phase (OD600 of 0.44) and plated on MRS agar plates in replicates. Plates were then incubated for 48 h in 5% CO2 atmosphere at 37 °C and the formed colonies were counted. Based on counting, an OD600 value of 0.44 was correlated to a cell concentration of 3.5 × 107 CFU mL−1.E. coli K12 was cultivated in Luria–Bertani (LB) medium (composed of 5 g of NaCl, 5 g of yeast extract and 10 g of tryptone in 1 L of deionized water) at 37 °C while shaking. Bacterial growth was monitored by OD measurements and bacteria concentration was calculated from OD600 value according to the correlation of 1 OD600 = 108 cells per mL.41
For biosensing experiments, L. acidophilus was grown overnight in MRS medium and a subculture was grown the next morning until a cell density corresponding to an OD600 value of 0.44. E. coli K12 was grown overnight in LB medium and a subculture was grown the next morning until an OD600 value of 0.5. Samples of the cultures (1 mL) were spun down in a standard lab centrifuge (10 min at 5000g), replacing the supernatant by 1 mL SB. Following the re-suspension of the cell pellet, the centrifugation and buffer replacement were repeated two more times before the final cell suspension was either further diluted or used directly for biosensing experiments.
For the biosensing experiments with mixed bacterial populations, upon the final centrifugation and supernatant replacement, a bacteria pellet was re-suspended in 1 mL SB, after which, another bacteria pellet was introduced to form a mixed culture suspension.
For biosensing experiments with non-viable bacteria, bacteria suspensions were heat-treated as suggested by Bunthof et al.48 Bacterial suspensions were placed on a dry heating block for 15 min at 70 °C and then stored on ice until further use.
Preparation and characterization of aptamer-conjugated PSiO2 biosensors
Si wafers were electrochemically etched in a 3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 (v/v) aqueous HF (48%)
1 (v/v) aqueous HF (48%)![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) ethanol solution. Two different etching conditions were used: (i) 30 s at 300 mA cm−2 current density and (ii) 375 s at 24 mA cm−2. CAUTION: HF is a highly corrosive liquid and must be handled with extreme care and under secured working conditions! A strip of aluminium foil was brought in contact with the backside of a Si wafer (exposed area 1.33 cm2) and mounted in an etching cell made from Teflon material. A platinum wire was used as the counter electrode. After etching, the wafer surface was rinsed with ethanol and subsequently dried under dry nitrogen gas flow. The obtained freshly etched PSi samples were thermally oxidized in a tube furnace (Thermolyne) at 800 °C for 1 h in ambient air, resulting in oxidized PSi (PSiO2) films.
ethanol solution. Two different etching conditions were used: (i) 30 s at 300 mA cm−2 current density and (ii) 375 s at 24 mA cm−2. CAUTION: HF is a highly corrosive liquid and must be handled with extreme care and under secured working conditions! A strip of aluminium foil was brought in contact with the backside of a Si wafer (exposed area 1.33 cm2) and mounted in an etching cell made from Teflon material. A platinum wire was used as the counter electrode. After etching, the wafer surface was rinsed with ethanol and subsequently dried under dry nitrogen gas flow. The obtained freshly etched PSi samples were thermally oxidized in a tube furnace (Thermolyne) at 800 °C for 1 h in ambient air, resulting in oxidized PSi (PSiO2) films.
For PSiO2 biofunctionalization, the porous film was incubated in a solution of 20 mM MPTMS in toluene for 1 h. After removal of the solution, the surface was rinsed with toluene, ethanol and acetone (for 2 min each) and dried under a stream of nitrogen. The silanized sample was rinsed with TE buffer prior to 1 h incubation with the aptamer solutions (50 μL) and successive rinsing with TE and PBS buffers. 2 mg mL−1 maleimide were dissolved in PBS, then applied to the aptamer-modified sample, and allowed to react for 1 h. Subsequently, the resulting biosensor was rinsed with PBS and finally incubated (30 min) in SB to allow proper aptamer folding.
High-resolution scanning electron microscopy (HRSEM) of PSiO2 films was performed with a Carl Zeiss Ultra Plus instrument at an accelerating voltage of 1 keV.
For gravimetric characterization, the silicon samples were weighed prior (m1) and after etching and oxidation (m2). Subsequently, the oxide layer was dissolved in a 3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 (v/v) solution of aqueous HF (48%) and ethanol and the remnant porous layer was removed by incubation in 1 M aqueous KOH solution. The remaining sample was weighed again (m3) and the porosity (P) was determined by the following equation:49
1 (v/v) solution of aqueous HF (48%) and ethanol and the remnant porous layer was removed by incubation in 1 M aqueous KOH solution. The remaining sample was weighed again (m3) and the porosity (P) was determined by the following equation:49
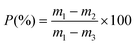 | (1) |
The SLIM method provides a nondestructive determination of thickness and porosity based on data from reflectance spectra obtained while different liquids infiltrate into the pores.49 The differences between the spectra are attributed to the change in optical thickness as the medium in the pores changes, under the assumption that all void spaces are filled equally.50 The optical thickness (2nL, where n is the average refractive index of the porous film and L is its thickness) of the PSiO2 is determined from the interferometric reflectance spectrum of the porous film in air and while immersed in ethanol and acetone, having refractive indices of 1.359 and 1.357, respectively. The refractive index of the SiO2 portion of the film is assumed to be 1.455. Values of 2nL are then fitted to a two-component Bruggeman model of refractive index for the composite-layer, which yields a unique solution for both the porosity and the thickness of the samples.42,50
Silanization of the samples was characterized by the Ellmann assay for free thiol groups.51 Herein, the silanized PSiO2 samples were incubated for 15 min in 2.5 mL Ellman buffer (100 mM sodium phosphate, pH 8.0, 1 mM EDTA) supplemented with 50 μl Ellman solution (4 mg mL−1 of DNTB in Ellman buffer). The supernatant was collected after the reaction and its absorbance was measured at 412 nm using a Varioskan flash plate reader (Thermo Scientific).
Bacteria biosensing
The biosensors were placed in fixed flow cell apparatus, as previously described44 and their interferometric reflectance spectra were collected using an Ocean Optics charge-coupled device (CCD) USB 4000 spectrometer fitted with a microscope objective lens coupled to a bifurcated fiber-optic cable. A tungsten light source was focused onto the center of the flow cell with a spot size of approximately 1–2 mm2. Reflectivity data were continuously recorded every minute in the wavelength range of 400–1000 nm, with a spectral acquisition time of 100 ms. As previous work on probiotic bacteria indicated their sensitivity to light,52,53 a shutter was used to block the light from reaching the sample surface between the measurements. This is to exclude possible effect of light on bacteria growth, behavior and interaction with the aptamer-conjugated surface. Both illumination of the surface and detection of the reflected light were performed along an axis coincident with the surface normal. The collected spectra were analyzed by applying fast Fourier transformation (FFT), as previously described by Massad-Ivanir et al.41 with the FFT intensity changes expressed as percentages and calculated using the following equation: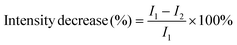 | (2) |
In a typical optical biosensing experiment, a freshly-prepared aptamer-functionalized PSiO2 sample was incubated with SB for 30 min to allow proper folding of the aptamer and to acquire a baseline signal. Thereafter, the respective bacteria suspensions (in SB) were introduced and allowed to incubate with the biosensor for 20 min. After removal of the bacteria suspension and thorough flushing of the cell with SB, the sample was incubated for 10 min in SB. Optical measurements were recorded every 1 min throughout the experiment. Note that during buffer exchange and rinsing steps, optical measurements were shortly paused to allow a thorough washing of the biosensor and the flow cell.
To confirm the bacteria capture on the biosensor surface, the biosensor was removed from the flow cell and immediately examined under a light microscope (ZEISS upright). Images were taken using the microscope camera (Axio Cam MRc, ZEISS).
Results and discussion
Biosensors fabrication and characterization
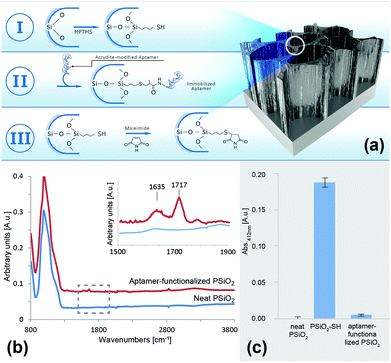
The first step in the biosensor preparation involves an anodization process of a Si wafer at a current density of 300 mA cm−2 for a duration of 30 s. These etching condition were adapted from our previous work on the design of aptamer-functionalized PSi for protein biosensing.44 Following anodization, the resulting porous films were thermally oxidized in order to passivate the silicon hydride surface and render it into a hydrophilic PSiO2 scaffold.50 The detailed nanostructure and physical properties of the resulting PSiO2 were characterized by HRSEM, gravimetry and SLIM and the results are summarized in Table S1 (ESI†). Detailed description of these techniques for the characterization of PSi films was previously reported.42,44,49 The PSiO2 films display a macroporous structure54 of cylindrical pores with a diameter ranging between 55–75 nm, the thickness of the porous layer was ∼5 μm, and a porosity of 79%.A simple three-step biofunctionalization route was used to immobilize the aptamers to the PSiO2 surface, as illustrated in Fig. 1a. The PSiO2 was first silanized with (3-mercaptopropyl)trimethoxysilane (MPTMS) to result in a thiolated surface (Fig. 1a-I), which was then reacted with the acrydite-modified aptamers to form thioether bonds55,56 (Fig. 1a-II). The latter are temperature and pH insensitive and provide stable conjugation of the aptamers to the porous nanostructure.56,57 The third and final step (Fig. 1a-III) involved blocking of the residual thiol groups with maleimide in order to minimize subsequent non-specific reaction with buffers or sample components.
Successful aptamer immobilization was confirmed by ATR-FTIR spectroscopy and the results are presented in Fig. 1b. For neat PSiO2, the typical –(OySiHx) vibration mode at 801 cm−1 and a peak at 1039 cm−1, ascribed to the Si–O–Si stretching mode, were observed clearly. The aptamer-functionalized surface depicted an additional characteristic DNA band at 1635 cm−1 (carbonyl) as well as a new peak at 1717 cm−1, which is attributed to the two C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O stretching frequencies of the maleimide blocking group.58,59 It should be noted that other typical DNA peaks below 1500 cm−1 (e.g., the phosphate diester bands) could not be observed due to the strong absorption of the silicon species, as well as the maleimide C–N–C stretching which overlaps with the Si–O–Si stretching modes (around 1180 cm−1).60–62
O stretching frequencies of the maleimide blocking group.58,59 It should be noted that other typical DNA peaks below 1500 cm−1 (e.g., the phosphate diester bands) could not be observed due to the strong absorption of the silicon species, as well as the maleimide C–N–C stretching which overlaps with the Si–O–Si stretching modes (around 1180 cm−1).60–62
Another validation of the immobilization scheme was performed by using the Ellman assay for thiol-groups. This assay is based on the reduction of 5,5′-dithio-bis-(2-nitrobenzoic acid) (Ellman's reagent, DTNB) in the presence of free sulfhydryls, resulting in the formation of 2-nitro-5-thiobenzoic acid (TNB),51 which can be easily quantified by absorbance measurements at 412 nm. Thus, following silanization (Fig. 1a-I), the resulting modified PSiO2 was incubated with DTNB and the collected supernatant absorbance was measured. The absorbance values are presented in Fig. 1c and compared to those obtained for neat PSiO2 and an aptamer-functionalized PSiO2. Negligible absorbance was observed for both the neat and the aptamer-functionalized PSiO2 samples, confirming that no free sulfhydryls were present on the surface, before silanization and following aptamer conjugation and blocking with maleimide, respectively. On the contrary, the measured absorbance for the silanized PSiO2 was drastically higher, indicative of successful thiolation of the scaffold. Note that aptamer conjugation to mesoporous PSiO2 was studied using the same methods and similar results as those presented above were observed (data not shown).
Optical biosensing experiments
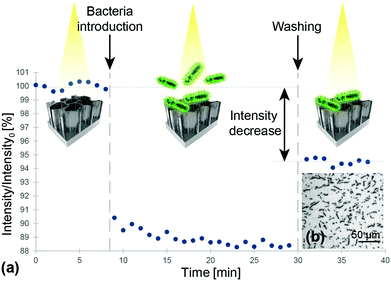
The Hemag1P-modified PSiO2 biosensors were exposed to L. acidophilus suspensions while the reflectivity spectra of the porous film was monitored in real time and the corresponding EOT intensity values were computed. In a typical biosensing experiment, see Fig. 2a, aptamer selection buffer (SB) was first introduced to assure correct folding of the aptamer and to establish the initial intensity baseline on the freshly prepared aptamer-functionalized biosensor. Introduction of L. acidophilus suspension (107 cells per mL in SB) induced a sharp decrease of approximately 10% in relative intensity due to light scattering by the bacteria cells on the biosensor surface; followed by incubation for 20 min with the suspension to allow proper interaction of the bacteria with the aptamer-decorated surface. The L. acidophilus cells targeted by the aptamer Hemag1P possess a typical size of 0.6–0.9 μm in width and 1.5–6 μm in length,20 thus, they are too large to penetrate into the porous nanostructure and they only reside on the biosensor surface influencing the intensity of reflected light. Subsequent washing of the biosensor with SB was performed to remove unbound species; after which the intensity increased and stabilized at a net intensity decrease value of 5.5%. These results are in agreement with our previous work on direct-capture of E. coli with conjugated antibodies41,43,63 and suggest successful capture of the bacteria onto the biosensor surface. However, the profound increase in the intensity during the washing step suggests that a fairly high amount of cells contained in the sample were not captured by the aptamers. A second washing step however, showed no further change in the optical signal, indicating that the remaining bacteria were tightly bound by the aptamers. Microscope images taken immediately after experiments (see Fig. 2b), reveal a large number of bacteria cells, with a typical morphology of L. acidophilus cells, captured onto the biosensor surface.Numerous replications of these biosensing experiments have demonstrated a similar behavior to that presented in Fig. 2 and a highly reproducible net intensity decrease value of 5.5% (± 0.07%) was achieved. However, attempts to detect 106 cells per mL of L. acidophilus demonstrated negligible optical response throughout the biosensing experiments.
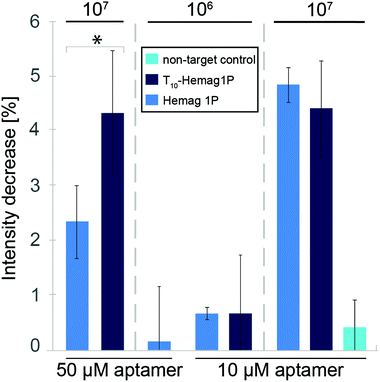
The aforementioned results demonstrate the successful construction of an aptamer-functionalized PSi biosensor and its successful operation in detecting high bacteria concentrations. While these concentrations are relevant for potential application of this biosensor in the food industry, we aimed to increase the dynamic range of the biosensor. Thus, we tuned the nanostructure of the PSi transducer by reducing the pore diameter to exhibit a mesoporous morphology54 with improved optical properties30 (see Table S1, ESI†). Moreover, the increased top surface area (see Fig. S1, ESI†) available for aptamer immobilization may facilitate the interaction between the target bacteria with the capture probe. For the mesoporous PSi transducer, noise-level was considerably reduced although the attained signal was lower (2.4% intensity decrease in comparison to 5.5%), the signal-to-noise ratio has improved significantly (see Fig. S2 and Table S2, ESI†). Based on these results, we have attempted to optimize the performance of the mesoporous PSi in terms of the aptamer surface coverage and spacing. Previous studies have revealed the vital role of the immobilized aptamer surface density, affecting the biosensor response,64,65 especially in systems where the amount of surface-immobilized capture probes directly correlates with the biosensor binding capacity (i.e. one capture probe can bind one target analyte). However, in this study, as the target is several orders of magnitude larger than the capture probe, it is likely that several aptamers bind one bacteria cell. As we used a relatively high aptamer concentration (i.e. 50 μM) to ensure a uniform coverage of the surface, the closely packed aptamers may hamper target cells from capture. The latter is ascribed to the target's limited access to the aptamer as well as to the aptamer's improper folding into secondary structures,66,67 leading to decreased binding capacity.7,68 Thus, two strategies for biosensor optimization were employed: lowering of the immobilization density (utilizing 10 μM instead of 50 μM aptamer solution) and optimization of the aptamer sequence, i.e. extension of the sequence by ten thymine bases positioned between the aptamer sequence and the 5′ acrydite modification (T10-Hemag1P).
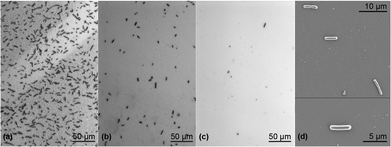
Fig. 3 summarizes the biosensing results for the different aptamer-functionalized mesoporous PSiO2. For the standard Hemag1P sequence as well as the elongated sequence, the optical signals increased at lower aptamer-density (4.9% and 4.5%, respectively) giving rise to the notion that a steric hindrance effect had occurred. At unchanged high aptamer immobilization density (50 μM employed concentration), utilization of T10-Hemag1P has a profound effect and the intensity signal increases from 2.4% (for the standard sequence) to 4.3%. The improved response of the biosensor may be attributed to the enhanced flexibility of the grafted aptamer and its proper folding.69 However, it should be noted that the unmodified Hemag1P sequence yields more reproducible results (see Fig. 3). The latter is observed for all studied bacteria concentrations. Accordingly, in order to allow reproducible and sensitive L. acidophilus detection, we have optimized the concentration of the Hemag1P. We found that lower aptamer concentrations (10 μM) facilitate the detection of 106 cells per mL L. acidophilus (a net intensity decrease of 0.68% was attained). As a negative control and to exclude possible effects of non-specific interactions between surface immobilized DNA and the bacteria, PSiO2 films conjugated with a non-target 6H7 aptamer, which is directed against his-tagged proteins, and exposed to 107 cells per mL L. acidophilus. These experiments resulted in inconsistent low intensity changes, indicative of minor adhesion of bacteria cells to the aptamer-decorated surface.43Fig. 4 presents micrographs of the biosensors’ surface, taken immediately after the biosensing experiments. Dense coverage of cells with a typical L. acidophilus morphology were observed (Fig. 4a). HRSEM images (see Fig. 4d) reveal the rod-shaped bacteria cells, with a characteristic length of ∼5 μm,20 nestled on the PSiO2. Bacteria coverage was observed to decrease profoundly upon exposure to lower bacteria concentration (i.e. 106 cells per mL, see Fig. 4b) in agreement with the lower optical biosensor response. Exposure of PSiO2, decorated with a non-target sequence (6H7 aptamer), to 107 cells per mL L. acidophilus, reveals a negligible number of cells bound to the surface (see Fig. 4c).
Although the change in pore size of the nanostructure improved the performance of the designed biosensor, the sensitivity is still lacking in comparison to similar biosensor systems. While Massad-Ivanir et al. achieved a limit of detection of 103 cells per mL of E. coli with their antibody-conjugated PSi biosensor;42 herein, a limit of detection of only 106 cells per mL was observed. Nevertheless, it should be taken into consideration that for L. acidophilus and other probiotics, a minimum therapeutic daily dose of 108–109 viable cells is suggested and an intake of 100 g of fermented bioproducts with a content of 106–107 viable cells per mL is recommended.20,70 Some probiotic products even claim much higher microorganism contents (e.g. 1011 cells per g in VSL#3 probiotic preparation71). Hence, with regard to its possible application, the sensitivity of the presented biosensor lies in the range of relevant concentrations and excels other systems by far in terms of its simplicity, cost, stability and speed of measurement. In the case of applications that necessitate higher sensitivity, integration of our biosensing scheme with a bacteria pre-concentration step,72 could facilitate detection.
Biosensing in mixtures and heat inactivated samples
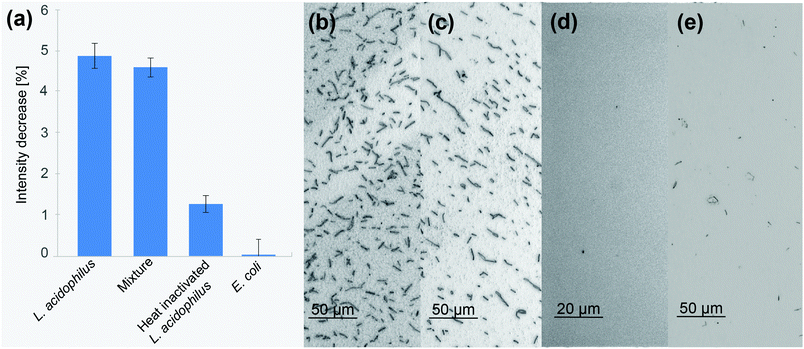
One of the main challenges of biosensors is the ability to perform in “real-time”, detecting the target analyte in its natural environment, surrounded by interfering molecules.44,73 To study the ability of the constructed biosensor to selectively bind its target bacteria in the presence of interfering microorganisms and molecules, the biosensor was exposed to a mixed suspension containing 107 cells per mL L. acidophilus and 107 cells per mL E. coli in SB. The latter was chosen as model Gram-negative bacteria, displaying a different membrane composition,74 which is also of high relevancy in the food industry. Fig. 5 presents the obtained optical signals and respective microscope images taken immediately after the biosensing experiments. Herein, all biosensors used were functionalized with 10 μM Hemag1P aptamer. Exposure of a biosensor to a mixture of 107 cells per mL L. acidophilus and 107 cells per mL E. coli yielded an average intensity decrease of 4.6%, which is in agreement with the results obtained for pure suspensions of 107 cells per mL L. acidophilus, see Fig. 5a. This result was also validated by the comparable amount of bound bacteria cells on the biosensors surface as shown in Fig. 5b and c. This demonstrates the ability of the biosensor to distinguish between the different bacteria species and selectively bind the target cells with high accuracy. As a control, the biosensor was also exposed to a suspension of 107 cells per mL E. coli, resulting in a negligible intensity decrease (Fig. 5a) and no cells were observed on its surface (Fig. 5d).Another important advantageous characteristic of a biosensor for bacteria detection would be its ability to distinguish between live and dead bacteria.75,76 To study the response of the Hemag1P-functionalized PSiO2 biosensor upon exposure to dead L. acidophilus, we have thermally treated the cell suspensions (15 minutes at 70 °C48) prior to biosensing experiments. Fig. 5a displays the biosensor's average intensity decrease upon introduction of these suspensions, revealing their significantly reduced response of 1.5% in comparison to 4.9% for a live culture. This result supports the hypothesis that the Hemag1P aptamer targets the membrane S-protein of the bacteria.19 When L. acidophilus is heat treated, these membrane proteins denature and as a result the aptamer-affinity to the bacteria cells drastically decreases. We suggest that the detected signal may be attributed to a combination of non-specific adsorption of denatured cells and some viable bacteria remaining in the suspensions, see Fig. 5e. This was confirmed by bacterial growth after medium addition to the biosensor. Thus, the biosensor can distinguish between live/dead populations due to the profound difference in its response.
Conclusions
In the presented work, a label-free optical biosensor based on aptamer-functionalized porous Si (Fabry–Pérot thin film) was designed, characterized and optimized. This demonstrates the first label-free detection of L. acidophilus with Hemag1P aptamer as capture probe at relevant concentrations for probiotic intake.20 Exposure to the target suspensions and mixed bacterial populations resulted in highly robust and reproducible changes in the optical reflectivity of the biosensor, given that the bacteria are viable. Control experiments revealed negligible binding of non-target species, confirming the excellent selectivity of this aptamer-based biosensor. Other important advantages of this biosensor are its ability to distinguish between live and dead target bacteria populations, as well as the short total assay time of less than one hour. Biosensing schemes for the rapid and label-free monitoring of live L. acidophilus are highly relevant for the functional food and pharmaceutical industry.70,77 These products become increasingly popular as the importance of the human microbiome and its influence on a variety of diseases is revealed.78,79 However, live bacteria cultures are essential for their probiotic activity.70,80 Finally, the availability of other species-targeting aptamers enables to implement this biosensing concept to facilitate the fast detection and identification of bacteria species in a simple and reliable manner, overcoming the need for time-consuming and unspecific culturing techniques as well as assays requiring highly sophisticated instruments.Acknowledgements
This work was funded by the German Research Foundation under the grant SCHE 279/32-1. ES, SA and KU acknowledge the core services and support from the Lorry I. Lokey Center for Life Science and Engineering.Notes and references
- C. Tuerk and L. Gold, Science, 1990, 249, 505–510 CAS.
- A. D. Ellington and J. W. Szostak, Nature, 1990, 346, 818–822 CrossRef CAS PubMed.
- S. Song, L. Wang, J. Li, C. Fan and J. Zhao, Trends Anal. Chem., 2008, 27, 108–117 CrossRef CAS.
- C. L. A. Hamula, H. Zhang, F. Li, Z. Wang, X. Chris Le and X.-F. Li, Trends Anal. Chem., 2011, 30, 1587–1597 CrossRef CAS.
- M. Lönne, G. Zhu, F. Stahl and J.-G. Walter, in Biosensors Based on Aptamers and Enzymes, ed. M. B. Gu and H.-S. Kim, Springer, Berlin, Heidelberg, 2014, ch. 231, vol. 140, pp. 121–154 Search PubMed.
- G. Zhu, M. Lübbecke, J. Walter, F. Stahl and T. Scheper, Chem. Eng. Technol., 2011, 34, 2022–2028 CrossRef CAS.
- J. G. Walter, O. Kökpinar, K. Friehs, F. Stahl and T. Scheper, Anal. Chem., 2008, 80, 7372–7378 CrossRef CAS PubMed.
- R. Nezlin, Mol. Immunol., 2016, 70, 149–154 CrossRef CAS PubMed.
- M. Ilgu and M. Nilsen-Hamilton, Analyst, 2016, 141, 1551–1568 RSC.
- K. Sefah, J. A. Phillips, X. Xiong, L. Meng, D. Van Simaeys, H. Chen, J. Martin and W. Tan, Analyst, 2009, 134, 1765–1775 RSC.
- R. A. Potyrailo, A. J. Murray, N. Nagraj, A. D. Pris, J. M. Ashe and M. Todorovic, Angew. Chem., Int. Ed., 2015, 54, 2174–2178 CrossRef CAS PubMed.
- A. C. A. Roque, C. R. Lowe and M. Â. Taipa, Biotechnol. Prog., 2004, 20, 639–654 CrossRef CAS PubMed.
- H. M. So, D. W. Park, E. K. Jeon, Y. H. Kim, B. S. Kim, C. K. Lee, S. Y. Choi, S. C. Kim, H. Chang and J. O. Lee, Small, 2008, 4, 197–201 CrossRef CAS PubMed.
- H. P. Dwivedi, R. D. Smiley and L. A. Jaykus, Appl. Microbiol. Biotechnol., 2010, 87, 2323–2334 CrossRef CAS PubMed.
- F. Chen, J. Zhou, F. Luo, A. B. Mohammed and X. L. Zhang, Biochem. Biophys. Res. Commun., 2007, 357, 743–748 CrossRef CAS PubMed.
- X. Cao, S. Li, L. Chen, H. Ding, H. Xu, Y. Huang, J. Li, N. Liu, W. Cao, Y. Zhu, B. Shen and N. Shao, Nucleic Acids Res., 2009, 37, 4621–4628 CrossRef CAS PubMed.
- G. Singh, P. Vajpayee, N. Rani, A. Jyoti, K. C. Gupta and R. Shanker, Ecotoxicol. Environ. Saf., 2012, 78, 320–326 CrossRef CAS PubMed.
- M. Citartan, E.-S. Ch'ng, T. S. Rozhdestvensky and T.-H. Tang, Microchem. J., 2016, 128, 187–197 CrossRef CAS.
- C. L. Hamula, H. Zhang, L. L. Guan, X. F. Li and X. C. Le, Anal. Chem., 2008, 80, 7812–7819 CrossRef CAS PubMed.
- A. M. P. Gomes and F. X. Malcata, Trends Food Sci. Technol., 1999, 10, 139–157 CrossRef CAS.
- N. P. Shah, Int. Dairy J., 2007, 17, 1262–1277 CrossRef.
- C. Zhang, X. Lv, H. Qing, L. Geng and Y. Deng, 2013 ICME International Conference on Complex Medical Engineering (CME), 2013.
- P. Zuo, X. Li, D. C. Dominguez and B.-C. Ye, Lab Chip, 2013, 13, 3921–3928 RSC.
- L. De Stefano, P. Arcari, A. Lamberti, C. Sanges, L. Rotiroti, I. Rea and I. Rendina, Sensors, 2007, 7, 214–221 CrossRef CAS.
- C. Pacholski, Sensors, 2013, 13, 4694–4713 CrossRef CAS PubMed.
- A. Jane, R. Dronov, A. Hodges and N. H. Voelcker, Trends Biotechnol., 2009, 27, 230–239 CrossRef CAS PubMed.
- K. A. Kilian, T. Bocking and J. J. Gooding, Chem. Commun., 2009, 0, 630–640 RSC.
- S. D. Alvarez, M. P. Schwartz, B. Migliori, C. U. Rang, L. Chao and M. J. Sailor, Phys. Status Solidi A, 2007, 204, 1439–1443 CrossRef CAS.
- K.-P. S. Dancil, D. P. Greiner and M. J. Sailor, J. Am. Chem. Soc., 1999, 121, 7925–7930 CrossRef CAS.
- A. Janshoff, K. P. S. Dancil, C. Steinem, D. P. Greiner, V. S. Y. Lin, C. Gurtner, K. Motesharei, M. J. Sailor and M. R. Ghadiri, J. Am. Chem. Soc., 1998, 120, 12108–12116 CrossRef CAS.
- M. M. Orosco, C. Pacholski, G. M. Miskelly and M. J. Sailor, Adv. Mater., 2006, 18, 1393–1396 CrossRef CAS.
- C. Pacholski, C. Yu, G. M. Miskelly, D. Godin and M. J. Sailor, J. Am. Chem. Soc., 2006, 128, 4250–4252 CrossRef CAS PubMed.
- M. P. Schwartz, A. M. Derfus, S. D. Alvarez, S. N. Bhatia and M. J. Sailor, Langmuir, 2006, 22, 7084–7090 CrossRef CAS PubMed.
- M. P. Schwartz, S. D. Alvarez and M. J. Sailor, Anal. Chem., 2006, 79, 327–334 CrossRef PubMed.
- J. Zhang, Y. Wu, B. Zhang, M. Li, S. Jia, S. Jiang, H. Zhou, Y. Zhang, C. Zhang and A. P. F. Turner, Anal. Lett., 2012, 45, 986–992 CrossRef CAS.
- B. Gupta, Y. Zhu, B. Guan, P. J. Reece and J. J. Gooding, Analyst, 2013, 138, 3593–3615 RSC.
- E. Tenenbaum, N. Ben-Dov and E. Segal, Langmuir, 2015, 31, 5244–5251 CrossRef CAS PubMed.
- C. K. Tsang, T. L. Kelly, M. J. Sailor and Y. Y. Li, ACS Nano, 2012, 6, 10546–10554 CAS.
- C. Pacholski, M. Sartor, M. J. Sailor, F. Cunin and G. M. Miskelly, J. Am. Chem. Soc., 2005, 127, 11636–11645 CrossRef CAS PubMed.
- G. Gaur, D. S. Koktysh and S. M. Weiss, Adv. Funct. Mater., 2013, 23, 3604–3614 CrossRef CAS.
- N. Massad-Ivanir, G. Shtenberg, A. Tzur, M. A. Krepker and E. Segal, Anal. Chem., 2011, 83, 3282–3289 CrossRef CAS PubMed.
- N. Massad-Ivanir, G. Shtenberg, T. Zeidman and E. Segal, Adv. Funct. Mater., 2010, 20, 2269–2277 CrossRef CAS.
- E. Tenenbaum and E. Segal, Analyst, 2015, 140, 7726–7733 RSC.
- K. Urmann, J.-G. Walter, T. Scheper and E. Segal, Anal. Chem., 2015, 87, 1999–2006 CrossRef CAS PubMed.
- K. Urmann, E. Tenenbaum, J. G. Walter and E. Segal, in Electrochemically Engineered Nanoporous Materials Methods, Properties and Applications, ed. D. Losic and A. Santos, Springer, 2015, pp. 93–116 Search PubMed.
- S. A. Doyle and M. B. Murphy, US Patent, 0142582A1, 2005 Search PubMed.
- S. P. Riley, M. E. Woodman and B. Stevenson, in Current Protocols Essential Laboratory Techniques, John Wiley & Sons, Inc., 2008, DOI:10.1002/9780470089941.et0402s00.
- C. J. Bunthof, K. Bloemen, P. Breeuwer, F. M. Rombouts and T. Abee, Appl. Environ. Microbiol., 2001, 67, 2326–2335 CrossRef CAS PubMed.
- M. J. Sailor, Porous silicon in practice: preparation, characterization and applications, John Wiley & Sons, 2012 Search PubMed.
- E. Segal, L. A. Perelman, F. Cunin, F. Di Renzo, J. M. Devoisselle, Y. Y. Li and M. J. Sailor, Adv. Funct. Mater., 2007, 17, 1153–1162 CrossRef CAS.
- G. L. Ellman, Arch. Biochem. Biophys., 1959, 82, 70–77 CrossRef CAS PubMed.
- G. Weiss, S. Rasmussen, L. H. Zeuthen, B. N. Nielsen, H. Jarmer, L. Jespersen and H. Frøkiær, Immunology, 2010, 131, 268–281 CrossRef CAS PubMed.
- K. Kiviharju, M. Leisola and N. von Weymarn, Biotechnol. Lett., 2004, 26, 539–542 CrossRef CAS PubMed.
- D. R. Thevenot, K. Toth, R. A. Durst and G. S. Wilson, Biosens. Bioelectron., 2001, 16, 121–131 CrossRef CAS PubMed.
- P. J. Knerr, A. Tzekou, D. Ricklin, H. Qu, H. Chen, W. A. van der Donk and J. D. Lambris, ACS Chem. Biol., 2011, 6, 753–760 CrossRef CAS PubMed.
- I. D. Technlogies, 2014, Technical report available from www.idtdna.com.
- Z. Li, Y. Chen, X. Li, T. I. Kamins, K. Nauka and R. S. Williams, Nano Lett., 2004, 4, 245–247 CrossRef CAS.
- J. Zhou and J. J. Rossi, Mol. Ther. –Nucleic Acids, 2014, 3, e169 CrossRef CAS PubMed.
- T. S. P. Cellet, M. R. Guilherme, R. Silva, G. M. Pereira, M. R. Mauricio, E. C. Muniz and A. F. Rubira, J. Colloid Interface Sci., 2012, 367, 494–501 CrossRef CAS PubMed.
- A. M. Peterson, R. E. Jensen and G. R. Palmese, Compos. Sci. Technol., 2011, 71, 586–592 CrossRef CAS.
- H. Ishida and S. Ohba, Polymer, 2005, 46, 5588–5595 CrossRef CAS.
- R. Voicu, R. Boukherroub, V. Bartzoka, T. Ward, J. T. C. Wojtyk and D. D. M. Wayner, Langmuir, 2004, 20, 11713–11720 CrossRef CAS PubMed.
- N. Massad-Ivanir, G. Shtenberg and E. Segal, J. Visualized Exp., 2013, e50805, DOI:10.3791/50805.
- R. J. White, N. Phares, A. A. Lubin, Y. Xiao and K. W. Plaxco, Langmuir, 2008, 24, 10513–10518 CrossRef CAS PubMed.
- T. Doneux, A. De Rache, E. Triffaux, A. Meunier, M. Steichen and C. Buess-Herman, ChemElectroChem, 2014, 1, 147–157 CrossRef.
- R. Zheng, B.-W. Park, D.-S. Kim and B. D. Cameron, Biomed. Opt. Express, 2011, 2, 2731–2740 CrossRef CAS PubMed.
- A. M. Giovannozzi, C. Renacco, M. Derosas, E. Enrico, A. Farano and A. M. Rossi, Phys. Status Solidi C, 2011, 8, 1878–1882 CrossRef CAS.
- Z. Yang, E. Castrignanò, P. Estrela, C. G. Frost and B. Kasprzyk-Hordern, Sci. Rep., 2016, 6, 21024 CrossRef CAS PubMed.
- C. Daniel, Y. Roupioz, D. Gasparutto, T. Livache and A. Buhot, PLoS One, 2013, 8, e75419 CAS.
- K. Kailasapathy and J. Chin, Immunol. Cell Biol., 2000, 78, 80–88 CrossRef CAS PubMed.
- A. Venturi, P. Gionchetti, F. Rizzello, R. Johansson, E. Zucconi, P. Brigidi, D. Matteuzzi and M. Campieri, Aliment. Pharmacol. Ther., 1999, 13, 1103–1108 CrossRef CAS PubMed.
- X. Xie, J. Bahnemann, S. Wang, Y. Yang and M. R. Hoffmann, Sci. Rep., 2016, 6, 20516 CrossRef CAS PubMed.
- G. A. Zelada-Guillén, S. V. Bhosale, J. Riu and F. X. Rius, Anal. Chem., 2010, 82, 9254–9260 CrossRef PubMed.
- W. W. Navarre and O. Schneewind, Microbiol. Mol. Biol. Rev., 1999, 63, 174–229 CAS.
- C. Lui, N. C. Cady and C. A. Batt, Sensors, 2009, 9, 3713–3744 CrossRef CAS PubMed.
- A. Singh, S. Poshtiban and S. Evoy, Sensors, 2013, 13, 1763 CrossRef CAS PubMed.
- D. C. Lin, Nutr. Clin. Pract., 2003, 18, 497–506 CrossRef PubMed.
- D. Knights, M. Silverberg, R. Weersma, D. Gevers, G. Dijkstra, H. Huang, A. Tyler, S. van Sommeren, F. Imhann, J. Stempak, H. Huang, P. Vangay, G. Al-Ghalith, C. Russell, J. Sauk, J. Knight, M. Daly, C. Huttenhower and R. Xavier, Genome Med., 2014, 6, 1–11 CrossRef PubMed.
- M. C. Cénit, V. Matzaraki, E. F. Tigchelaar and A. Zhernakova, Biochim. Biophys. Acta, 2014, 1842, 1981–1992 CrossRef PubMed.
- P. S. Yeung, M. E. Sanders, C. L. Kitts, R. Cano and P. S. Tong, J. Dairy Sci., 2002, 85, 1039–1051 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c6an00810k |
| This journal is © The Royal Society of Chemistry 2016 |