Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Spatial regulation of a common precursor from two distinct genes generates metabolite diversity†
Chun-Jun
Guo
a,
Wei-Wen
Sun
a,
Kenneth S.
Bruno
b,
Berl R.
Oakley
c,
Nancy P.
Keller
d and
Clay C. C.
Wang
*ae
aDepartment of Pharmacology and Pharmaceutical Sciences, School of Pharmacy, University of Southern California, Los Angeles, CA 90089, USA. E-mail: clayw@usc.edu
bChemical and Biological Process Development Group, Energy and Environment Directorate, Pacific Northwest National Laboratory, Richland, WA 99352, USA
cDepartment of Molecular Biosciences, University of Kansas, Lawrence, KS 66045, USA
dDepartment of Medical Microbiology and Immunology, University of Wisconsin-Madison, Madison, WI 53706, USA
eDepartment of Chemistry, College of Letters, Arts, and Sciences, University of Southern California, Los Angeles, CA 90089, USA
First published on 13th July 2015
Abstract
In secondary metabolite biosynthesis, core synthetic genes such as polyketide synthase genes usually encode proteins that generate various backbone precursors. These precursors are modified by other tailoring enzymes to yield a large variety of different secondary metabolites. The number of core synthesis genes in a given species correlates, therefore, with the number of types of secondary metabolites the organism can produce. In our study, heterologous expression of all the A. terreus NRPS-like genes showed that two NRPS-like proteins, encoded by atmelA and apvA, release the same natural product, aspulvinone E. In hyphae this compound is converted to aspulvinones whereas in conidia it is converted to melanin. The genes are expressed in different tissues and this spatial control is probably regulated by their own specific promoters. Comparative genomics indicates that atmelA and apvA might share a same ancestral gene and the gene apvA is located in a highly conserved region in Aspergillus species that contains genes coding for life-essential proteins. Our data reveal the first case in secondary metabolite biosynthesis in which the tissue specific production of a single compound directs it into two separate pathways, producing distinct compounds with different functions. Our data also reveal that a single trans-prenyltransferase, AbpB, prenylates two substrates, aspulvinones and butyrolactones, revealing that genes outside of contiguous secondary metabolism gene clusters can modify more than one compound thereby expanding metabolite diversity. Our study raises the possibility of incorporation of spatial, cell-type specificity in expression of secondary metabolites of biological interest and provides new insight into designing and reconstituting their biosynthetic pathways.
Introduction
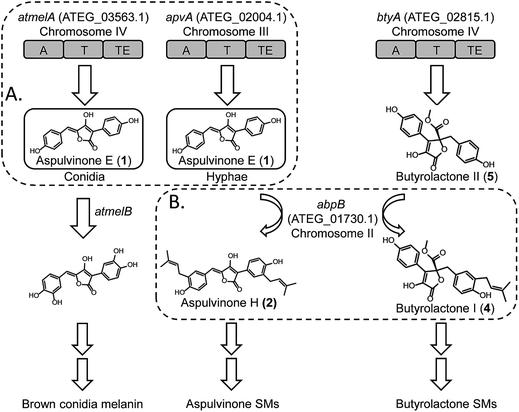
Filamentous fungi, such as species found within the genus Aspergillus, are well known producers of secondary metabolites (SMs) with interesting biological activities. The genome sequencing of Aspergillus species revealed that the number of putative SM genes greatly exceeds the number of identified SMs, suggesting many novel types of SMs still remain to be discovered.1 In general, these SM genes are clustered in the genome.2 A typical cluster contains one key gene required for the synthesis of the precursor skeleton. These genes usually encode large multidomain enzymes belonging to the polyketide synthase (PKS) or non-ribosomal peptide synthetase (NRPS) class.3 Other adjacent genes encode enzymes that are involved in tailoring modifications, transport of the product, or co-regulation of the cluster genes.4 In a given fungal species, the SM arsenal is usually governed by the number of key synthetic genes in the genome, and this diversity is multiplied by various tailoring enzymes that give rise to a considerable number of different natural products.5 For instance, the fungus A. nidulans has 27 PKS genes, 12 NRPS genes, 14 NRPS-like genes and one PKS/NRPS hybrid gene.6 In a previous study, Ahuja et al. systematically characterized the polyketide products of eight non reducing PKSs (NR-PKSs) in A. nidulans.5 In combination with the six previously characterized NR-PKS genes, the study demonstrated that the 14 NR-PKS genes in A. nidulans could be divided into seven groups based on phylogenetic analysis. More importantly, each of these NR-PKSs produces a unique product which can be modified by other tailoring enzymes and incorporate into various SM biosynthetic pathways, indicating the potential of A. nidulans for biosynthesizing a large variety of different PKS-derived SMs.5Recently, application of an efficient gene targeting system enabled us to link two NRPS-like genes apvA and btyA to their corresponding SMs, aspulvinones and butyrolactones, respectively, in A. terreus strain NIH 2624.7 In this study, we demonstrated that one NRPS-like gene, atmelA, is involved in the synthesis of a brown conidial melanin in this fungus.7 Compared to typical NRPSs, NRPS-like genes encode single module (A-T-TE) proteins missing the condensation (C) domain.3,8 The adenylation (A) domain is responsible for aryl acid substrate recognition and activation. The activated substrate is loaded onto the thiolation (T) domain. The thioesterase (TE) domain is suggested to be involved in the condensation and releasing of the final product.8 Therefore, the diversity of NRPS-like products is expanded by a combination of different A domains (substrate selection and activation) and TE domains (various cyclization and release mechanisms). The A. terreus genome contains 14 NRPS-like genes with predicted A-T-TE or similar domain architecture and the SM products for the majority of these are unknown.
Here we report our efforts to systematically characterize the products of these 14 NRPS-like genes. We heterologously expressed each gene in A. nidulans under the control of the inducible alcA promoter. Surprisingly, our study reveals that two NRPS-like genes, apvA and atmelA, are responsible for the formation of the same intermediate, aspulvinone E (1, Fig. 1A). The aspulvinone E produced by AtmelA is further modified by a tyrosinase, AtmelB, and is incorporated into the brown melanin biosynthesis pathway in A. terreus (Fig. 1A). In parallel, the aspulvinone E synthesized by ApvA is further prenylated by a trans-prenyltransferase (AbpB) to produce aspulvinones (Fig. 1B). AbpB also prenylates butyrolactones, and this reveals that modifying genes outside of specific clusters can be responsible for modifying compounds from more than one SM gene cluster, thus expanding the diversity of SMs produced by an organism (Fig. 1B).
Our results suggest that the promoter of apvA drives expression in hyphae resulting in the production of aspulvinone E (1), which is modified to produce aspulvinone variants. The atmelA promoter drives expression in conidia, also resulting in the production of aspulvinone E (1) but in this cell type it is converted to melanin. Further genetic analysis of apvA and atmelA indicates that these two genes may share a common ancestral gene. The gene apvA, which may result from duplication of the ancestral gene, is inserted in a genomic region consisting of genes that codes for life-essential proteins. Our study suggests an unprecedented pathway for conidial pigment biosynthesis in A. terreus that incorporates an NRPS-like product aspulvinone E (1) as its substrate (Fig. 1). Our data also demonstrated how the SM diversity can be expanded by (1) allocating the same natural product in different fungal compartments to produce molecules with different functions; (2) encoding tailoring enzyme that is capable of chemically modifying more than one type of SMs.
Results
Heterologous expression of the NRPS-like genes apvA and atmelA in A. nidulans both result in aspulvinone E production
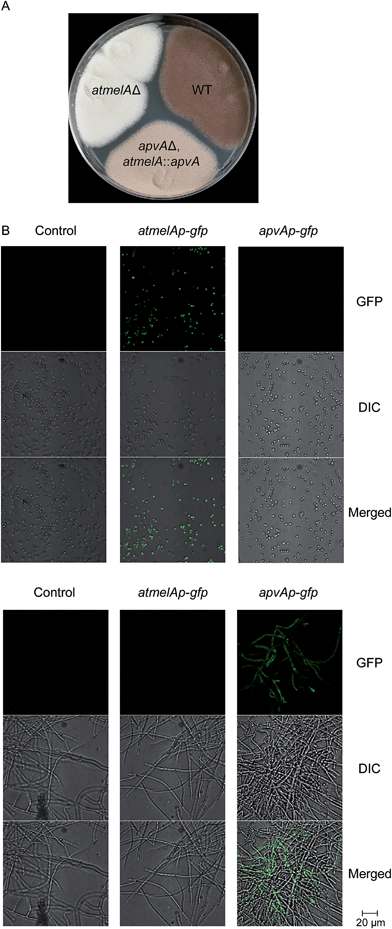
Using a recently reported efficient heterologous expression (HE) system in A. nidulans,9 the individual NRPS-like genes identified in the A. terreus genome were expressed at either the wA or yA locus of A. nidulans under regulation of the inducible alcA promoter (Fig. S1A†). In a previous study, targeted deletion of apvA depleted production of aspulvinones in A. terreus indicating that this NRPS-like gene is responsible for the biosynthesis of the aspulvinone core.7 As expected, heterologous expression of apvA results in accumulation of aspulvinone E (1) which is speculated to be the first intermediate in the aspulvinone pathway (Fig. 1 and 2A and B).7 Unexpectedly, our HE experiments revealed that AtmelA produces the same compound (Fig. 2A and B). The gene apvA is responsible for aspulvinone biosynthesis while atmelA is involved in the synthesis of the brown conidial pigment.7 The brown conidial melanin is still produced in the apvA deletant strain as shown in Fig. 3A. In contrast, deletion of atmelA generates an albino mutant that is still capable of synthesizing aspulvinones.7 Together, these data reveal that despite having the same activity (i.e. synthesis of aspulvinone E), ApvA and AtmelA function in different roles in the fungus.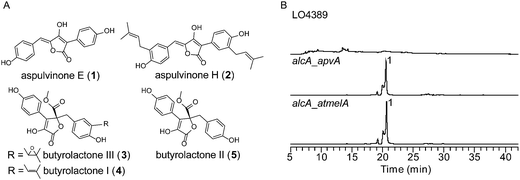 | ||
| Fig. 2 (A) Compounds related to this study. (B) HPLC profiles of extracts of HE strains as detected by total scan UV. | ||
 | ||
| Fig. 3 (A) Phenotype of A. terreus wild type and other A. terreus mutant strains growing on LCMM for 5 days. Morphological change of the fungal conidia can be observed if the deleted genes are involved in the biosynthesis of the brown conidial pigment. Total extracts (B), conidial extracts (C) and hyphal extracts (D) of A. terreus wild type and mutant strains as detected by UV at 370 nm. Aspulvinone related natural products are labeled in red. In B–D, the black box represents the top layer agar as described in tissue-specific extraction. The green box represents the bottom layer agar in tissue-specific extraction. The numbering of the peaks corresponds to the natural products shown in Fig. 2A. The aspulvinone E (1) and its related natural products can be detected in LC/MS traces Bi, Bii, Biii, Ciii, Di, Dii, and Diii. Trace amount of 1 identified in D iii is due to the diffusion of this compound synthesized in the conidia of the mutant strain. *This metabolite is related with aspulvinones according to its UV absorption and MS spectrum. | ||
Aspulvinone E occurs as a precursor in both aspulvinone and melanin pathways
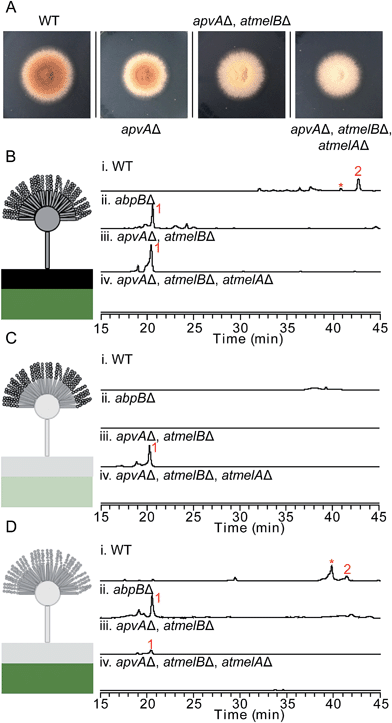
To test our hypothesis that the aspulvinone E is an intermediate in two different pathways, the aspulvinone pathway and melanin pathway, we wished to delete the first tailoring gene in each pathway to accumulate the precursor produced by the gene responsible for the backbone metabolite. Since we have established the genetic linkage between apvA and aspulvinones,7 we set out to identify the first tailoring enzyme, which we presumed was responsible for prenylating 1 to give 2 (Fig. 2A). However, we could not locate a prenyl transferase (PT) gene proximal to apvA.7 We then targeted each of the putative PT genes in the A. terreus genome for deletion (Fig. 4). The individual genes were knocked out using fragments created by fusion PCR.7 The SM profiles of the correct mutants were examined by LC-MS, and of the 11 different mutants, only the ATEG_01730.1Δ strain accumulated aspulvinone E (1) (Fig. 4). Unexpectedly, removal of ATEG_01730.1 leads to the accumulation of butyrolactone II (5) as well (Fig. 4). A previous study showed that the NRPS-like gene btyA is responsible for the biosynthesis of butyrolactone core.7 The expression of three genes, apvA, btyA, and abpB were also analyzed using real-time quantitative reverse transcription PCR (qRT-PCR). (Fig. S2C†) Our data showed that these three genes are co-expressed under aspulvinone and butyrolactone producing conditions. These pieces of evidence suggest that this single PT is responsible for the prenylation of two different metabolites, aspulvinone E (1) and butyrolactone II (5). Thus, we name the gene a (aspulvinone) b (butyrolactone) p (PT) B.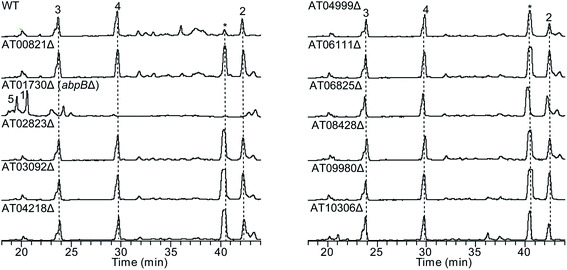 | ||
| Fig. 4 HPLC profiles of extracts of A. terreus prenyltransferase (PT) genes deletants as detected by UV at 330 nm and 370 nm. The numbering of the peaks corresponds to the natural products shown in Fig. 2A. Compounds 3 and 4 are prenylated derivatives of compound 5. The “*” compound is an aspulvinone derivative according to its UV absorption spectrum. ATXXXXX is abbreviated for “ATEG_XXXXX.1”. Deletion of the gene ATEG_00702.1, a homolog of tdiB involved in the asterriquinone biosynthesis, was not achieved due to the unsuccessful PCR amplification of its flanking region. | ||
Definitively establishing the role of AtmelA in the synthesis of aspulvinone E (1) required additional gene deletion experiments. We first deleted the gene apvA using the direct repeat (DR) strategy10 followed by the recycling of the AfpyrG marker (Fig. S1B†). This ensures that aspulvinone E (1) identified in the later mutants is not from ApvA. However, limited information is available about the biosynthesis of the brown conidial melanin in A. terreus. Our study suggests that the melanin pathway originates from the NRPS-like product aspulvinone E (1), which is unlike the two main precursors [di-hydroxyphenylalanin (DOPA) and dihydroxynaphthalene (DHN)] of currently known melanins.11 The biosynthesis of DOPA melanin starts with a tyrosine that is oxidized to either DOPA or dopaquinone (DAQ) by a tyrosinase.11 Considering that aspulvinone E (1) shares the same phenol moiety as tyrosine, we speculate that a similar tyrosinase might catalyze the hydroxylation of 1 at the ortho position to give a dihydroxylated intermediate that becomes incorporated into the brown melanin (Fig. 1). Following the cluster paradigm, we examined the genes surrounding atmelA and identified one gene ATEG_03564.1 (atmelB) that encodes a putative tyrosinase. Removal of this gene in the apvAΔ background changed the strain's phenotype: the brown melanin was no longer synthesized and the conidia became bright yellow (Fig. 3A). As expected, the yellow material is aspulvinone E (1), which accumulated in the metabolite profiles of the apvA and atmelB double deletion strain (Fig. 3B). We next deleted the gene atmelA in the double apvAΔ, atmelBΔ strain. The LC-MS profile of this triple deletion mutant shows the abrogation of the aspulvinone E (1) which had reappeared in the double mutant (Fig. 3B), in accord with our HE results that AtmelA was also capable of producing Aspulvinone E (1).
Aspulvinone E (1) produced by ApvA and AtmelA accumulates in different fungal tissues
Our studies indicate that ApvA and AtmelA synthesize the same product aspulvinone E (1). We next asked how the fungus is able to allocate the same chemical synthesized by different proteins into their own specific pathway without cross-interference. Previous literature reports have shown that the production of SMs and/or their precursors can be specific to both cellular organelle and fungal tissue.12,13 We speculated that the aspulvinone E (1) from the two genes might be generated in different fungal tissues. Since removal of atmelA or atmelB changes the phenotype of A. terreus conidia (Fig. 3A), it is likely that aspulvinone E (1) from AtmelA might be produced in conidia. Likewise aspulvinones, derived from 1 that is produced by ApvA, might be produced inside the hyphae and secreted into the medium.To test this hypothesis, we performed tissue-specific extraction12,13 to reinvestigate the SM profiles of the four strains ((1) wild type; (2) abpBΔ; (3) apvAΔ, atmelBΔ; (4) apvAΔ, atmelBΔ, atmelAΔ). Fungal cultures were divided into three fractions: (1) conidial (mostly conidia and minor conidiophore), (2) top layer agar (mixture of conidiophore, vegetative hyphae, minor invasive hyphae), (3) lower layer agar (mostly invasive hyphae) (Fig. 3C and D). Compared to the SM profiles of total extracts (Fig. 3B), extraction of the conidia showed the accumulation of 1 only in the apvAΔ, atmelBΔ strain (Fig. 3C), not in strains carrying abpBΔ or atmelAΔ. This result indicates that aspulvinone E (1) from AtmelA is produced in conidia. Extraction of the hyphal (mostly invasive) fraction showed the production of 1 in the abpBΔ strain and compound 2 in wild type, indicating that aspulvinone E (1) from ApvA and its derivative 2 are specifically produced in hyphae.
Exchanging atmelA with apvA, under control of the atmelA promoter, restores melanin production in A. terreus
Next, we asked about the molecular mechanism for regulating the tissue-specific production of 1. We first examined the expression profiling of atmelA and apvA in different tissues using Real-Time qRT-PCR (Fig. S2, see ESI† for experimental details). As expected, the gene atmelA is specifically expressed in conidia compartment (Fig. S2B†) while the gene apvA is locally expressed in hyphae (Fig. S2C†). We then probed this question by determining whether ApvA could replace AtmelA in brown melanin biosynthesis. We assumed that tailoring enzymes like AtmelB could still recognize aspulvinone E (1) generated from either ApvA or AtmelA. Another study suggests that the products of two tubulin genes, benA and tubC, are functionally interchangeable. The method they used was to disrupt benA and then put tubC under control of the benA promoter.14,15 Herein we implemented a similar strategy by replacing the coding region of atmelA with that of apvA, placing apvA under control of the atmelA promoter (atmelAp) in the apvAΔ background (Fig. S1Ci†). As shown in Fig. 5A, the mutant stain (apvAΔ, atmelA::apvA) produces brownish conidia indicating that the brown conidial pigment is produced in the mutant strain (apvAΔ, atmelA::apvA). As anticipated, the production of aspulvinones (1 or 2) were not detected (Fig. S3†). The pigment is not produced as much as in the wild type (Fig. 5A) probably because atmelAp, after all, is not the native promoter of ApvA. Quantitative analysis of their expression using Real-Time qRT-PCR reveals that the expression level of apvA, which is under the control of atmelA promoter, is lower than that of atmelA. (Fig. S2B†) This could be one of the reasons for reduced production of the conidial melanin in the mutant strain (apvAΔ, atmelA::apvA). But more importantly, our experiment shows that compound 1 from apvA can be incorporated into the melanin pathway when apvA is regulated by atmelAp. As mentioned earlier, the production of 1 from atmelA is conidia-specific. Thus, this experiment shows that the product of ApvA can also be produced inside conidia and incorporated into melanin, suggesting that the tissue specific allocation of their products may be due to cell-type specific expression of the genes atmelA and apvA under regulation of their specific promoters.Tissue specific expression of gfp occurs when the coding sequence of gfp replaces atmelA and apvA
To test our hypothesis, we generated two mutants (atmelAp-gfp, apvAp-gfp) in which the coding regions of apvA and atmelA were replaced by the green fluorescent protein coding sequence (gfp), placing gfp under control of their specific promoters (Fig. S1Cii†). Both the melanin pathway and aspulvinones biosynthetic pathways are active when A. terreus is cultivated on LCMM agar. Under this culture condition, we hypothesized that atmelAp would turn on the expression of gfp specifically inside the conidia while the gfp regulated by apvAp would display a green fluorescent signal in the hyphae. As expected, we were able to visualize localization of GFP within the conidia only in the strain carrying atmelAp-gfp but not in the parental strain (used as control) or the strain apvAp-gfp. (Fig. 5B) In comparison, hyphal localization of GFP fluorescence was observed only in strain apvAp-gfp. (Fig. 5B) Thus, the tissue specific accumulation of aspulvinone E (1) is probably due to cell-type-specific expression of the two genes apvA and atmelA.Discussion
Our study suggests that metabolite diversity can be expanded by spatial regulation of the same precursor biosynthesized by two distinct genes. Melanin plays an important role in fungal pathogenesis as well as in the protection of the producing organisms from ultraviolet radiation.11,16,17 No definite structures for melanins have been elucidated due to their large molecular weight, insolubility in aqueous or organic solvents, and heterogeneity.11 The characterized conidial melanin of most Aspergillus species belongs to the dihydroxynaphthalene (DHN) melanin class.11 Our study reports a unique and unprecedented pathway of conidial pigment biosynthesis in A. terreus that originates from the NRPS-like product aspulvinone E (1) (Fig. 1). We also identified one putative tyrosinase encoded by the gene atmelB that is involved in a tailoring modification of aspulvinone E (1) to yield the brown conidial pigment (Fig. 1). We speculate that the function of the tyrosinase, AtmelB, might resemble that of its homolog which participates in dihydroxyphenylalanine (DOPA)-melanin biosynthesis11 and catalyzes the ortho hydroxylation of the phenol moiety in the aspulvinone E (1) core (Fig. 1). It is entirely possible that other genes are involved in the polymerization of the dual hydroxylated aspulvinone E and more efforts will be needed to decipher the complete biosynthetic pathway of the brown conidial melanin in A. terreus.Compounds in the aspulvinone family have been found to have anti-influenza A viral (H1N1) activity and are potent inhibitors of firefly luciferase.18,19 The production of aspulvinones, as revealed in a previous study, is a general stress response of A. terreus.20 The biological functions of aspulvinones, and what role they play in the biology of A. terreus, remain elusive. AtmelA shares 73% sequence similarity with ApvA. We noted that there is a 63 bp sequence in atmelA, corresponding to the TE domain of AtmelA, that has strong nucleotide identity with a portion of apvA, indicating that these two genes may share a common origin (Fig. S5C†). Interestingly, homology analysis of apvA and its surrounding genes showed that apvA is inserted into a conserved genome region consisting of genes that are essential for fungal survival (Fig. S4†). It is possible that the insertion of apvA is due to the duplication and translocation of its ancestral homolog. The production of aspulvinone-type SMs by A. terreus, in response to general stress, could be related to the chromosomal location of its producing gene apvA. However, more experiments are definitely necessary to elucidate the exact role the aspulvinone-type metabolites play in the growth, survival or metabolic processes of the fungus A. terreus.
Previous literature suggested that p-hydroxylphenylpyruvate (HPP), which originates from the shikimate pathway, is the biosynthetic precursor of aspulvinones.21,22 The shikimate pathway, highly conserved in bacteria, fungi, and plant species, generates carbon skeletons for the aromatic amino acids including tryptophan, tyrosine, and phenylalanine.23 HPP is proposed to be an intermediate in the conversion of prephenate to L-tyrosine in the shikimate pathway. Limited information has been reported regarding the tissue localization of HPP in fungi. Our study suggests that the distribution of HPP is not limited to certain types of fungal compartments since both the biosynthesis of brown conidial melanin and hyphae specific aspulvinones requires the presence of HPP. Our data indicate that the tissue-specific expression of apvA and atmelA are due to the promoters of the two genes. The promoter of apvA drives expression specifically in hyphae while the promoter of atmelA drives expression in conidia. There are many examples of hyphal versus spore specific gene regulation. For example, the expression of the melanin synthesis gene atmelA might regulated by spore specific transcriptional regulators as demonstrated in a recent study showing that BrlA, the transcription factor required to initiate conidiophore development in Aspergillus spp., is necessary for fumiquinazoline gene expression and product production.13 The fact that atmelAp is capable of turning on the expression of both apvA (SM gene) and the gfp (reporter gene), suggests that it might be a useful tool for directing the expression of genes specifically inside of conidia.
Besides spatial regulation of the intermediate, the SM diversity can be further expanded, as shown in our study, through tailoring enzymes that are capable of modifying SMs with different chemical scaffolds. Early literature reported the enzymatic characterization of aspulvinone dimethylallyltransferase in A. terreus. This enzyme is capable of catalyzing the mono or dual prenylation of aspulvinone E (1).24 The substrate promiscuity of some PTs has also been tested in vitro by feeding experiments.25 Our study reveals the in vivo versatility of AbpB, as it accepts substrates with different chemical scaffolds. Interestingly, the three genes are dispersed in the A. terreus genome (abpB on chromosome II; apvA on chromosome III; btyA on chromosome IV), representing another deviation from the SM gene cluster paradigm. We have discovered in A. nidulans that in some cases, the corresponding PT genes are not located in the same cluster as the core PKS genes.26,27 Further comparison of the butyrolactones with aspulvinones shows that the chemical modifications after prenylation, including epoxidation and dehydrogenation, are very similar (Fig. S6†). Thus, these two natural product families may share the same set of tailoring enzymes that specifically modifies the prenyl groups attached by AbpB.
Fungi are capable of producing a large variety of SMs with unique chemical scaffolds. It is usually expected that core synthetic genes, in a given species, are behind the biosynthesis of different core intermediates, as demonstrated in a study showing that all individual NRPKS in A. nidulans generate a unique PKS product.5 Phylogenetic analysis, using the protein sequences of those NRPS-like homologs obtained from the Broad Institute Aspergillus comparative database, revealed several other characterized NRPS-like homologs including TdiA (terrequinone A biosynthesis in A. nidulans),8 RalA (ralfuranone biosynthesis in Ralstonia solanacearum),28 MicA (microperfuranone biosynthesis in A. nidulans),29 AtqA (asterriquinone biosynthesis in A. terreus),7 BtyA (butyrolactone biosynthesis in A. terreus),7 and EchA (echosides biosynthesis in Streptomyces sp. LZ35)30 (Fig. S5†). These NRPS-like enzymes, with A-T-TE domain architecture, fall within Clade I of the phylogenetic tree (Fig. S5†). They are capable of synthesizing natural products with various chemical scaffolds using similar substrates like phenylpyruvic acid. Previous literature suggested that these pyruvic acid substrates could be produced via the shikimate pathway.21,31,32 The TE domains are proposed to catalyze various condensation, cyclization and releasing reactions to yield different chemical backbones.8 In comparison, another characterized NRPS-like protein (encoded by ATEG_03630.1), with A-T-R domain arrangement, belongs to Clade V (Fig. S5†).33 In this case, the aryl acid substrate, generated by an adjacent PKS (encoded by ATEG_03629.1), is loaded onto the A domain and is reduced to its aryl-aldehyde precursor by the R domain. Thus, SM diversity is enriched from these disparate starting points. It is also common to identify two highly homologous core synthetic genes, in different species, that produces the same precursor. For example, in the biosynthesis of the meroterpenoids austinol (A. nidulans) and terretonin (A. terreus), the two PKS genes ausA and trt4 synthesize the same intermediate 3, 5-dimethylorsellinic acid.34
Conclusion
Our work indicates that chemical diversity in SM biosynthesis can be expanded via multiple dimensions. Prior to our work, the richness of the fungal SM pool was presumed to be determined by the number of SM gene clusters while the clustered genes are associated with the biosynthesis of a distinct type of SM, with some exceptions.12,13,35 In our study, we have shown that A. terreus deploys an additional strategy to enrich its natural product pool. Although the same precursor aspulvinone E (1) is shared in two pathways (the aspulvinone pathway, a typical SM pathway and the melanin pathway, producing a self-protection pigment that might also be involved in the pathogenicity of this fungus), the tissue specific expression of their biosynthetic genes results in the production of the same compound in different fungal tissues and allows it to be modified into two different products that, we assume, confer selective advantages in the specific tissues in which they are produced. The localization of production is possibly regulated by their specific promoters, but it is entirely possible that a more complex regulatory mechanism underlies this phenomenon. This expands our insight into spatial regulation of SMs in fungi.36 More investigation into these promoters might provide a means to the cell-type-directed biosynthesis of SMs or manipulating the location of the expression of some genes responsible for products with interesting biological activities. Finally, our data demonstrate that AbpB prenylates compounds in two pathways revealing that two pathways may share the same tailoring genes. It will be of interest to determine if this characteristic is common to, and specific to, prenyl transferases.Materials and methods
Strains and molecular manipulations
Primers used in this study are listed in Table S1.† The fungal strains used in this study are listed in Table S2.† The scheme for heterologously expressing the NRPS-like genes of A. terreus in A. nidulans is shown in Fig. S1.† The direct repeat (DR) deletion and AfpyG marker recycling experimental design is shown in Fig. S1.† The construction of fusion PCR products, protoplast generation, and transformation were carried out as previously described.7,9 The scheme of diagnostic PCR is shown in Fig. S8.†For real time qRT-PCR, the A. terreus wild type strain and the mutant strain (apvAΔ, atmelA::apvA) were cultivated on LCMM agar for mRNA extraction from conidia. The A. terreus wild type strain was cultivated in LCMM liquid broth for mRNA extraction from hyphae. Total mRNA was extracted by using the Qiagen RNeasy Plant Mini Kit. The cDNA was made from the equal amount of mRNA. The expression of every gene was analyzed with the ABI 7900HT Fast Real-Time PCR system (see ESI for experimental details†).
Fermentation and LC-MS analysis
(1) A. nidulans strain LO4389 and other HE mutant strains were cultivated at 37 °C in 50 ml LMM liquid medium (6 g l−1 NaNO3, 0.52 g l−1 KCl, 0.52 g l−1 MgSO4·7H2O, 1.52 g l−1 KH2PO4, 20 g l−1 lactose supplemented with 1 ml l−1 of trace element solution) at 1 × 106 spore per ml per 125 ml flask with shaking at 180 rpm. The nutrients uridine, uracil, pyridoxine and riboflavin were supplemented if necessary. As previously reported,9 44 μl (10 mM) cyclopentanone was added into the medium after 18 h of incubation. The incubator temperature was then changed to 30 °C and the culture medium was collected 72 h after cyclopentanone induction. The medium was filtrated and extracted twice with ethyl acetate (EtOAc, 50 ml). Preparation of the HPLC-MS samples and the condition for MS analysis were as previously described.7(2) For the prenyltransferase screening experiment, A. terreus NIH 2624 and the mutant strains were point inoculated at 30 °C on LCMM agar plates (6 g l−1 NaNO3, 0.52 g l−1 KCl, 0.52 g l−1 MgSO4·7H2O, 1.52 g l−1 KH2PO4, 10 g l−1D-glucose, 20 g l−1 lactose, 15 g l−1 agar supplemented with 1 ml l−1 of trace element solution) per plate (D = 10 cm). After 5 days, agar was chopped into small pieces and extracted with 50 ml MeOH followed by 60 ml 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 CH2Cl2/MeOH. The extract was evaporated in vacuo to yield a water residue, which was suspended in 25 ml H2O and partitioned with 25 ml EtOAc, twice. Preparation of the HPLC-MS samples and the condition for MS analysis were the same as previously described.7
1 CH2Cl2/MeOH. The extract was evaporated in vacuo to yield a water residue, which was suspended in 25 ml H2O and partitioned with 25 ml EtOAc, twice. Preparation of the HPLC-MS samples and the condition for MS analysis were the same as previously described.7
(3) For the extraction of different fungal tissues for aspulvinone production, the procedure was similar to a previously reported procedure.12,13 Plates containing 20 ml LCMM agar (1.5% agar) were overlaid with another 10 ml of LCMM agar (0.75% agar). Plates were point inoculated with wild type or the DR mutant strains and grown at 30 °C for 4 days. Conidia were harvested by adding 7 ml salt solution (8.5 g l−1 NaCl) followed by gently scraping with a sterile spreader. The conidial solution was inspected under the microscope to be largely free of hyphae and conidiophores. The conidial fraction was sonicated for 1 h and extracted twice with an equal volume of EtOAc. The top layer was then washed with 10 ml sterile water twice and removed with a sterile spatula. The bottom agar layer was then chopped into small pieces and extracted as previously described. Preparation of the HPLC-MS samples from different tissues and the conditions for MS analysis were as previously described.7
Isolation of secondary metabolites
For scale up, A. nidulans HE strains were cultivated at 37 °C in 1 liter LMM liquid medium (∼100 ml per flask) at 1 × 106 spores per ml with shaking at 180 rpm. The nutrients uridine, uracil, pyridoxine and riboflavin were supplemented if necessary. To induce expression, 88 μl cyclopentanone was added into the medium after 18 h of incubation. The incubator temperature was then changed to 30 °C and the culture medium was collected 72 h after cyclopentanone induction. The medium was filtrated and extracted three times with equal volume of EtOAc. The combined EtOAc layers were evaporated to a crude extract. Further purification of fractions with targeted compounds was carried out by gradient HPLC on a C18 reverse phase column [Phenomenex Luna 5 μm C18, 250 × 10 mm] with a flow rate of 5.0 ml min−1 and measured by a UV detector at 254 nm.Fluorescence microscopy
The wild type and two mutant strains (atmelAp-gfp, apvAp-gfp) were cultivated on the two-layer agar plates as mentioned. Plates were point inoculated with wild type or the mutant strains and grown at 30 °C for 5 days. The conidia were collected as previously described and the conidial solution was diluted 10 times with sterile water. A 10 μl portion of the diluted solution was placed on a pre-cleaned microscope slide and covered with a coverslip. For imaging of the hyphal GFP fluorescence signal, the top layer was removed as previously described. A small portion of hyphae containing agar (<10 μl) was placed on a pre-cleaned microscope slide and covered with a coverslip. All the samples were examined under an optical microscope to confirm that the conidia and hyphae could be clearly visualized. Images were taken with a Zeiss LSM 510 Meta NLO (Thornwood, NY) confocal imaging system equipped with Argon, red HeNe, and green HeNe lasers and a Coherent Chameleon Ti–Sapphire laser mounted on a vibration-free table.Acknowledgements
We thank Dr James F. Sanchez, Dr Yi-Ming Chiang, and Dr Shu-Lin Chang for their insightful discussion. Chun-Jun Guo was supported by the National Science Foundation – Emerging Frontiers in Research and Innovation-MIKS (Grant 1136903). Research conducted in the lab of Clay C. C. Wang is supported by the National Institute of General Medical Sciences GM084077, the National Science Foundation – Emerging Frontiers in Research and Innovation-MIKS (Grant 1136903), the Department of Energy, Bioenergy Technologies office, the Department of Defense Strategic Environmental Research and Development program (SERDP), and the National Aeronautics and Space Administration Space Biology program.References
- N. Khaldi, F. T. Seifuddin, G. Turner, D. Haft, W. C. Nierman, K. H. Wolfe and N. D. Fedorova, Fungal Genet. Biol., 2010, 47, 736 CrossRef CAS PubMed.
- N. P. Keller, G. Turner and J. W. Bennett, Nat. Rev. Microbiol., 2005, 3, 937 CrossRef CAS PubMed.
- M. A. Fischbach and C. T. Walsh, Chem. Rev., 2006, 106, 3468 CrossRef CAS PubMed.
- D. W. Brown, J. H. Yu, H. S. Kelkar, M. Fernandes, T. C. Nesbitt, N. P. Keller, T. H. Adams and T. J. Leonard, Proc. Natl. Acad. Sci. U. S. A., 1996, 93, 1418 CrossRef CAS.
- M. Ahuja, Y.-M. Chiang, S.-L. Chang, M. B. Praseuth, R. Entwistle, J. F. Sanchez, H.-C. Lo, H.-H. Yeh, B. R. Oakley and C. C. C. Wang, J. Am. Chem. Soc., 2012, 134, 8212 CrossRef CAS PubMed.
- H. von Döhren, Fungal Genet. Biol., 2009, 46, S45 CrossRef PubMed.
- C. J. Guo, B. P. Knox, J. F. Sanchez, Y. M. Chiang, K. S. Bruno and C. C. Wang, Org. Lett., 2013, 15, 3562 CrossRef CAS PubMed.
- C. J. Balibar, A. R. Howard-Jones and C. T. Walsh, Nat. Chem. Biol., 2007, 3, 584 CrossRef CAS PubMed.
- Y. M. Chiang, C. E. Oakley, M. Ahuja, R. Entwistle, A. Schultz, S. L. Chang, C. T. Sung, C. C. Wang and B. R. Oakley, J. Am. Chem. Soc., 2013, 135, 7720 CrossRef CAS PubMed.
- J. B. Nielsen, M. L. Nielsen and U. H. Mortensen, Fungal Genet. Biol., 2008, 45, 165 CrossRef CAS PubMed.
- K. Langfelder, M. Streibel, B. Jahn, G. Haase and A. A. Brakhage, Fungal Genet. Biol., 2003, 38, 143 CrossRef CAS.
- E. Berthier, F. Y. Lim, Q. Deng, C. J. Guo, D. P. Kontoyiannis, C. C. Wang, J. Rindy, D. J. Beebe, A. Huttenlocher and N. P. Keller, PLoS Pathog., 2013, 9, e1003289 CAS.
- F. Y. Lim, B. Ames, C. T. Walsh and N. P. Keller, Cell. Microbiol., 2014, 16, 1267 CrossRef CAS PubMed.
- G. S. May, J. Cell Biol., 1989, 109, 2267 CrossRef CAS.
- B. R. Oakley, Fungal Genet. Biol., 2004, 41, 420 CrossRef CAS PubMed.
- H. C. Eisenman and A. Casadevall, Appl. Microbiol. Biotechnol., 2012, 93, 931 CrossRef CAS PubMed.
- Q. Gao and F. Garcia-Pichel, Nat. Rev. Microbiol., 2011, 9, 791 CrossRef CAS PubMed.
- H. Gao, W. Guo, Q. Wang, L. Zhang, M. Zhu, T. Zhu, Q. Gu, W. Wang and D. Li, Bioorg. Med. Chem. Lett., 2013, 23, 1776 CrossRef CAS PubMed.
- P. G. Cruz, D. S. Auld, P. J. Schultz, S. Lovell, K. P. Battaile, R. MacArthur, M. Shen, G. Tamayo-Castillo, J. Inglese and D. H. Sherman, Chem. Biol., 2011, 18, 1442 CrossRef CAS PubMed.
- A. Hanlon and S. O'Connor, 2006, http://hdl.handle.net/1721.1/37692.
- K. Nitta, N. Fujita, T. Yoshimura, K. Arai and Y. Yamamoto, Chem. Pharm. Bull., 1983, 31, 1528 CrossRef CAS.
- P. M. Dewick, Nat. Prod. Rep., 1984, 1, 451 RSC.
- T. Tohge, M. Watanabe, R. Hoefgen and A. R. Fernie, Front. Plant Sci., 2013, 4, 62 Search PubMed.
- I. Takahashi, N. Ojima, K. Ogura and S. Seto, Biochemistry, 1978, 17, 2696 CrossRef CAS.
- X. Yu, X. Xie and S. M. Li, Appl. Microbiol. Biotechnol., 2011, 92, 737 CrossRef CAS PubMed.
- J. F. Sanchez, R. Entwistle, J.-H. Hung, J. Yaegashi, S. Jain, Y.-M. Chiang, C. C. C. Wang and B. R. Oakley, J. Am. Chem. Soc., 2011, 133, 4010 CrossRef CAS PubMed.
- H.-C. Lo, R. Entwistle, C.-J. Guo, M. Ahuja, E. Szewczyk, J.-H. Hung, Y.-M. Chiang, B. R. Oakley and C. C. C. Wang, J. Am. Chem. Soc., 2012, 134, 4709 CrossRef CAS PubMed.
- B. Wackler, P. Schneider, J. M. Jacobs, J. Pauly, C. Allen, M. Nett and D. Hoffmeister, Chem. Biol., 2011, 18, 354 CrossRef CAS PubMed.
- H.-H. Yeh, Y.-M. Chiang, R. Entwistle, M. Ahuja, K.-H. Lee, K. Bruno, T.-K. Wu, B. Oakley and C. C. Wang, Appl. Microbiol. Biotechnol., 2012, 96, 739 CrossRef CAS PubMed.
- J. Zhu, W. Chen, Y.-Y. Li, J.-J. Deng, D.-Y. Zhu, J. Duan, Y. Liu, G.-Y. Shi, C. Xie, H.-X. Wang and Y.-M. Shen, Gene, 2014, 546, 352 CrossRef CAS PubMed.
- A. R. Knaggs, Nat. Prod. Rep., 2003, 20, 119 RSC.
- K. Arai and Y. Yamamoto, Chem. Pharm. Bull., 1990, 38, 2929 CrossRef CAS.
- M. Wang, M. Beissner and H. Zhao, Chem. Biol., 2014, 21, 257 CrossRef CAS PubMed.
- C.-J. Guo, B. P. Knox, Y.-M. Chiang, H.-C. Lo, J. F. Sanchez, K.-H. Lee, B. R. Oakley, K. S. Bruno and C. C. C. Wang, Org. Lett., 2012, 14, 5684 CrossRef CAS PubMed.
- P. Wiemann, C. J. Guo, J. M. Palmer, R. Sekonyela, C. C. Wang and N. P. Keller, Proc. Natl. Acad. Sci. U. S. A., 2013, 110, 17065 CrossRef CAS PubMed.
- F. Y. Lim and N. P. Keller, Nat. Prod. Rep., 2014, 31, 1277 RSC.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c5sc01058f |
| This journal is © The Royal Society of Chemistry 2015 |