Selection, identification, and characterization of aptamers for pro-gastrin-releasing peptide (31–98), a tumor marker for small cell lung cancer†
Hui-Fang Cui*,
Ya-Jun Li,
Jia Wang,
Xiao-Jia Li,
Qiong-Lin Wang and
Yan-Feng Bai
Department of Bioengineering, School of Life Sciences, Zhengzhou University, 100# Science Avenue, Zhengzhou, 450001, P. R. China. E-mail: hfcui@zzu.edu.cn
First published on 17th December 2015
Abstract
Pro-gastrin-releasing peptide (31–98) (ProGRP31–98) is a highly reliable, sensitive, and specific tumor marker for small cell lung cancer (SCLC). DNA aptamers for ProGRP31–98 were selected in this study from an 89-mer DNA library with a random region of 48 nucleotides (nt) flanked with two primer hybridization sites, by using the systematic evolution of ligands by the exponential enrichment (SELEX) method. The DNA sequences binding to ProGRP31–98 immobilized magnetic beads were selected by running 12 SELEX cycles and 3 negative selection cycles. The selected DNA sequences were separated by cell based DNA cloning, and then sequence analyzed. The DNA sequences obtained were screened and investigated for binding specificity to ProGRP31–98 by using electrochemiluminescence (ECL) measurement with [Ru(bpy)2dppz]2+, a molecular light-switch, as the ECL probe. One DNA sequence (89 nt) and its random region (48 nt) were identified as specific aptamers for ProGRP31–98. Based on the secondary structures of the obtained DNA aptamers, two other truncated DNA aptamers, 40 nt and 15 nt long, respectively, were identified. The obtained aptamers have strong affinities to ProGRP31–98, with a Kd value of 16 nM, and can detect ProGRP31–98 with a detection limit of 17 nM using the label-free ECL measurement.
Introduction
Aptamers are single-stranded DNA or RNA molecules that can bind to target molecules with high affinity and specificity, similar to antibodies. In addition, aptamers possess a number of advantages over antibodies, such as being easily synthesised, having a wide range of molecular targets, low production cost and time, easily modified, strong chemical and thermal stability, and long storage life.1–5 Since their discovery, aptamers have become increasingly important molecular tools for target validation, diagnostics, therapeutics, and drug discovery.1,3,4Since the first aptamers against bacteriophage T4 DNA polymerase and several organic dyes were selected by using the method of systematic evolution of ligands by exponential enrichment (SELEX) in 1990,6,7 a lot of specific aptamers affinitive to various targets, such as sialyl Lewis X,2 HIV gp 120,3 L-arginine,8 adenosine triphosphate (ATP),9 human thrombin,10 prostate-specific antigen,11 have been reported. It is highly desirable to select aptamers for more targets, to expand their applications.
Gastrin-releasing peptide (GRP) is a neuroendocrine peptide, secreted from the dense neurosecretory granules of small cell lung cancer (SCLC) cells.12 GRP is rarely detected in the serum of patients with lung cancer, because of its short half life. It is generated by the proteolytic cleavage of pro-gastrin-releasing peptide (ProGRP), the more stable precursor of GRP. The proteolytic cleavage of ProGRP occurs after the Lys–Lys sequence at the position 29–30 to release the C-terminal fragment.13 The area between amino acids 43 and 97 is conserved and found only in mammals.14 The 31–98 segment of ProGRP (ProGRP31–98) has been clearly identified as a highly reliable, sensitive, and specific tumor marker for SCLC,15–17 which grows quickly and spread early in the course of the disease, and accounts for about twenty-five percent of lung cancer. Measurement of serum ProGRP31–98 levels enables diagnosis, prediction of prognosis, and treatment monitoring in SCLC.18,19 Recently, a 25-mer DNA aptamer for ProGRP1–98 with dissociation constant (Kd) of ∼1.2 μM was selected.20 ProGRP1–98 is the 1–98 segment of ProGRP. To our knowledge, no aptamer selection against ProGRP31–98, the 31–98 segment of ProGRP, has been reported. Compared to ProGRP1–98, ProGRP31–98 is a more reliable, sensitive, and specific tumor marker for SCLC.14–17
Aptamers usually fold into unique three dimensional structures to ensure their specific binding to targets. [Ru(bpy)2dppz]2+ (bpy = 2,2′-bipyridine, dppz = dipyrido[3,2-a:2′,3′-c] phenazine), a molecular light switch, shows negligible electrochemiluminescence (ECL) in aqueous solution, but exhibits significantly enhanced ECL intensity when it intercalates into the base pairs of the aptamer three-dimensional structures.21,22 The aptamer ligand, e.g. ATP, can cause the three-dimensional structure changes to the aptamer/[Ru(bpy)2dppz]2+ complex, leading to ECL intensity changes.21 This technique has been applied for DNA interaction study and a label-free ATP aptasensor.21
This paper reports the selection of DNA aptamers for ProGRP31–98 from an 89-mer DNA library with a random region of 48 nucleotides (nt) flanked with two primer hybridization sites using the SELEX technique. The DNA sequences binding to ProGRP31–98 immobilized magnetic beads were selected by running 12 SELEX cycles. The selected DNA sequences were amplified by polymerase chain reaction (PCR), and then cloned into pEASY-T3 cloning vector, followed by transformation into Escherichia Coli (E. coli) Trans5α chemically competent cells. The DNA sequences of some positive clones were determined. The binding activity and selectivity of the selected DNA sequences towards ProGRP31–98 were evaluated by using the ECL technique with [Ru(bpy)2dppz]2+ as the probe. The most active DNA sequences were truncated according to their secondary structure; and the affinity and the selectivity of the truncated DNA sequences were also evaluated. The obtained affinitive and selective DNA aptamers were applied in label-free aptasensors using the ECL technique.
Experimental
Materials
Silica coated magnetic beads modified with carboxylic groups (SM3-P100, diameter: 1150 nm) were purchased from Shanghai Allrun Nano Science & Technology (Shanghai, China). Streptavidin was obtained from, and all DNA sequences were custom synthesized by Sangon Shanghai Biotech (Shanghai, China). Concanavalin A (ConA), bovine serum albumin (BSA), bovine insulin, and 1-ethyl-3-(3-dimethylaminopropyl)carbodiimide hydrochloride (EDC) were obtained from Sigma-Aldrich (St. Louis, MO). ProGRP31–98 (Mw: ∼7.5 kD) was kindly provided by Meike BioMeditech Wuxi (Wuxi, China). TransTaq-T DNA polymerase, deoxynucleotide triphosphates (dNTPs), Trans 2K Plus DNA marker, ampicillin (Amp), isopropyl β-D-thiogalactoside (IPTG), 5-bromo-4-chloro-3-indolyl β-D-galactopyranoside (X-gal), EcoR I, pEASY-T3 cloning vector, and E. coli Trans5α chemically competent cells were purchased from Beijing TransGen Biotech (Beijing, China).[Ru(bpy)2dppz]2+ was synthesized according to the method reported.23 Briefly, [Ru(bpy)2]Cl2·2H2O (1.54 g, 2.97 mmol) and dipyridophenazine (0.93 g, 3.19 mmol) were dissolved in methanol–water mixture (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2, v/v, 380 mL) and refluxed for 4.5 h. The reaction mixture was then filtrated and the filtrate was precipitated with the addition of 10% NaBF4. A deep red product was finally recrystallized from ethanol. Dipyridophenazine was synthesized from o-phenylenediamine and l,10-phenanthroline-5,6-dione;23,24 and l,10-phenanthroline-5,6-dione was synthesized from 1,10-phenanthroline monohydrate.23
2, v/v, 380 mL) and refluxed for 4.5 h. The reaction mixture was then filtrated and the filtrate was precipitated with the addition of 10% NaBF4. A deep red product was finally recrystallized from ethanol. Dipyridophenazine was synthesized from o-phenylenediamine and l,10-phenanthroline-5,6-dione;23,24 and l,10-phenanthroline-5,6-dione was synthesized from 1,10-phenanthroline monohydrate.23
[Ru(bpy)2]Cl2·2H2O, o-phenylenediamine, and 1,10-phenanthroline were obtained from Aladdin Industrial (Shanghai, China). Other chemicals, such as 4-morpholine ethanesulfonic acid (MES), were obtained from Sinopharm Chemical Reagent (Shanghai, China). All chemicals were of analytical grade and used as received. Deionized water obtained from a Millipore water system was used throughout the experiment.
Instrumentation
HB-1000 hybridizer (UVP, Upland, CA, USA) was used for conjugating proteins/peptides onto the magnetic beads. UV 2450 UV-Vis spectrometer (Shimadzu, Japan) and F-4500 fluorescence spectrophotometer (Hitachi, Japan) were employed for protein/peptide and DNA concentration measurement, respectively. PCR was performed using Eppendorf PCR instrument (Mastercycler Gradient, Germany). Electrophoresis experiments were realized using DYY-6C electrophoresis power supply (Beijing Liuyi Instrument, China) and JY04S-3C gel imaging system (Beijing Junyi-Dongfang Electrophoresis Equipment, China). The magnetic beads were processed using MS-12 Otzi magnetic separator (Shanghai Allrun Nano Science & Technology, China). A conventional three-electrode system consisting of an Ag/AgCl/(3 M KCl) reference electrode, a platinum wire counter electrode and a glassy carbon working electrode (GCE) was used in the ECL experiment. Prior to the experiment, the GCE was sequentially polished with aqueous slurries of 1 μm, 0.3 μm and 0.05 μm alumina powders respectively, and then sonicated in water and ethyl alcohol bath each for 5 min. ECL intensities were measured using BPCL-1-TIC Ultra-Weak luminescence analyzer (Beijing Jianxin Lituo Science & Technology, China) equipped with Ingsens 1010s electrochemical workstation (Guangzhou Ingsens Sensor Technology, China). The PMT in the ECL measurement was biased at 1200 V. All the measurements were performed at room temperature (∼25 °C).Conjugation of proteins/peptides onto the magnetic beads
The proteins/peptides, including ProGRP31–98, streptavidin, ConA, BSA, or insulin were conjugated onto the magnetic beads through covalent reaction between –COOH on the magnetic beads and –NH2 of the proteins/peptides, using EDC and N-hydroxysuccinimide (NHS) as the coupling agents. In brief, the magnetic beads (2 mg) were washed with 0.01 M MEST buffer (2.13 g L−1 MES·H2O, 0.05% Tween 20, pH = 6.0) twice, and then incubated in a 400 μL solution containing 2.5 mg mL−1 EDC and 5 mg mL−1 NHS at 37 °C for 1 h to activate the –COOH group. The magnetic beads were then washed with 0.01 M MES buffer (2.13 g L−1 MES·H2O, 0.31 g L−1 boric acid, pH = 9.0) twice, and then mixed with 100 μL 0.05 M phosphate buffer solution (PBS) (pH = 7.4) containing 1 mg mL−1 protein/peptide. The reaction mixture was adjusted to a total volume of 500 μL with the MES buffer and agitated in the HB-1000 hybridizer at 37 °C overnight. The amount of conjugated proteins/peptides was determined as the protein/peptide diminishment in the supernatant after the reaction by using Bradford method.In vitro selection of aptamers using the SELEX method
For vitro selection of aptamers for ProGRP31–98, the 89-mer DNA library (sequence: 5′-CTTCTGCCCGCCTCCTTCC-48 nt-GGAGACGAGATAGGCGGACACT-3′) (2.4 nmol) was dissolved in 200 μL binding buffer (100 mM NaCl, 2 mM MgCl2, 5 mM KCl, 1 mM CaCl2, 20 mM Tris–HCl, 0.02% Tween 20, pH = 7.6) and then denatured at 95 °C for 10 min. After being cooled in ice bath for 10 min, the DNA library was incubated with 300 μL 0.1 mg mL−1 ProGRP31–98 coated magnetic beads in the binding buffer at 37 °C for 2 h in the HB-1000 hybridizer. The beads were then washed with sterilized water and taken as the template for PCR (94 °C for 5 min; 94 °C for 30 s, 58 °C for 30 s, 72 °C for 30 s, 15–30 cycles; 72 °C for 10 min) using F-FAM (5′-FAM-CTTCTGCCCGCCTCCTTCC-3′) and R-biotin (5′-biotin-AGTGTCCGCCTATCTCGTCTCC-3′) as the forward and the reverse primer, respectively, in the presence of TransTaq-T DNA polymerase and dNTPs. FAM is a fluorescence molecule. The double-stranded DNA (dsDNA) (250 μL) obtained from the PCR was verified using agarose gel electrophoresis (AGE) analysis and then incubated with 200 μL 10 mg mL−1 streptavidin coated magnetic beads in TE–NaCl solution (10 mM Tris–HCl, 1 mM EDTA, 2 M NaCl, pH = 8.0) at room temperature for 30 min, when the dsDNA bound to the streptavidin coated magnetic beads' surface. The bound dsDNA was then denatured by incubating the beads in 100 μL 0.15 M NaOH for 10 min. The supernatant was neutralized with 0.3 M HCl, and then diluted to 200 μL with the binding buffer, producing a FAM-labeled DNA library.The above SELEX procedure was repeated for 12 cycles. From the second SELEX cycle, the fluorescence intensity of the ProGRP31–98 coated magnetic beads after incubation with the FAM-labeled DNA library was measured to monitor the DNA enrichment. To enhance the specificity of the selected aptamers for ProGRP31–98, 3 negative selection cycles of incubating the DNA library with insulin or BSA coated magnetic beads were inserted after the 6th SELEX cycle, and repeated every 1 to 3 SELEX cycles.
Separation and sequence analysis of the DNA aptamers
The obtained final DNA library was incubated with ProGRP31–98 coated magnetic beads, and then the beads were served as PCR template (94 °C for 5 min; 94 °C for 30 s, 58 °C for 30 s, 72 °C for 30 min, 15 cycles; 72 °C for 20 min) using F (5′-CTTCTGCCCGCCTCCTTCC-3′) and R (5′-AGTGTCCGCCTATCTCGTCTCC-3′) as the forward and the reverse primer, respectively. Finally, the PCR product was purified using AGE (1.5%) and spin column separation methods, and verified using AGE analysis. The purified dsDNA product was cloned into pEASY-T3 cloning vector and then transformed into Trans5α E. coli chemically competent cells. The cells were then cultured on solid LB medium (10 g L−1 tryptone, 5 g L−1 yeast extract, 10 g L−1 NaCl, 15 g L−1 agar, pH = 7.4) containing 100 μg mL−1 Amp and coated with IPTG and X-gal, at 37 °C overnight to screen positive clones. The white colonies were chosen and subjected to colony PCR (94 °C for 10 min; 94 °C for 30 s, 58 °C for 30 s, 72 °C for 2 min, 30 cycles; 72 °C for 10 min) using M13F (5′-TGTAAAACGACGGCCAGT-3′) and M13R (5′-CAGGAAACAGCTATGACC-3′) as the forward and the reverse universal primer, respectively. After the PCR product was verified by AGE analysis, the selected positive recombinants were inoculated into liquid LB medium (10 g L−1 tryptone, 5 g L−1 yeast extract, 10 g L−1 NaCl, pH = 7.4) containing 100 μg mL−1 Amp. The culture was shaken at 37 °C overnight. The plasmids were then extracted from the positive recombinants and digested by restriction endonuclease EcoR I. The digested plasmids were subjected to AGE analysis to confirm the positivity of the recombinants. The positive clones were sent to Beijing Liuhe Greatness Technology Co. for DNA sequence analysis using M13F (5′-TGTAAAACGACGGCCAGT-3′) and M13R (5′-CAGGAAACAGCTATGACC-3′) as the universal forward and reverse primer, respectively. The secondary structures of the selected DNA sequences were analyzed using OligoAnalyzer.Binding specificity assay of the selected aptamers
The DNA aptamers selected using the SELEX method were custom-synthesized and verified for their binding specificity to ProGRP31–98 by ECL measurement with [Ru(bpy)2dppz]2+ as the probe. Briefly, 10 μL 100 μM DNA aptamer in TE buffer (10 mM Tris–HCl, 1 mM EDTA, pH = 8.0) was denatured at 95 °C for 10 min, and then added to 1 mL 20 μM [Ru(bpy)2dppz]2+ in 5 mM oxalate/oxalic acid solution (pH = 5.5). The ECL intensity was monitored until the ECL peak value (denoted as ECL0) was stabilized in ∼50 min. With the addition of 50 μL 1.2 mg mL−1 protein/peptide, the ECL intensity was continuously monitored in ∼40 min and the stabilized ECL peak value was denoted as ECL1. The (ECL0 − ECL1)/ECL0 value was calculated.Affinity assay of the selected aptamers
The affinity of the selected DNA aptamers to ProGRP31–98 was investigated by measuring the amount of the bound aptamers onto the ProGRP31–98 coated magnetic beads in various concentrations (18–244 nM, denoted as C0) of the aptamers in the binding buffer. The aptamers were labeled with fluorescence molecule FAM at their 5′ ends. The aptamer solution (400 μL) was denatured at 95 °C for 10 min, and then incubated with 10 μL 1 mg mL−1 ProGRP31–98 coated magnetic beads for 2 h at 37 °C in the HB-1000 hybridizer. The aptamer concentration was estimated by measuring the fluorescence intensity at the emission wavelength of 520 nm. The concentration of the bound aptamer (Cb) was calculated based on the aptamer concentrations before and after the binding. Following the Lineweaver–Burk equation (eqn (1)):| 1/Cb = Kd/BmaxC0 + 1/Bmax, | (1) |
Aptamer-based ProGRP31–98 assay by ECL titration
The selected aptamers were applied in label-free aptasensors using the ECL measurement with [Ru(bpy)2dppz]2+ as the probe in 5 mM oxalate/oxalic acid solution (pH = 5.5). Similar with the binding specificity assay experiment, a [Ru(bpy)2dppz]2+/aptamer complex was formed and its ECL peak value (denoted as ECL0) was recorded. The ProGRP31–98 solution was then added gradually in a batch mode. The ECL peak value (ECL1) was recorded for various ProGRP31–98 concentrations after the chemiluminescence was stabilized in ∼40 min. The changes of the ECL peak values (ΔECL = ECL0 − ECL1) with the addition of ProGRP31–98 was calculated as the signal.Results and discussion
In vitro selection and sequence analysis of DNA aptamers
The conjugation of proteins/peptides onto the magnetic beads and the DNA enrichment in the SELEX procedure were confirmed using the UV-Vis and the fluorescence measurement, respectively (data not shown). The agarose electrophoresis gel image (Fig. 1) of the PCR product amplified from the final SELEX cycle showed an ∼100 base pair (bp) DNA band, a length slightly longer than the synthesized DNA length (i.e., 89 bp), because the DNA polymerase TransTaq-T added poly-nucleotides A to the DNA ends. The final DNA library and the PCR product had an expected and specific DNA length.After the PCR product from the final SELEX cycle was purified, cloned and then transformed into the E. coli chemically competent cells, the white colonies were subjected to colony PCR. Most of the white colonies had positive PCR products of 345 bp, the right size from the positive vectors (Fig. 2). The map and the construct of the pEASY-T3 cloning vector are shown in Fig. S-1.†
The plasmids of the positive colonies were extracted and then digested by EcoR I. The agarose electrophoresis gel analysis results of some digestion products are illustrated in Fig. 3. Most of the digested plasmids clearly exhibited two electrophoresis bands with the size of ∼126 bp and 3004 bp respectively, consistent with theoretic values. Twenty-nine of the positive colonies with confirmed plasmid digestion products were subjected to DNA sequence analysis. Eight colonies were successfully sequenced with the right length of random region (i.e. 48 nt). The obtained DNA sequences, denoted as DNA aptamer candidates for ProGRP31–98, are listed in Table 1. The DNA aptamer candidates were named as A2, A12, etc., followed after their corresponding colony number C2, C12, respectively. Among the 8 DNA aptamer candidates obtained, A19 and A21 have exactly the same oligonucleotide sequence (i.e., the sequence occurrence is 2).
| Name | Sequence |
|---|---|
| A2 | 5′-AATGCCGTGGAGACGAGATAGGCGGACACTAACCCCTCTAGGACGAGA-3′ |
| A12 | 5′-TGGGTCATCTAGCCTTCTCTGCGTCGCAGCACTATTGCTTCGGAGTTT-3′ |
| A16 | 5′-AAACTCTTGTAATCATAACGGTTTTATCAACGGAGCTCCTACGCACAT-3′ |
| A18 | 5′-CATGCGGAGTAGATTCGAGCCCAGATAGTCCCTGGTTATTTCCTTAGG-3′ |
| A19 | 5′-ATCCTCTGTCGCCTTTCTACACGCGCGCTGGAGGCACCATTATTTGCC-3′ |
| A21 | 5′-ATCCTCTGTCGCCTTTCTACACGCGCGCTGGAGGCACCATTATTTGCC-3′ |
| A31 | 5′-TACCATACCTTGGTATTCGCAGTTATAAAGCGTCTGGGTCTCCCGGTG-3′ |
| A41 | 5′-CCCGGTTCTATACGACTGGCTAGATTTTACTTTGACCATACAGTGACG-3′ |
Binding specificity screening of the DNA aptamer candidates
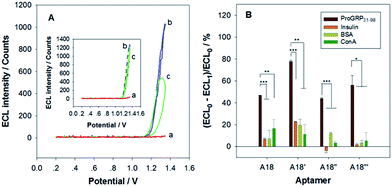
The obtained DNA aptamer candidates were chemically synthesized and screened for their binding specificity to ProGRP31–98 by ECL measurement with [Ru(bpy)2dppz]2+ as the probe. [Ru(bpy)2dppz]2+ is a remarkable luminescent reporter of DNA structure, described as a molecular light switch for DNA, exhibiting a negligible ECL intensity in aqueous solution. The ECL intensity of the molecular light switch enhances for at least 104 times when it intercalates into the base pairs of double-strand DNA, such as the hairpin DNA stem.21,22 Aptamers usually fold into unique three-dimensional structures to bind to targets specifically. The folded structures of the aptamers allow [Ru(bpy)2dppz]2+ to intercalate to produce strong ECL.21 Introduction of target molecules may compete with [Ru(bpy)2dppz]2+ to bind to aptamer,21 or causing three-dimensional structure changes to the aptamer, leading to the ECL intensity changes.Among the 8 DNA aptamer candidates, A18 can specifically interact with ProGRP31–98. The ECL intensity peak value decreased for 46.9% (from 1023 counts to 543 counts) with the presence of ProGRP31–98 in the oxalate/oxalic acid solution containing 20 μM [Ru(bpy)2dppz]2+ and 1 μM A18 (Fig. 4A). In contrast, the ECL intensity had no obvious changes upon the addition of the interference protein BSA (inset in Fig. 4A). The interference proteins/peptides tested, including insulin, BSA, and ConA, caused only ∼6.9%, 7.2%, and 16.6% ECL intensity changes respectively, significantly lower than ProGRP31–98 did (Fig. 4B), indicating that the A18 sequence is a specific DNA aptamer for ProGRP31–98.
Sequence modification of DNA aptamers
The A18 sequence flanked with the two primer hybridization sites, a DNA sequence of 89 nt long (denoted as A18′, Table 2) was then verified for binding specificity to ProGRP31–98. Similarly, the ECL peak value of the [Ru(bpy)2dppz]2+/aptamer complex decreased for ∼77.6% with the addition of ProGRP31–98, significantly higher than those with the introduction of insulin, BSA, and ConA (∼22.7%, 19.7%, and 11.3%, respectively) (Fig. 4B), indicating that the A18′ sequence is also a specific DNA aptamer for ProGRP31–98.| Aptamer | Sequence |
|---|---|
| A18 | 5′-CATGCGGAGTAGATTCGAGCCCAGATAGTCCCTGGTTATTTCCTTAGG-3′ |
| A18′ | 5′-CTTCTGCCCGCCTCCTTCC-CATGCGGAGTAGATTCGAGCCCAGATAGTCCCTGGTTATTTCCTTAGG-GGAGACGAGATAGGCGGACACT-3′ |
| A18′′ | 5′-CATGCGGAGTAGATTCGAGCCCAGATAGTCCCTGGTTATT-3′ |
| A18′′′ | 5′-CCAGATAGTCCCTGG-3′ |
The secondary structure of the A18 aptamer contains two stem-loop hairpins (Fig. 5A), with 4 bp for one hairpin stem, and 2 bp for the other. In contrast, the A18′ aptamer has four stem-loop hairpins (Fig. 5B), with one hairpin the same with the 4 bp stem one of A18. Assuming that the hairpin areas are the main binding domains, the A18 aptamer sequence was truncated at their two ends to a DNA sequence of 40 nt long (A18′′, Table 2), without changing its secondary structure (Fig. 5C). The ECL peak value of the [Ru(bpy)2dppz]2+/aptamer complex of A18′′ decreased for ∼44.1% with the addition of ProGRP31–98, but only for about −3.8%, 12.3%, and 3.4% with the addition of insulin, BSA, and ConA respectively (Fig. 4B), indicating that the A18′′ sequence is a specific DNA aptamer for ProGRP31–98.
The A18, A18′, and A18′′ aptamers have a common 4 bp stem hairpin (the circled area in Fig. 5, 15 nt long), denoted as A18′′′ (Table 2). Assuming that the 4 bp stem hairpin is a main domain in binding with the target, the binding specificity of the hairpin DNA sequence to ProGRP31–98 was verified. The ECL peak value of the [Ru(bpy)2dppz]2+/aptamer complex of A18′′′ decreased for ∼56.1% with the introduction of ProGRP31–98, significantly larger than those with the introduction of insulin, BSA, and ConA (∼1.8%, 3.2%, and 5.5%, respectively) (Fig. 4B), indicating that the A18′′′ sequence is also a specific DNA aptamer for ProGRP31–98, and is indeed a main binding domain.
Affinity assay and aptamer-based ProGRP31–98 assay
The affinities of the aptamer A18, A18′, A18′′, and A18′′′ to ProGRP31–98 were investigated. The calibration plot of the fluorescence intensity of the FAM-labeled A18 versus the aptamer concentration, and the plot of 1/Cb versus 1/C0 of A18 are illustrated in the ESI (Fig. S-2 and S-3, respectively).† The Kd value of the aptamer A18 binding to ProGRP31–98 was estimated to be 49 nM, and those of the aptamer A18′, A18′′, and A18′′′ were estimated to be 306, 87, and 16 nM, respectively. The Kd values of the aptamers to ProGRP31–98 are in the middle-nanomolar range, 2 order of magnitude smaller than the 25-mer one reported for ProGRP1–98 (Kd: ∼1.2 μM),20 and similar to those of antibody–antigen interactions (Kd = 0.01–1000 nM), indicating that these aptamers have high affinity to ProGRP31–98. It should be noted that the selected aptamers in this work are totally different in sequence with the 25-mer one reported for ProGRP1–98, a different target from ProGRP31–98.20The obtained aptamers were applied in label-free aptasensors using ECL measurement with [Ru(bpy)2dppz]2+ as the probe. The ECL intensity–potential curves of the [Ru(bpy)2dppz]2+/aptamer complex under the titration with ProGRP31–98 were recorded, and those for A18 and A18′ are illustrated in Fig. 6. Those for A18′′, and A18′′′ are shown in the ESI (Fig. S-4).† The ECL intensity decreased monotonically with the increase of ProGRP31–98 concentration. The ECL peak value changes (i.e. ΔECL) versus the ProGRP31–98 concentration for A18 was linear with 0.48–3.36 μM target (regression coefficient = 0.9969), with a limit of detection (LOD) of 17 nM (signal to noise ratio = 3). The LOD values for the aptamer A18′, A18′′, and A18′′′ were obtained to be 440, 770, and 210 nM, respectively. The LOD value of the A18 aptamer-based ProGRP31–98 assay was in the middle-nanomolar range, ∼2 order of magnitude smaller than that of a surface plasmon resonance aptasensor reported for ProGRP1–98 (LOD: 1 μM), a different target from ProGRP31–98.20 To our knowledge, no aptasensors for ProGRP31–98 have been reported.
Conclusions
One 48 nt DNA aptamer and one 89 nt DNA aptamer, specifically binding to ProGRP31–98, a highly reliable, sensitive, and specific tumor marker for SCLC, have been selected and characterized. By analyzing the secondary structures of the two DNA aptamers, other two highly specific DNA aptamers with 40 nt and 15 nt long respectively, have been obtained. The aptamers exhibit strong affinity to ProGRP31–98, with Kd values in the middle-nanomolar range. By using [Ru(bpy)2dppz]2+ as a probe, the ECL intensity of the [Ru(bpy)2dppz]2+/aptamer complex decreased significantly and specifically with the addition of ProGRP31–98. The aptamers can sensitively and selectively response to ProGRP31–98 in a label-free mode in the presence of the [Ru(bpy)2dppz]2+ probe, by using the ECL measurement. The obtained aptamers may be promisingly applicable in clinical diagnosis.Acknowledgements
This work was supported by National Natural Science Foundation of China (NSFC21345007), Plan for Scientific Innovation Talent of Henan Province, Open-up and Collaboration Program of Science and Technology of Henan Province (132106000070), Key Science and Technology Program of Henan Province (132102310042), and Key Project of Science and Technology of The Education Department Henan Province (14A180017). We thank Meike BioMeditech Wuxi (Wuxi, China) for kindly providing ProGRP31–98.Notes and references
- K. Abe and K. Ikebukuro, in Biosensors – Emerging Materials and Applications, ed. Prof. Pier A. S., InTech Publisher: Croatia, 2011, pp. 227–242 Search PubMed.
- S. Jeong, T. Y. Eom and S. J. Kim, Biochem. Biophys. Res. Commun., 2001, 281, 237–243 CrossRef CAS PubMed.
- J. Zhou, P. Swiderski, H. Li, J. Zhang, C. P. Neff, R. Akkina and J. J. Rossi, Nucleic Acids Res., 2009, 37, 3094–3109 CrossRef CAS PubMed.
- J. R. Cole Jr, L. W. Dick, E. J. Morgan and L. B. McGown, Anal. Chem., 2007, 79, 273–279 CrossRef PubMed.
- D. Shangguan, L. Meng, Z. C. Cao, Z. Xiao, X. Fang, Y. Li, D. Cardona, R. P. Witek, C. Liu and W. Tan, Anal. Chem., 2008, 80, 721–728 CrossRef CAS PubMed.
- C. Tuerk and L. Gold, Science, 1990, 249, 505–510 CAS.
- A. D. Ellington and J. W. Szostak, Nature, 1990, 346, 818–822 CrossRef CAS PubMed.
- A. Geiger, P. Burgstaller, H. Vondereltz, A. Roeder and M. Famulok, Nucleic Acids Res., 1996, 24, 1029–1036 CrossRef CAS PubMed.
- M. Sassanfar and J. W. Szostak, Nature, 1993, 364, 550–553 CrossRef CAS PubMed.
- L. C. Bock, L. C. Griffin, J. A. Latham, E. H. Vermaas and J. J. Toole, Nature, 1992, 355, 564–566 CrossRef CAS PubMed.
- N. Savory, K. Abe, K. Sode and K. Ikebukuro, Biosens. Bioelectron., 2010, 26, 1386–1391 CrossRef CAS PubMed.
- E. A. Sausville, A. M. Lebacq-Verheyden, E. R. Spindel, F. Cuttitta, A. F. Gasdar and J. F. Battey, J. Biol. Chem., 1986, 261, 2451–2457 CAS.
- J. Ischia, O. Patel, A. Shulkes and G. S. Baldwin, BioFactors, 2009, 35, 69–75 CrossRef CAS PubMed.
- G. S. Baldwin, O. Patel and A. Shulkes, Regul. Pept., 2007, 143, 1–14 CrossRef CAS PubMed.
- R. Molina, X. Filella and J. M. Auge, Clin. Biochem., 2004, 37, 505–511 CrossRef CAS PubMed.
- P. Stieber, H. Dienemann, A. Schalhorn, U. M. Schmitt, J. Reinmeidl, K. Hofmann and K. Yamaguchi, Anticancer Res., 1999, 19, 2673–2678 CAS.
- J. J. Holst, M. Hansen, E. Bork and T. W. Schwartz, J. Clin. Oncol., 1989, 7, 1831–1838 CAS.
- Y. Miyake, T. Kodama and K. Yamaguchi, Cancer Res., 1994, 54, 2136–2140 CAS.
- K. Aoyagi, M. Yoshio, U. Kenichi, K. Tomiko, K. Tetsuro and Y. Ken, Clin. Chem., 1995, 41, 537–543 CAS.
- M. Mie, T. Kai, T. Le, A. E. G. Cass and E. Kobatake, Appl. Biochem. Biotechnol., 2013, 169, 250–255 CrossRef CAS PubMed.
- L. Hu, Z. Bian, H. Li, S. Han, Y. Yuan, L. Gao and G. Xu, Anal. Chem., 2009, 81, 9807–9811 CrossRef CAS PubMed.
- R. M. Hartshorn and J. K. Barton, J. Am. Chem. Soc., 1992, 114, 5919–5925 CrossRef CAS.
- E. Amouyal, A. Homsi, J. C. Chambron and J. P. Sauvage, J. Chem. Soc., Dalton Trans., 1990, 6, 1841–1845 RSC.
- J. E. Dickeson and L. A. Summers, Aust. J. Chem., 1970, 23, 1023–1027 CrossRef CAS.
Footnote |
| † Electronic supplementary information (ESI) available: additional information and figures. See DOI: 10.1039/c5ra24703a |
| This journal is © The Royal Society of Chemistry 2016 |