Synthesis, antimalarial activity, and target binding of dibenzazepine-tethered isoxazolines†
Koravangala S. Vinay Kumara,
Gejjalagere S. Lingarajua,
Yadaganahalli K. Bommegowdaa,
Ajjampura C. Vinayakaa,
Pritesh Bhatb,
Challanayakanahally S. Pradeepa Kumaraa,
Kanchugarakoppal S. Rangappa*a,
D. Channe Gowda*c and
Maralinganadoddi P. Sadashiva*ac
aDepartment of Chemistry, University of Mysore, Mysuru, Karnataka, India. E-mail: mpsadashiva@gmail.com; rangappaks@yahoo.com
bManipal College of Pharmaceutical Sciences, Manipal University, Madhav Nagar, Manipal, India
cDepartment of Biochemistry and Molecular Biology, Pennsylvania State University College of Medicine, Hershey, PA 17033, USA. E-mail: gowda@psu.edu
First published on 15th October 2015
Abstract
Malaria, a complex and deadly parasitic infectious disease, is a huge public health problem in many endemic countries around the globe. The prevailing extensive resistance of malaria parasites to traditional drugs and emergence of resistance to the currently used frontline artemisinin-based chemotherapy calls for the development of new drugs. Towards this objective and since compounds containing the dibenzazepine moiety are effective in treating both gametocyte and asexual stage malaria parasites, including multi drug resistant parasites, a library of dibenzazepine tethered 3,5-disubstituted isoxazolines was synthesised via 1,3-dipolar cycloaddition reaction. An additional diversified group of dibenzazepine derivatives were accessed by Suzuki coupling of one of the above dibenzazepine derivatives with various organoboronic acids. All compounds were structurally characterized and were evaluated for their antimalarial activity. They exhibited good to excellent inhibitory activity against the growth of drug-sensitive Plasmodium falciparum 3D7 strain with IC50 values ranging from 0.2 to 7.7 μM. About 50% of the compounds were either minimally or not toxic to human cell lines. Five of the compounds (6j, 6k, 8c, 8k and 8l) that highly inhibited the parasite growth were further assessed for antimalarial activity using an additional chloroquine-sensitive (D6) and two chloroquine-resistant (W2 and 7G8) P. falciparum strains. These compounds were effective against all four strains (3D7, D6, W2 and 7G8), exhibiting IC50 values of 0.1 to 1.75 μM. The dibenzazepines were identified to target the metalloamino-peptidase of parasites. Molecular docking and dynamics simulation studies were performed to understand the binding mode and binding strengths of the selected compounds with the enzyme. In agreement with their excellent antimalarial activity, the data suggested that the compounds can strongly bind to the active site of the enzyme.
Introduction
Malaria is a deadly infectious disease in many tropical and subtropical countries of the world. Approximately 50% of the global population is at risk of contracting the disease. According to a recent WHO report, in 2013, ∼200 million clinical cases of malaria were diagnosed, and ∼600![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 deaths have occurred worldwide, most of which were in young children and pregnant women.1 In addition to the huge death toll it exerts, malaria vastly curtails socioeconomic progress of the population in endemic areas, which are mostly underdeveloped, by causing frequent debilitating clinical conditions and thus, contributing to the loss of enormous productive manpower. Although five species of Plasmodium family of protozoan parasites can infect humans to cause malaria, P. falciparum and P. vivax are responsible for almost all malarial deaths. While P. falciparum is most prevalent in Africa, both P. vivax (majorly) and P. falciparum (to variable degrees) are prevalent in Southeast Asia including Indian subcontinent, some parts of Africa, and South America. Further, P. falciparum, the most virulent among malaria parasites that can infect human, accounts for >90% of all malarial deaths, whereas P. vivax is responsible for most of the remainder of deaths.
000 deaths have occurred worldwide, most of which were in young children and pregnant women.1 In addition to the huge death toll it exerts, malaria vastly curtails socioeconomic progress of the population in endemic areas, which are mostly underdeveloped, by causing frequent debilitating clinical conditions and thus, contributing to the loss of enormous productive manpower. Although five species of Plasmodium family of protozoan parasites can infect humans to cause malaria, P. falciparum and P. vivax are responsible for almost all malarial deaths. While P. falciparum is most prevalent in Africa, both P. vivax (majorly) and P. falciparum (to variable degrees) are prevalent in Southeast Asia including Indian subcontinent, some parts of Africa, and South America. Further, P. falciparum, the most virulent among malaria parasites that can infect human, accounts for >90% of all malarial deaths, whereas P. vivax is responsible for most of the remainder of deaths.
Traditionally chloroquine and other quinine derivatives have been the most commonly used drugs to treat malaria. However, parasites have developed widespread resistance against these drugs.2 To overcome this problem, artemisinin-based combination therapy (ACT) has been developed as a frontline treatment.3,4 Although ACT has been effective in treating malaria and is still being widely used, resistance has emerged against this treatment in several Southeast Asian countries.5,6 Hence, development of new, effective antimalarial drugs is urgently needed. Towards this goal, our research group has recently reported the potential utility of 1,2-disubstituted-4-quinolones and abietane diterpenoids as new antimalarial agents,7,8 and we have been continuing efforts to identify new classes of antimalarial agents.
The blood stage infection of Plasmodium parasites is responsible for the clinical symptoms and severe illnesses of malaria. The initial infection involves the entry of merozoites released from the matured liver stage parasites into erythrocytes, where they grow into trophozoites, which upon maturation undergo schizogony to form merozoites. At the end of 48 h intraerythrocytic life cycle of parasites, the merozoites released from infected erythrocytes invade new erythrocytes to start the next 48 h life cycle.9,10 During their growth in erythrocytes, parasites depends on the host products for nutritional need. For example, amino acids formed by the degradation of host cell haemoglobin by aminopeptidases are the main source for parasite protein synthesis.11,12 P. falciparum M1 alanyl aminopeptidase (PfA-M1), a terminal zinc-metalloamino-peptidase of hemoglobin digestion converts the cleaved hemoglobin peptides into an amino acid pool, which is used by parasites for its protein synthesis.13–15 Since hemoglobin-derived amino acids are critical for parasite growth, PfA-M1 is considered as a potential target for the development of antimalarial agents.16–19
Compounds containing dibenzazepine 1 scaffold exhibit a wide range of biological activity and hence, a variety of derivatives have been synthesized and evaluated as antimicrobial and antifungal,20 antioxidant,21,22 anticancer,23 antiepileptic,24 anticonvulasent,25–27 and serotonin agents.28 Some examples of clinically important dibenzazepine-based drugs are carbamazepine,25,26,29 opipramol,30 and oxacarbazepine24 that used for treating epilepsy, anxiety, and mood disorders. A recent report describes dibenzazepine derivatives as efficient antimalarial agents for both the gametocyte stage and the blood stage P. falciparum.31 The dibenzazepine moiety has also been used for the development of agents against multi drug resistant Plasmodium parasites.32 Given the potential usefulness of dibenzazepines as target-specific antimalarial agents and in view of exploring various new classes of compounds for antimalarial activity, we synthesized a library of dibenzazepines having substituted five-membered heterocyclic rings at the N-position of the azepine ring and evaluated their antimalarial activity.
Results and discussion
Chemistry
At first, a series of 3,5-disubstituted isoxazolines (6a–6k) bearing aryl, heteroaryl and alicyclic group at the C-3 position of isoxazoline ring and having dibenzazepine moiety at the C-5 position were synthesized36 through the reactions depicted in Scheme 1. The synthon, dipolarophile 5-allyl-5H-dibenzazepine 2 was prepared by N-allylation of 5H-dibenzo[b,f]azepine 1 using allyl bromide in presence of tetra-n-butylammonium bromide (TBAB) as a phase transfer catalyst, sodium hydroxide as a base in the biphasic mixture of toluene–water in 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 ratio at 55 °C. The dipolar aryl nitrile oxides (5a–5k) was generated in situ in two-step processes by the addition of hydroxylamine hydrochloride to aryl, heteroaryl or alicyclic aldehydes (3a–3k) in the presence of sodium acetate in methanol followed by the action of N-chlorosuccinimide (NCS) on oximes formed (4a–4k) in the presence of triethyl amine base (Scheme 1). Coupling of synthon 2 with aryl, heteroaryl or alicyclic nitrile oxides in the presence of triethyl amine afforded dibenzazepine tethered 3,5-disubstituted isoxazolines (6a–6k). The compounds were characterised by LC-MS, 1H and 13C NMR, and elemental analyses (see the ESI file†).
1 ratio at 55 °C. The dipolar aryl nitrile oxides (5a–5k) was generated in situ in two-step processes by the addition of hydroxylamine hydrochloride to aryl, heteroaryl or alicyclic aldehydes (3a–3k) in the presence of sodium acetate in methanol followed by the action of N-chlorosuccinimide (NCS) on oximes formed (4a–4k) in the presence of triethyl amine base (Scheme 1). Coupling of synthon 2 with aryl, heteroaryl or alicyclic nitrile oxides in the presence of triethyl amine afforded dibenzazepine tethered 3,5-disubstituted isoxazolines (6a–6k). The compounds were characterised by LC-MS, 1H and 13C NMR, and elemental analyses (see the ESI file†).
Dibenzazepine derivatives carrying substituents on the dibenzazepine ring were found to exhibit effective biological and pharmacological properties with minimal side effects.27,32 Therefore, these class of compounds have been of special interest for exploiting as drug candidates. Considering this, an additional series of compounds (8a–8l) were prepared by introducing various substituents on the para-position of the C-3 phenyl moiety of isoxazoline ring in 6d. This was accomplished via Suzuki coupling of 6d with various organoboronic acids (7a–7l) in the presence of Pd(dppf)Cl2·CH2Cl2 catalyst37 using Cs2CO3 as base in dioxane–water biphasic solvent under nitrogen atmosphere (Scheme 2). The products were purified by column chromatography on silica gel (60–120 mesh) using mixtures of ethyl acetate and hexane in suitable ratios and were characterised by LC-MS, 1H and 13C NMR, and elemental analyses (see the ESI file†). Additionally, compound 6d was characterized by single crystal XRD studies and the crystallographic data have been deposited at the CCDC No. 1049585 (ref. 38) (Fig. S1, see the ESI file†).
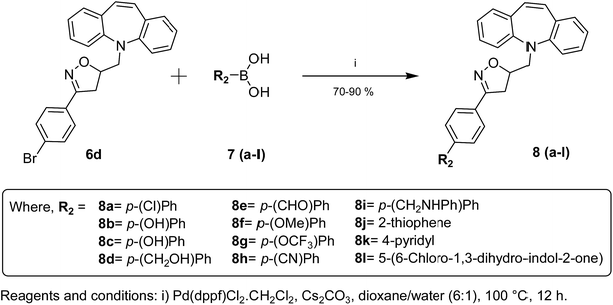 | ||
| Scheme 2 Suzuki coupling of 5-[3-(4-bromo-phenyl)-4,5-dihydro-isoxazol-5-ylmethyl]-5H-dibenzo[b,f]azepine 6d with various boronic acids. | ||
Biology
All the synthesized compounds (6a–6k and 8a–8l) were initially examined for antimalarial activity against chloroquine-sensitive P. falciparum 3D7 strain. The parasite growth inhibition was assessed at concentrations ranging from 0.1 to 100 μM by a SYBR Green assay.39,40 All compounds showed good to excellent antimalarial activity with IC50 values of 0.2 to 7.7 μM (Table 1). The cytotoxicity of the compounds was examined using human kidney embryonic (HEK 293) cells and human lung tumor epithelial cell (A549) by the MTS assay. Many of the compounds exhibited either minimal or no cytotoxicity (IC50 > 40 μM), and others displayed low cytotoxicity (20 to 40 μM) (Table 1). The compounds, 6j, 6k, 8c, 8k, and 8l that showed inhibitory activity less than 2 μM were further assessed for their ability to inhibit an additional chloroquine-sensitive parasite (D6) strain and two chloroquine-resistant (W2 and 7G8) strains at concentrations ranging from 0.10 μM to 1.75 μM. Each of the five compounds inhibited both chloroquine-sensitive and chloroquine-resistant parasites to similar extents (Table 2). Further, we tested 6j, 6k, 8c, 8k and 8l for lysis of red blood cells at concentration ranging from 2.5 μM to 40 μM. The compounds were not lytic at or below 20 μM, suggesting that the antimalarial activity of dibenzazepines is not due to the lysis of red blood cells.| Product | Parasite growth inhibition IC50 (μM ± SD) | |||
|---|---|---|---|---|
| R1 | 3D7 straina | A549 cellsb | HEK293 cellsb | |
| a The fluorescence intensity was measured in a microliter plate reader.b The absorbance was measured in a microliter plate spectrophotometer. | ||||
| 6a |  |
2.65 ± 0.95 | >100 | >100 |
| 6b | 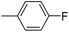 |
2.11 ± 0.74 | 18.40 ± 2.61 | 21.60 ± 1.51 |
| 6c |  |
1.69 ± 0.28 | 18.60 ± 2.30 | 16.60 ± 3.14 |
| 6d | 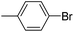 |
1.42 ± 0.96 | 26.80 ± 4.51 | 31.60 ± 4.53 |
| 6e |  |
3.35 ± 0.95 | >100 | >100 |
| 6f |  |
2.27 ± 0.88 | 43.20 ± 3.66 | 40.16 ± 6.23 |
| 6g |  |
2.27 ± 0.88 | 24.10 ± 3.16 | 23.60 ± 4.36 |
| 6h |  |
4.17 ± 1.20 | 42.10 ± 5.03 | 43.40 ± 5.20 |
| 6i |  |
2.23 ± 0.85 | 23.40 ± 4.41 | 26.60 ± 3.22 |
| 6j |  |
0.58 ± 0.18 | 28.40 ± 3.17 | 22.10 ± 3.22 |
| 6k | 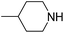 |
0.19 ± 0.06 | 45.60 ± 5.62 | 44.80 ± 6.11 |
| 8a |  |
7.70 ± 1.46 | 20.10 ± 3.94 | 36.40 ± 5.27 |
| 8b |  |
2.69 ± 1.49 | 36.70 ± 4.68 | 42.30 ± 5.09 |
| 8c |  |
1.75 ± 0.84 | 41.60 ± 3.13 | 31.90 ± 6.73 |
| 8d | 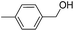 |
2.83 ± 1.06 | 33.60 ± 4.43 | 41.20 ± 3.50 |
| 8e |  |
5.67 ± 1.63 | 21.20 ± 3.82 | 31.20 ± 3.61 |
| 8f | 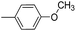 |
2.70 ± 0.94 | 56.20 ± 6.22 | >100 |
| 8g | 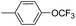 |
4.79 ± 0.93 | 18.60 ± 4.20 | 29.10 ± 5.36 |
| 8h |  |
2.63 ± 1.22 | 39.50 ± 4.63 | 44.30 ± 5.03 |
| 8i |  |
5.20 ± 1.10 | 46.40 ± 6.63 | 41.30 ± 5.15 |
| 8j |  |
3.14 ± 0.93 | 42.40 ± 4.93 | 58.40 ± 4.66 |
| 8k |  |
0.995 ± 0.21 | >100 | >100 |
| 8l | 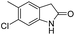 |
1.15 ± 0.67 | 56.6 ± 6.72 | 50.10 ± 4.70 |
| Parasite growth inhibition (IC50, μM ± SEM) | ||||
|---|---|---|---|---|
| Compound | 3D7 | D6 | W2 | 7G8 |
| 6j | 0.58 ± 0.18 | 0.40 ± 0.08 | 0.36 ± 0.09 | 0.42 ± 0.06 |
| 6k | 0.19 ± 0.06 | 0.16 ± 0.04 | 0.12 ± 0.09 | 0.10 ± 0.06 |
| 8c | 1.75 ± 0.84 | 1.66 ± 0.16 | 1.22 ± 0.09 | 1.30 ± 0.12 |
| 8k | 0.99 ± 0.21 | 1.66 ± 0.04 | 1.10 ± 0.12 | 1.20 ± 0.02 |
| 8l | 1.15 ± 0.67 | 1.06 ± 0.13 | 0.92 ± 0.09 | 0.93 ± 0.15 |
| Chloroquine | 0.13 ± 0.06 | 0.12 ± 0.08 | 0.32 ± 0.09 | 0.28 ± 0.03 |
| Mefloquine | 0.04 ± 0.02 | 0.03 ± 0.06 | 0.16 ± 0.07 | 0.11 ± 0.03 |
The compound 6d showed docking score of −6.68 kcal mol−1. It exhibited combination of both electrostatic and hydrophobic interactions resulting in strong binding affinity. The oxygen and nitrogen atom of isoxazoline ring formed co-ordination bond with the Zn2+ ion with the bond length 2.2 Å and 2.5 Å, respectively; the dibenzazepine ring formed π–π parallel stacking interactions with His496; the terminal aromatic ring formed π–π stacking interactions with Tyr575; the bromine formed week halogen interactions with backbone carbonyl of Glu319; the residues Val493, Val523, Tyr580, Tyr575, Val459, Met462 formed hydrophobic interaction with ligand (Fig. 1, Table 3).
| Compound | Docking score (kcal mol−1) | Metal coordinate bond | π–π stacking or π–cation interaction | H-bond forming residues | Hydrophobic interaction residues |
|---|---|---|---|---|---|
| 6d | −6.68 | With Zn2+ | His496, Tyr575 | — | Val493, Val523, Tyr580, Tyr575, Val459, Met462 |
| 6j | −8.06 | With Zn2+ | His496, Tyr575 | Tyr580 | Val493, V523, Tyr580, Tyr575, Val459, Met462, Met1034, Ala320 |
| 6k | −7.90 | With Zn2+ | His496, Tyr575 | Glu572 | Val493, Tyr580, Tyr575, Val459, Met462, Met1034, Ala320 |
| 8k | −7.80 | With Zn2+ | His496, Tyr575 | Tyr580 | Val493, Tyr580, Tyr575, Val459, Met462, Met1034, Ala320 |
| 8l | −7.30 | With Zn2+ | His496, Tyr575 | Asn458 | Val493, Val523, Tyr580, Tyr575, Val459, Met462, Met1034, Ala320 |
The compound 6j exhibited docking score of −8.06 kcal mol−1; the oxygen and nitrogen atom of isoxazoline ring of 6j formed co-ordination bond with the Zn2+ ion with the bond length 2.3 Å and 2.6 Å, respectively; the nitrogen atom of isoxazoline ring formed week H-bond with hydroxyl of Tyr580; the dibenzazepine ring formed π–π interactions with His496; the terminal pyridine ring formed edge to face type of π–π interactions with Tyr575; the residues Val493, V523, Tyr580, Tyr575, Val459, Met462, Met1034, Ala320 formed hydrophobic interaction with ligand (Table 3).
The compound 6k exhibited potent in vitro antimalarial activity. The docking pose of 6k with protein showed strong combination of electrostatic and hydrophobic interactions with active site amino acid residues of PfA-M1, the observed docking score was −7.90 kcal mol−1. The oxygen and nitrogen atom of isoxazoline ring formed co-ordination bond with the Zn2+ ion with the bond length 2.3 Å and 2.8 Å, respectively; the dibenzazepine ring formed π–π interactions with His496; the terminal piperidine ring formed strong ionic interaction (like salt bridge) with Glu572 and the protonated nitrogen of the piperidine ring formed π–cation interaction with Tyr575 resulting in high gain in binding affinity; the residues Val493, Tyr580, Tyr575, Val459, Met462, Met1034, Ala320 formed hydrophobic interaction with ligand (Fig. 1, Table 3).
To improve the mode of interaction and binding ability with the target, the structural modification on compound 6d was carried out and obtained several additional compounds (8 series) and tested for antimalarial activity. Of these, two compounds, 8k and 8l, that showed excellent activity were analysed for binding to PfA-M1. The docking score for compound 8k was −7.80 kcal mol−1. The oxygen and nitrogen atom of isoxazoline ring formed co-ordination bond with the Zn2+ ion with the bond length 2.1 Å and 2.5 Å, respectively; the nitrogen atom of isoxazoline ring formed weak H-bond with hydroxyl group of Tyr580; the dibenzazepine ring formed π–π interactions with His496; the terminal pyridine ring and the penultimate phenyl ring formed edge to face π–π interaction with Tyr575; the residues V523, Val493, Tyr580, Tyr575, Val459, Met462, Met1034, Ala320 formed hydrophobic interaction with ligand (Fig. 1, Table 3).
The compound 8l showed a docking score of −7.30 kcal mol−1. The oxygen and nitrogen atom of isoxazoline ring formed co-ordination bond with the Zn2+ ion with the bond length 2.2 Å and 2.3 Å respectively; the dibenzazepine ring formed π–π interactions with His496; the phenyl ring formed π–π stacking interactions with Tyr575, the terminal carbonyl of 2-indolone formed H-bond with Asn458 and formed π–π stacking interactions with Tyr575; the residues Val493, Val523, Tyr580, Tyr575, Val459, Met462, Met1034, Ala320 formed hydrophobic interaction with ligand (Fig. 1, Table 3). The ligand displayed combination of electrostatic and hydrophobic interactions, but the binding pose bumped on van der Waals radii of the protein atoms, which could be the reason for observed decreased activity of 8l compared to 8k (Fig. 1, Table 3).
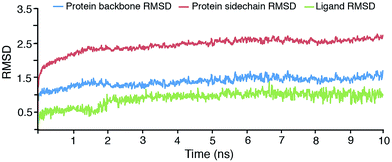
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 244 water molecules. The system was simulated for 50 ps for equilibration. The final simulation run was for a total of 10 ns, during which 1000 structures enumerated were saved in the trajectory. To understand the stability of the complex during MD simulation, the protein backbone frames were aligned to the backbone of initial frame and then the RMS deviation was calculated with respect to the initial frame. The RMS deviations between the original structure and the structure enumerated during MD simulation were plotted (Fig. 2). The protein backbone RMS deviation recorded during simulation shows large deviation for the initial 500 ps due to the initial protein structural stabilization; after about 500 ps the system showed a steady state dynamics. The backbone structural deviations observed in protein for the latter phase of 2 to 10 ns was in the range of 1.28 Å to 1.69 Å compared to that of original structure. From 2 ns to till the end of simulation, the total RMS deviation of protein was within a range of 0.41 Å. This clearly suggests that 10 ns of simulation are sufficient for stabilizing this complex. To further confirm the structure stability of the protein, the RMS deviations between the side chain conformations of the amino acid residues was measured and found minimal RMS deviations of 0.43 Å after 2 ns (Fig. 2).
244 water molecules. The system was simulated for 50 ps for equilibration. The final simulation run was for a total of 10 ns, during which 1000 structures enumerated were saved in the trajectory. To understand the stability of the complex during MD simulation, the protein backbone frames were aligned to the backbone of initial frame and then the RMS deviation was calculated with respect to the initial frame. The RMS deviations between the original structure and the structure enumerated during MD simulation were plotted (Fig. 2). The protein backbone RMS deviation recorded during simulation shows large deviation for the initial 500 ps due to the initial protein structural stabilization; after about 500 ps the system showed a steady state dynamics. The backbone structural deviations observed in protein for the latter phase of 2 to 10 ns was in the range of 1.28 Å to 1.69 Å compared to that of original structure. From 2 ns to till the end of simulation, the total RMS deviation of protein was within a range of 0.41 Å. This clearly suggests that 10 ns of simulation are sufficient for stabilizing this complex. To further confirm the structure stability of the protein, the RMS deviations between the side chain conformations of the amino acid residues was measured and found minimal RMS deviations of 0.43 Å after 2 ns (Fig. 2).
Further the RMS deviation of the ligand as shown in the Fig. 2, drastically reduced to 0.52 Å after 2 ns simulation. This strongly demonstrates that the ligand is well stabilized in binding site of the protein. Various inter-molecular interactions, including H-bond, hydrophobic, ionic, and electrostatic interactions were formed between ligand and protein during the MD simulation, making the ligand well stabilized in the binding pocket. The Fig. 3 shows the summary of the total interactions observed during the MD simulation. The stacked bar charts are normalized over the course of the trajectory. Values over 1.0 are possible as some amino acid residues could make multiple contacts of same subtype with the ligand. All the ligand–protein interactions found in the docking study were retained throughout the MD simulation. Fig. 4 shows a schematic diagram of the ligand interacting with the amino acid residues of protein structure evolved during MD simulation. Residues with the ligand interactions that occurred more than 25% of the simulation time in the trajectory are shown. The total number of specific contacts the protein made with the ligand over the course of the trajectory is shown in Fig. 5. From the structure evolved from the MD simulation, it is evident that ionic interaction with Glu572, co-ordination bond formation with Zn2+, the π–cation interaction with Tyr575 and hydrophobic interaction with Tyr580 of ligand is contributing strongly for the binding affinity; the π–cation interaction with Arg489 and π–π interactions with His496 were also observed. The water mediated bridged interaction between the amino acid residues Glu319 and Glu463 further stabilizing the ligand in the pocket.
 | ||
| Fig. 4 Schematic diagram of ligand interaction with the amino acid residues of protein evolved during MD simulation. Interactions that occur more than 25% of the simulation time are shown. | ||
Experimental section
Materials and methods
The starting chemicals used here were purchased from commercial sources. The solvents were of analytical grade and were used without further purification. Reactions were monitored by thin-layer chromatography (TLC) using pre-coated sheets of silica gel 60 (Merck 60F254, 0.25 mm thickness) and visualization under UV light. The specific molecule purified by preparative HPLC, waters SPD-4245, USA. The melting points were determined on SELACO melting point apparatus and are uncorrected. 1H NMR (400 MHz) and 13C NMR (100 MHz) spectra were obtained using Agilent and Bruker NMR spectrometer, respectively. Chemical shifts (δ) are given in parts per million (ppm) using the residue solvent (CDCl3 and DMSO-d6) peaks as reference relative to TMS. Coupling constant (J) values are given in Hz. Electrospray ionization (ESI) mass spectral analysis was performed using Waters-Synapt G2 mass spectrometer. The C, H, and N analysis were performed using CE-400 CHN analyser. Infrared spectra were recorded on Shimadzu FT-IR model 8300 spectrophotometer.Experimental procedure for the synthesis of 5-allyl-5H-dibenzo[b,f]azepine (2)
To 5H-dibenzazepine (1, 25.8 mmol) in a mixture of toluene and water in the ratio of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 was added sodium hydroxide (51.7 mmol) followed by TBAB (2.5 mmol) at room temperature. After 15 minutes, allyl bromide was added drop wise (38.8 mmol) at room temperature. The reaction mixture was heated at 55 °C for 4 h. After completion of the reaction as monitored by TLC, the reaction mixture was diluted with water (100 mL) and the aqueous layer was extracted with ethyl acetate (3 × 50 mL). The combined ethyl acetate layer was washed with 0.1 N hydrochloric acid (2 × 50 mL), followed by brine solution (2 × 50 mL). The organic layer was dried over anhydrous sodium sulphate, filtered, and concentrated under reduced pressure. The crude compound 2, was purified by column chromatography over silica gel (60–120 mesh) using hexane
1 was added sodium hydroxide (51.7 mmol) followed by TBAB (2.5 mmol) at room temperature. After 15 minutes, allyl bromide was added drop wise (38.8 mmol) at room temperature. The reaction mixture was heated at 55 °C for 4 h. After completion of the reaction as monitored by TLC, the reaction mixture was diluted with water (100 mL) and the aqueous layer was extracted with ethyl acetate (3 × 50 mL). The combined ethyl acetate layer was washed with 0.1 N hydrochloric acid (2 × 50 mL), followed by brine solution (2 × 50 mL). The organic layer was dried over anhydrous sodium sulphate, filtered, and concentrated under reduced pressure. The crude compound 2, was purified by column chromatography over silica gel (60–120 mesh) using hexane![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) ethyl acetate mixture in 9.5
ethyl acetate mixture in 9.5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0.5 ratios as eluent. The crude compound, allyl-5H-dibenzo[b,f]azepine (2), was crystallized in ethyl acetate and hexane mixture to obtain yellow crystals.
0.5 ratios as eluent. The crude compound, allyl-5H-dibenzo[b,f]azepine (2), was crystallized in ethyl acetate and hexane mixture to obtain yellow crystals.
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/i_char_e001.gif) CH2), 5.10 (dd, J1–2 = 10.4 Hz, J1–3 = 1.2 Hz, 1H, N–CH), 5.30 (dd, J1–2 = 17.2 Hz, J1–3 = 1.2 Hz, 1H, N–CH), 5.74–5.84 (m, 1H, –C
CH2), 5.10 (dd, J1–2 = 10.4 Hz, J1–3 = 1.2 Hz, 1H, N–CH), 5.30 (dd, J1–2 = 17.2 Hz, J1–3 = 1.2 Hz, 1H, N–CH), 5.74–5.84 (m, 1H, –C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/i_char_e001.gif) C–H), 6.75 (s, 2H, ArH), 6.98 (t, J = 6.26 Hz, 4H, ArH), 7.06 (dd, J1–2 = 8 Hz, J1–3 = 1.6 Hz, 2H, ArH), 7.24 (td, J1–2 = 8 Hz, J1–3 = 1.2 Hz, 2H, ArH); 13C NMR (100 MHz, CDCl3): δ 53.54, 117.5, 120.5, 123.3, 128.6, 129.1, 132.2, 133.7, 135.1, 150.7; MS (ESI): m/z = 234.1 [M + H]+; elemental composition calculated for C17H15N: C, 87.52; H, 6.48; N, 6.00; found: C, 87.56; H, 6.55; N, 6.04.
C–H), 6.75 (s, 2H, ArH), 6.98 (t, J = 6.26 Hz, 4H, ArH), 7.06 (dd, J1–2 = 8 Hz, J1–3 = 1.6 Hz, 2H, ArH), 7.24 (td, J1–2 = 8 Hz, J1–3 = 1.2 Hz, 2H, ArH); 13C NMR (100 MHz, CDCl3): δ 53.54, 117.5, 120.5, 123.3, 128.6, 129.1, 132.2, 133.7, 135.1, 150.7; MS (ESI): m/z = 234.1 [M + H]+; elemental composition calculated for C17H15N: C, 87.52; H, 6.48; N, 6.00; found: C, 87.56; H, 6.55; N, 6.04.General procedure for the synthesis of isoxazolines
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) ethyl acetate mixture in 8
ethyl acetate mixture in 8![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2 ratios as eluent. The compounds (6a–6k) were crystallized in ethyl acetate and hexane mixture.
2 ratios as eluent. The compounds (6a–6k) were crystallized in ethyl acetate and hexane mixture.
5-[3-(4-Nitro-phenyl)-4,5-dihydro-isoxazol-5-ylmethyl]-5H-dibenzo[b,f]azepine (6a). Yield 72%; yellow crystal; mp 166–168 °C; FT-IR (cm−1): 2993, 1727, 1519, 1484, 1269, 1169, 909, 764, 704; 1H NMR (400 MHz, CDCl3): δ 3.21 (dd, J1–2 = 16.8 Hz, J1–1 = 10.8 Hz, 1H, C4–H isoxazoline), 3.46 (dd, J1–2 = 17.2 Hz, J1–1 = 6.4 Hz, 1H, N–CH), 3.58 (dd, J1–2 = 12.8 Hz, J1–1 = 8.8 Hz, 1H, C4–H isoxazoline), 4.32 (dd, J1–2 = 13.2 Hz, J1–1 = 4.4 Hz, 1H, N–CH), 4.94–5.02 (m, 1H, C5–H isoxazoline), 6.70 (q, J = 11.6 Hz, 2H, ArH), 6.98–7.04 (m, 4H, ArH), 7.09 (d, J = 6.4 Hz, 1H, ArH), 7.18 (d, J = 8.0 Hz, 1H, ArH), 7.23–7.31 (m, 2H, ArH), 7.73 (d, J = 8.8 Hz, 2H, ArH), 8.20 (d, J = 8.8 Hz, 2H, ArH); 13C NMR (100 MHz, CDCl3): δ 37.7, 53.5, 79.7, 120.7, 120.9, 123.8, 124.0, 124.1, 127.3, 129.0, 129.2, 129.4, 131.8, 132.1, 133.5, 134.2, 135.7, 148.3, 148.4, 151.1, 154.9; MS: m/z = 397.4 (calculated), m/z = 398.8 [M + H]+ (found); elemental composition calculated for C24H19N3O3: C, 72.53; H, 4.82; N, 10.57; O, 12.08; found: C, 72.56; H, 4.89; N, 10.59.
5-[3-(4-Fluoro-phenyl)-4,5-dihydro-isoxazol-5-ylmethyl]-5H-dibenzo[b,f]azepine (6b). Yield 76%; yellow solid; mp 98–100 °C; FT-IR (cm−1): 2980, 1731, 1523, 1489, 1260, 1153, 901, 764; 1H NMR (400 MHz, CDCl3): δ 3.19 (dd, J1–2 = 16.8 Hz, J1–1 = 10.4 Hz, 1H, N–CH), 3.83–3.52 (m, 2H, C4–2H isoxazoline), 4.35 (dd, J1–2 = 12.8 Hz, J1–1 = 4.0 Hz, 1H, N–CH), 4.87–4.94 (m, 1H, C5–H isoxazoline), 6.73 (d, J = 2.4 Hz, 2H, ArH), 7.10–6.98 (m, 7H, ArH), 7.18–7.23 (m, 2H, ArH), 7.27–7.32 (m, 1H, ArH), 7.56–7.60 (m, 2H, ArH); 13C NMR (100 MHz, DMSO-d6): δ 38.3, 54.2, 78.7, 115.9, 116.3, 121.3, 121.4, 124.0, 126.4, 126.5, 129.1, 129.2, 129.4, 129.5, 132.3, 132.5, 133.9, 150.4, 150.7, 155.9, 162.1, 164.6; MS: m/z = 370.4 (calculated), m/z = 371.5 [M + H]+ (found); elemental composition calculated for: C24H19FN2O: C, 77.82; H, 5.17; F, 5.13; N, 7.56; O, 4.32; found: C, 77.84; H, 5.21; N, 7.58.
5-[3-(4-Chloro-phenyl)-4,5-dihydro-isoxazol-5-ylmethyl]-5H-dibenzo[b,f]azepine (6c). Yield 82%; yellow solid; mp 155–157 °C; FT-IR (cm−1): 2925, 1593, 1494, 1485, 1456, 1434, 1355, 1238, 1179; 1H NMR (400 MHz, CDCl3): δ 3.18 (dd, J1–2 = 16.8 Hz, J1–1 = 10.4 Hz, 1H, C4–H isoxazoline), 3.40 (dd, J1–2 = 16.8 Hz, J1–1 = 6.0 Hz, 1H, C4–H isoxazoline), 3.51 (dd, J1–2 = 12.8 Hz, J1–1 = 9.2 Hz, 1H, N–CH), 4.33 (dd, J1–2 = 12.8 Hz, J1–1 = 4.4 Hz, 1H, N–CH), 4.87–4.95 (m, 1H, C5–H isoxazoline), 6.73 (q, J = 11.2 Hz, 2H, ArH), 6.98– 7.05 (m, 4H, ArH), 7.09 (d, J = 7.6 Hz, 1H, ArH), 7.18–7.23 (m, 2H, ArH), 7.27–7.33 (m, 3H, ArH), 7.51–7.54 (m, 2H, ArH); 13C NMR (100 MHz, DMSO-d6): δ 38.1, 54.2, 79.0, 121.3, 124.1, 128.6, 128.8, 129.2, 129.5, 132.2, 132.5, 133.9, 134.9, 156.0; MS: m/z = 386.8 (calculated), m/z = 387.5 [M + H]+ (found); elemental composition calculated for: C24H19ClN2O: C, 74.51; H, 4.95; Cl, 9.16; N, 7.24; O, 4.14; found: C, 74.53; H, 4.99; N, 7.25.
5-[3-(4-Bromo-phenyl)-4,5-dihydro-isoxazol-5-ylmethyl]-5H-dibenzo[b,f]azepine (6d). Yield 83%; brown solid; mp 138–140 °C; FT-IR (cm−1): 2923, 1590, 1486, 1457, 1398, 1347, 1235, 1158, 1114; 1H NMR (400 MHz, CDCl3): δ 3.17 (dd, J1–2 = 17.2 Hz, J1–1 = 10.8 Hz, 1H, C4–H isoxazoline), 3.39 (dd, J1–2 = 16.8 Hz, J1–1 = 6.0 Hz, 1H, N–CH), 3.50 (dd, J1–2 = 12.8 Hz, J1–1 = 9.2 Hz, 1H, C4–H isoxazoline), 4.31 (dd, J1–2 = 12.8 Hz, J1–1 = 4.4 Hz, 1H, N–CH), 4.89–4.91 (m, 1H, C5–H isoxazoline), 6.72 (d, J = 3.2 Hz, 2H, ArH), 7.04 (q, J = 15.6 Hz, 3H, ArH), 7.09 (d, J = 6.4 Hz, 1H, ArH), 7.18 (d, J = 8.0 Hz, 1H, ArH), 7.22–7.30 (m, 3H, ArH), 7.43–7.49 (m, 4H, ArH); 13C NMR (100 MHz, CDCl3): δ 38.2, 53.6, 78.8, 120.7, 120.9, 123.9, 124.0, 124.2, 128.1, 128.5, 129.0, 129.1, 129.2, 129.4, 131.7, 131.8, 132.2, 133.5, 134.2, 148.4, 151.4, 155.6; MS: m/z = 431.3 (calculated), m/z = 431.9 [M + H]+ (found); elemental composition calculated for: C24H19BrN2O: C, 66.83; H, 4.44; Br, 18.53; N, 6.49; O, 3.71; found: C, 66.86; H, 4.49; N, 6.71.
5-[3-(2,4-Dimethyl-phenyl)-4,5-dihydro-isoxazol-5-ylmethyl]-5H-dibenzo[b,f]azepine (6e). Yield 78%; yellow gummy solid; FT-IR (cm−1): 2935, 1590, 1484, 1481, 1453, 1424, 1359, 1231, 1199; 1H NMR (400 MHz, CDCl3): δ 2.23 (S, 3H, CH3), 2.37 (S, 3H, CH3), 3.18 (dd, J1–2 = 16.8 Hz, J1–1 = 10.4 Hz, 1H, C4–H isoxazoline), 3.36–3.42 (m, 2H, N–CH and C4–H isoxazoline), 4.25 (dd, J1–2 = 12.8 Hz, J1–1 = 4.4 Hz, 1H, N–CH), 4.75 (t, J = 4.8 Hz, 1H, C5–H isoxazoline), 6.65 (S, 2H, ArH), 6.90–7.02 (m, 7H, ArH), 7.08–7.22 (m, 4H, ArH); 13C NMR (100 MHz, CDCl3): δ 21.1, 22.9, 41.1, 53.5, 77.0, 120.7, 120.9, 123.8, 124.0, 125.7, 126.3, 128.9, 129.02, 129.09, 129.1, 129.4, 131.9, 132.1, 132.3, 133.5, 134.2, 137.8, 139.2, 148.5, 151.7, 157.3; MS: m/z = 380.48 (calculated), m/z = 381.3 [M + H]+ (found); elemental composition calculated for C26H24N2O: C, 82.07; H, 6.36; N, 7.36; O, 4.21; found: C, 82.09; H, 6.40; N, 4.23.
5-[3-(4-Methoxy-phenyl)-4,5-dihydro-isoxazol-5-ylmethyl]-5H-dibenzo[b,f]azepine (6f). Yield 75%; orange gummy solid; FT-IR (cm−1): 2953, 1575, 1491, 1466, 1376, 1339, 1237, 1160, 1101; 1H NMR (400 MHz, CDCl3): δ 3.26 (dd, J1–2 = 16.4 Hz, J1–1 = 10.0 Hz, 1H, C4–H isoxazoline), 3.44–3.49 (m, 2H, C4–H isoxazoline and N–CH), 3.81 (s, 3H, OCH3), 4.33 (dd, J1–2 = 12.8 Hz, J1–1 = 4.4 Hz, 1H, N–CH), 6.73 (s, 2H, ArH), 4.80–4.85 (m, 1H, C5–H isoxazoline), 6.97–7.03 (m, 7H, ArH), 7.09 (dd, J1–2 = 7.6 Hz, J1–3 = 1.6 Hz, 1H, ArH), 7.15–7.23 (m, 3H, ArH), 7.26–7.32 (m, 1H, ArH); 13C NMR (100 MHz, CDCl3): δ 38.3, 55.5, 55.6, 79.7, 114.5, 120.3, 120.6, 120.7, 122.0, 123.7, 129.0, 129.1, 130.5, 132.1, 133.5, 146.5, 150.1, 159.6; MS: m/z = 382.4 (calculated), m/z = 383.3 [M + H]+ (found); elemental composition calculated for: C25H22N2O2: C, 78.51; H, 5.80; N, 7.32; O, 8.37; found: C, 78.54; H, 5.88; N, 7.35.
4-(5-Dibenzo[b,f]azepin-5-ylmethyl-4,5-dihydro-isoxazol-3-yl)-phenol (6g). Yield 70%; buff white solid; mp 169–171 °C; FT-IR (cm−1): 3210, 2924, 1598, 1521, 1485, 1459, 1437, 1360, 1276, 1188, 1128; 1H NMR (400 MHz, CDCl3): δ 3.23 (dd, J1–2 = 16.8 Hz, J1–1 = 10.8 Hz, 1H, C4–H isoxazoline), 3.42–3.54 (m, 2H, C4–H isoxazoline and N–CH), 4.35 (dd, J1–2 = 12.8 Hz, J1–1 = 4.4 Hz, 1H, N–CH), 4.89–4.96 (m, 1H, C5–H isoxazoline), 6.74 (d, J = 1.2 Hz, 2H, ArH), 6.99–7.06 (m, 4H, ArH), 7.10 (dd, J1–2 = 7.6 Hz, J1–3 = 1.6 Hz, 1H, ArH), 7.19–7.25 (m, 4H, ArH), 7.28–7.33 (m, 1H, ArH), 7.49–7.53 (m, 2H, ArH), 9.20 (s, 1H, ArH); 13C NMR (100 MHz, CDCl3): δ 38.2, 54.5, 77.2, 120.2, 120.3, 121.5, 123.8, 129.1, 129.2, 129.7, 132.1, 133.5, 134.2, 135.5, 147.0, 149.9; MS: m/z = 368.43 (calculated), m/z = 369.0 [M + H]+ (found); elemental composition calculated for: C24H20N2O2: C, 78.24; H, 5.47; N, 7.60; O, 8.69; found: C, 78.25; H, 5.49; N, 7.61.
Benzoic acid 4-(5-dibenzo[b,f]azepin-5-ylmethyl-4,5-dihydro-isoxazol-3-yl)-phenyl ester (6h). Yield 80%; yellow solid; mp 140–142 °C; FT-IR (cm−1): 2990, 1727, 1484, 1269, 1169, 909, 764, 704; 1H NMR (400 MHz, CDCl3): δ 3.30–3.43 (m, 2H, C4–2H isoxazoline), 3.82 (dd, J1–2 = 13.6 Hz, J1–1 = 6.4 Hz, 1H, N–CH), 4.05 (dd, J1–2 = 13.6 Hz, J1–1 = 6.4 Hz, 1H, N–CH), 4.75–4.80 (m, 1H, C5–H isoxazoline), 6.77 (d, J = 1.6 Hz, 2H, ArH), 7.03 (d, J = 5.6 Hz, 2H, ArH), 7.10–7.23 (m, 4H, ArH), 7.30–7.37 (m, 4H, ArH), 7.59–7.69 (m, 4H, ArH), 7.76 (t, J = 7.2 Hz, 1H, ArH), 8.14 (d, J = 7.2 Hz, 2H, ArH); 13C NMR (100 MHz, DMSO-d6): δ 38.3, 54.2, 78.8, 121.3, 121.4, 122.8, 124.1, 127.7, 128.2, 129.1, 129.4, 129.5, 130.3, 132.3, 132.5, 133.9, 134.6, 150.4, 150.7, 152.1, 156.1, 164; MS: m/z = 472.5 (calculated), m/z = 473.6 [M + H]+ (found); elemental composition calculated for: C31H24N2O3: C, 78.79; H, 5.12; N, 5.93; O, 10.16; found: C, 78.80; H, 5.17; N, 5.95.
5-[3-(4-Trifluoromethyl-phenyl)-4,5-dihydro-isoxazol-5-ylmethyl]-5H-dibenzo[b,f]azepine azepine (6i). Yield 83%; yellow solid; mp 143–145 °C; FT-IR (cm−1): 2940, 1593, 1485, 1457, 1457, 1410, 1323, 1229, 1163, 1127; 1H NMR (400 MHz, CDCl3): δ 3.21 (dd, J1–2 = 17.2 Hz, J1–1 = 10.8 Hz, 1H, N–CH), 3.42–3.57 (m, 2H, C4–2H isoxazoline), 4.33 (dd, J1–2 = 12.8 Hz, J1–1 = 4.4 Hz, 1H, N–CH), 4.92–4.99 (m, 1H, C5–H isoxazoline), 6.72 (q, J = 11.2 Hz, 2H, ArH), 6.90–7.05 (m, 4H, ArH), 7.10 (d, J = 6.4 Hz, 1H, ArH), 7.19 (d, J = 8.0 Hz, 1H, ArH), 7.23–7.32 (m, 2H, ArH), 7.61 (d, J = 8.4 Hz, 2H, ArH), 7.70 (d, J = 8.4 Hz, 2H, ArH); 13C NMR (100 MHz, DMSO-d6): δ 37.9, 54.1, 79.4, 121.3, 121.4, 123.1, 124.1, 125.8, 126.03, 126.07, 126.1, 127.6, 129.3, 129.5, 130.0, 130.3, 130.6, 132.2, 132.5, 133.8, 133.9, 150.4, 150.6, 156.0; MS: m/z = 420.4 (calculated), m/z = 421.5 [M + H]+ (found); elemental composition calculated for: C25H19F3N2O: C, 71.42; H, 4.56; F, 13.56; N, 6.66; O, 3.81; found: C, 71.45; H, 4.59; N, 6.67.
5-[3-(4-Pyridin-3-yl-phenyl)-4,5-dihydro-isoxazol-5-ylmethyl]-5H-dibenzo[b,f]azepine (6j). Yield 76%; pale brown solid; mp 162–164 °C; FT-IR (cm−1): 2922, 2853, 1732, 1583, 1583, 1571, 1484, 1433, 1407, 1358, 1334, 1298, 1240, 1225, 1162; 1H NMR (400 MHz, CDCl3): δ 3.26 (dd, J1–2 = 16.8 Hz, J1–1 = 10.4 Hz, 1H, N–CH), 3.45–3.57 (m, 2H, C4–2H isoxazoline), 4.35 (dd, J1–2 = 12.8 Hz, J1–1 = 4.4 Hz, 1H, N–CH), 4.90–4.97 (m, 1H, C5–H isoxazoline), 6.73 (d, J = 2.8 Hz, 2H, ArH), 6.98–7.05 (m, 4H, ArH), 7.10 (d, J = 7.6 Hz, 1H, ArH), 7.22 (t, J = 8.4 Hz, 2H, ArH), 7.30 (t, J = 7.2 Hz, 1H, ArH), 7.02–7.84 (m, 5H, ArH), 8.00 (d, J = 8.4 Hz, 2H, ArH), 8.34 (d, J = 4.4 Hz, 1H, ArH); 13C NMR (100 MHz, CDCl3): δ 38.1, 54.2, 78.2, 120.2, 120.3, 120.4, 123.8, 124.0, 127.8, 129.1, 129.2, 132.1, 133.5, 133.6, 141.4, 147.4, 149.6, 149.9; MS: m/z = 429.5 (calculated), m/z = 430.0 [M + H]+ (found); elemental composition calculated for: C29H23N3O: C, 81.09; H, 5.40; N, 9.78; O, 3.73; found: C, 81.10; H, 5.46; N, 9.79.
5-(3-Piperazin-1-yl-4,5-dihydro-isoxazol-5-ylmethyl)-5H-dibenzo[b,f]azepine (6k). Yield 67%; buff white solid; mp 126–128 °C; FT-IR (cm−1): 3400, 2712, 1593, 1483, 1458, 1308, 1224, 1160; 1H NMR (400 MHz, CDCl3): δ 1.85–2.01 (m, 4H, 2CH2 piperidine), 2.5–2.52 (m, 1H, CH piperidine), 2.78 (dd, J1–2 = 17.2 Hz, J1–1 = 10.8 Hz, 1H, C4–H isoxazoline), 2.90–3.01 (m, 3H, CH2 piperidine and N–CH), 3.32 (S, 2H, CH2 piperidine), 3.41 (dd, J1–2 = 12.8 Hz, J1–1 = 9.2 Hz, 1H, C4–H isoxazoline), 4.22 (dd, J1–2 = 12.8 Hz, J1–1 = 4.0 Hz, 1H, N–CH), 4.67–4.73 (m, 1H, C5–H isoxazoline), 5.18 (br, 1H, NH), 6.72 (d, J = 2.0 Hz, 2H, ArH), 7.04 (t, J = 6.4 Hz, 3H, ArH), 7.05–7.07 (m, 2H, ArH), 7.12 (d, J = 8.4 Hz, 1H, ArH), 7.23–7.28 (m, 2H, ArH); 13C NMR (100 MHz, CDCl3): δ 26.0, 29.6, 33.1, 38.8, 43.0, 53.4, 77.6, 120.7, 121.0, 123.9, 124.0, 129.0, 129.1, 129.2, 129.4, 131.9, 132.1, 133.5, 134.2, 148.5, 151.3, 159.2; MS: m/z = 359.464 (calculated), m/z = 360.189 [M + H]+ (found); elemental composition calculated for C23H25N3O: C, 76.85; H, 7.01; N, 11.69; O, 4.45; found: C, 76.88; H, 7.05; N, 11.70; O, 4.49.
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) ethyl acetate mixture in 7
ethyl acetate mixture in 7![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3 ratios as eluent.
3 ratios as eluent.
5-[3-(4′-Chloro-biphenyl-4-yl)-4,5-dihydro-isoxazol-5-ylmethyl]-5H-dibenzo[b,f]azepine (8a). Yield 82%; yellow solid; mp 165–167 °C; FT-IR (cm−1): 2986, 2873, 1762, 1576, 1573, 1561, 1474, 1433, 1360, 1278, 1230, 1152; 1H NMR (400 MHz, DMSO-d6): δ 3.25 (dd, J1–2 = 17.2 Hz, J1–1 = 6.8 Hz, 1H, C4–H isoxazoline), 3.37 (dd, J1–2 = 17.2 Hz, J1–1 = 10.8 Hz, 1H, C4–H isoxazoline), 3.75 (dd, J1–2 = 14.0 Hz, J1–1 = 6.4 Hz, 1H, N–CH), 4.01 (dd, J1–2 = 19.6 Hz, J1–1 = 6.4 Hz, 1H, N–CH), 4.69–4.74 (m, 1H, C5–H isoxazoline), 6.72 (d, J = 2.4 Hz, 2H, ArH), 6.97–7.27 (m, 8H, ArH), 7.41 (d, J = 8.4 Hz, 2H, ArH), 7.61 (d, J = 8.0 Hz, 2H, ArH), 7.66–7.69 (m, 4H, ArH); 13C NMR (100 MHz, DMSO-d6): δ 38.2, 54.2, 78.7, 121.4, 124.0, 127.3, 127.5, 128.8, 129.2, 129.4, 129.5, 129.5, 132.2, 133.2, 133.9, 138.4, 140.4, 150.3, 150.7, 156.4; MS: m/z = 462.97 (calculated), m/z = 463.4 [M + H]+ (found); elemental composition calculated for C30H23ClN2O: C, 77.83; H, 5.01; Cl, 7.66; N, 6.05; O, 3.46; found: C, 77.85; H, 5.08; N, 6.06; O, 3.47.
4′-(5-Dibenzo[b,f]azepin-5-ylmethyl-4,5-dihydro-isoxazol-3-yl)-biphenyl-4-ol (8b). Yield 79%; yellow solid; mp 119–121 °C; FT-IR (cm−1): 2985, 1585, 1482, 1306, 1200, 1047; 1H NMR (400 MHz, DMSO-d6): δ 3.25 (dd, J1–2 = 16.0 Hz, J1–1 = 5.6 Hz, 1H, C4–H isoxazoline), 3.40 (d, J = 3.2 Hz, 1H, C4–H isoxazoline), 3.75 (dd, J1–2 = 12.8 Hz, J1–1 = 5.6 Hz, 1H, N–CH), 4.02 (dd, J1–2 = 13.6 Hz, J1–1 = 5.2 Hz, 1H, N–CH), 4.72–4.74 (m, 1H, C5–H isoxazoline), 6.75 (s, 2H, ArH), 6.85 (d, J = 7.6 Hz, 2H, ArH), 7.00–7.36 (m, 8H, ArH), 7.51 (d, J = 7.6 Hz, 2H, ArH), 7.60 (d, J = 8.1 Hz, 4H, ArH), 9.63 (s, 1H, ArH); 13C NMR (100 MHz, DMSO-d6): δ 38.3, 54.2, 78.5, 116.0, 116.2, 121.3, 124.0, 126.4, 127.4, 127.7, 128.2, 129.4, 129.5, 130.2, 132.2, 132.4, 133.8, 141.9, 156.5, 158.0; MS: m/z = 444.52 (calculated), m/z = 445.1 [M + H]+ (found); elemental composition calculated for C30H24N2O2: C, 81.06; H, 5.44; N, 6.30; O, 7.20; found: C, 81.09; H, 5.50; N, 6.32; O, 7.21.
4′-(5-Dibenzo[b,f]azepin-5-ylmethyl-4,5-dihydro-isoxazol-3-yl)-biphenyl-3-ol (8c). Yield 89%; pale brown solid; mp 108–110 °C; FT-IR (cm−1): 3419, 2988, 2254, 1659, 1481, 1244, 1050, 1023; 1H NMR (400 MHz, DMSO-d6): δ 3.23 (dd, J1–2 = 17.2 Hz, J1–1 = 6.4 Hz, 1H, C4–H isoxazoline), 3.36 (dd, J1–2 = 16.8 Hz, J1–1 = 10.4 Hz, 1H, C4–H isoxazoline), 3.73 (dd, J1–2 = 11.2 Hz, J1–1 = 6.4 Hz, 1H, N–CH), 4.00 (dd, J1–2 = 13.2 Hz, J1–1 = 6.0 Hz, 1H, N–CH), 4.68–4.72 (m, 1H, C5–H isoxazoline), 6.73 (d, J = 1.6 Hz, 2H, ArH), 6.80–6.83 (m, 2H, ArH), 6.98–7.27 (m, 8H, ArH), 7.47–7.50 (m, 12H, ArH), 7.54–7.61 (m, 6H, ArH), 9.32 (s, 1H, ArH); 13C NMR (100 MHz, DMSO-d6): δ 38.3, 54.2, 78.5, 115.9, 116.2, 121.3, 124.0, 126.4, 127.4, 127.7, 128.1, 129.5, 130.3, 131.6, 132.2, 133.9, 141.9, 150.3, 150.7, 156.5, 156.6, 158.0; MS: m/z = 444.52 (calculated), m/z = 445.1 [M + H]+ (found); elemental composition calculated for C30H24N2O2: C, 81.06; H, 5.44; N, 6.30; O, 7.20; found: C, 81.09; H, 5.50; N, 6.32; O, 7.21.
[4′-(5-Dibenzo[b,f]azepin-5-ylmethyl-4,5-dihydro-isoxazol-3-yl)-biphenyl-4-yl]-methanol (8d). Yield 90%; yellow solid; mp 145–147 °C; FT-IR (cm−1): 3445, 2250, 2135, 1672, 1055, 1001; 1H NMR (400 MHz, DMSO-d6): δ 3.28 (dd, J1–2 = 16.8 Hz, J1–1 = 6.4 Hz, 1H, C4–H isoxazoline), 3.40 (dd, J1–2 = 16.8 Hz, J1–1 = 10.8 Hz, 1H, C4–H isoxazoline), 3.75 (dd, J1–2 = 13.6 Hz, J1–1 = 6.4 Hz, 1H, N–CH), 4.01 (dd, J1–2 = 13.6 Hz, J1–1 = 6.4 Hz, 1H, N–CH), 4.54 (d, J = 5.6 Hz, 2H, OCH2), 4.70–4.74 (m, 1H, C5–H isoxazoline), 5.22 (t, J = 11.6 Hz, 1H, ArH), 6.73 (d, J = 2.4 Hz, 2H, ArH), 6.98–7.31 (m, 9H, ArH), 7.39 (t, J = 8.4 Hz, 1H, ArH), 7.51 (d, J = 7.6 Hz, 1H, ArH), 7.62 (d, J = 8.4 Hz, 3H, ArH), 7.68 (d, J = 8.8 Hz, 2H, ArH); 13C NMR (100 MHz, DMSO-d6): δ 38.2, 54.2, 63.2, 78.6, 121.3, 124.0, 125.0, 125.2, 125.4, 126.4, 127.3, 127.5, 128.8, 129.2, 129.4, 129.5, 132.2, 132.4, 139.4, 133.9, 142.0, 143.8, 150.3, 150.7, 156.4; MS: m/z = 458.55 (calculated), m/z = 459.1 [M + H]+ (found); elemental composition calculated for C31H26N2O2: C, 81.20; H, 5.72; N, 6.11; O, 6.98; found: C, 81.23; H, 5.79; N, 6.13; O, 6.99.
4′-(5-Dibenzo[b,f]azepin-5-ylmethyl-4,5-dihydro-isoxazol-3-yl)-biphenyl-4-carbaldehyde (8e). Yield 78%; yellow solid; mp 119–121 °C; FT-IR (cm−1): 2251, 2125, 1661, 1052, 1024, 1005, 821, 758, 624; 1H NMR (400 MHz, DMSO-d6): δ 3.27 (dd, J1–2 = 17.2 Hz, J1–1 = 4.4 Hz, 1H, C4–H isoxazoline), 3.39–3.43 (m, 1H, C4–H isoxazoline), 3.78 (dd, J1–2 = 13.2 Hz, J1–1 = 6.0 Hz, 1H, N–CH), 4.01 (dd, J1–2 = 13.6 Hz, J1–1 = 6.4 Hz, 1H, N–CH), 4.72–4.76 (m, 1H, C5–H isoxazoline), 6.73 (d, J = 2.8 Hz, 2H, ArH), 6.97–7.31 (m, 8H, ArH), 7.67 (t, J = 7.2 Hz, 3H, ArH), 7.79 (dd, J1–2 = 6.8 Hz, J1–1 = 1.6 Hz, 2H, ArH), 7.87–7.90 (m, 1H, ArH), 8.00–8.03 (m, 1H, ArH), 8.20 (t, J = 1.6 Hz, 1H, ArH), 10.06 (s, 1H, CHO); 13C NMR (100 MHz, DMSO-d6): δ 38.2, 54.2, 78.8, 121.4, 124.1, 127.5, 127.6, 128.3, 128.8, 129.5, 130.3, 132.2, 132.4, 132.9, 133.9, 137.3, 140.4, 140.5, 150.3, 150.6, 156.4, 193.6; MS: m/z = 456.53 (calculated), m/z = 457.0 [M + H]+ (found); elemental composition calculated for C31H24N2O2: C, 81.56; H, 5.30; N, 6.14; O, 7.01; found: C, 81.58; H, 5.37; N, 6.16; O, 7.02.
5-[3-(4′-Methoxy-biphenyl-4-yl)-4,5-dihydro-isoxazol-5-ylmethyl]-5H-dibenzo[b,f]azepine (8f). Yield 87%; pale yellow solid; mp 187–189 °C; FT-IR (cm−1): 2926, 2853, 1777, 1551, 1529, 1424, 1369, 1258, 1222, 1172; 1H NMR (400 MHz, DMSO-d6): δ 3.28 (dd, J1–2 = 16.8 Hz, J1–1 = 6.4 Hz, 1H, C4–H isoxazoline), 3.40 (dd, J1–2 = 16.8 Hz, J1–1 = 10.4 Hz, 1H, C4–H isoxazoline), 3.75–3.80 (m, 4H, N–CH and OCH3), 4.05 (dd, J1–2 = 13.6 Hz, J1–1 = 6.0 Hz, 1H, N–CH), 4.72–4.77 (m, 1H, C5–H isoxazoline), 6.77 (d, J = 1.6 Hz, 2H, ArH), 7.03 (d, J = 8.8 Hz, 4H, ArH), 7.10–7.32 (m, 6H, ArH), 7.61–7.69 (m, 6H, ArH); 13C NMR (100 MHz, DMSO-d6): δ 38.2, 54.2, 55.6, 78.5, 114.9, 124.0, 126.7, 127.4, 128.1, 128.2, 129.5, 131.9, 132.2, 132.5, 133.9, 141.5, 156.5, 159.7; MS: m/z = 458.55 (calculated), m/z = 459.10 [M + H]+ (found); elemental composition calculated for C31H26N2O2: C, 81.20; H, 5.72; N, 6.11; O, 6.98; found: C, 81.23; H, 5.79; N, 6.13; O, 6.99.
5-[3-(4′-Trifluoromethoxy-biphenyl-4-yl)-4,5-dihydro-isoxazol-5-ylmethyl]-5H-dibenzo[b,f]azepine (8g). Yield 80%; yellow solid; mp 165–167 °C; FT-IR (cm−1): 2902, 2260, 2128, 1662, 1075, 1021, 987; 1H NMR (400 MHz, DMSO-d6): δ 3.25 (dd, J1–2 = 17.2 Hz, J1–1 = 6.8 Hz, 1H, C4–H isoxazoline), 3.37 (dd, J1–2 = 17.2 Hz, J1–1 = 10.9 Hz, 1H, C4–H isoxazoline), 3.75 (dd, J1–2 = 14.0 Hz, J1–1 = 6.4 Hz, 1H, N–CH), 4.01 (dd, J1–2 = 13.6 Hz, J1–1 = 6.4 Hz, 1H, N–CH), 4.70–4.75 (m, 1H, C5–H isoxazoline), 6.73 (d, J = 2.4 Hz, 2H, ArH), 6.96–7.29 (m, 8H, ArH), 7.41 (d, J = 8 Hz, 2H, ArH), 7.62 (d, J = 8.4 Hz, 2H, ArH), 7.70 (d, J = 8 Hz, 2H, ArH), 7.77 (d, J = 8.8 Hz, 2H, ArH); 13C NMR (100 MHz, DMSO-d6): δ 38.2, 54.1, 78.7, 119.2, 121.3, 121.3, 121.8, 121.9, 124.0, 127.5, 127.6, 129.0, 129.3, 129.4, 129.5, 132.2, 132.4, 133.9, 138.9, 140.3, 148.5, 150.3, 150.7, 156.4; MS: m/z = 512.525 (calculated), m/z = 513.10 [M + H]+ (found); elemental composition calculated for C31H23F3N2O2: C, 72.65; H, 4.52; F, 11.12; N, 5.47; O, 6.24; found: C, 72.67; H, 4.57; N, 5.48; O, 6.25.
4′-(5-Dibenzo[b,f]azepin-5-ylmethyl-4,5-dihydro-isoxazol-3-yl)-biphenyl-4-carbonitrile (8h). Yield 84%; pale yellow solid; mp 190–193 °C; FT-IR (cm−1): 2971, 1483, 1457, 1226, 1052, 907; 1H NMR (400 MHz, DMSO-d6): δ 3.29 (dd, J1–2 = 16.8 Hz, J1–1 = 6.4 Hz, 1H, C4–H isoxazoline), 3.40 (dd, J1–2 = 12.0 Hz, J1–1 = 6.8 Hz, 1H, C4–H isoxazoline), 3.78 (dd, J1–2 = 13.2 Hz, J1–1 = 6.0 Hz, 1H, N–CH), 4.04 (dd, J1–2 = 14.0 Hz, J1–1 = 6.4 Hz, 1H, N–CH), 4.73 (m, 1H, C5–H isoxazoline), 6.72 (d, J = 2.8 Hz, 2H, ArH), 6.99–7.28 (m, 8H, ArH), 7.64–7.76 (m, 2H, ArH), 7.76–7.85 (m, 2H, ArH), 7.85–7.91 (m, 4H, ArH); 13C NMR (100 MHz, DMSO-d6): δ 38.1, 54.2, 78.9, 110.8, 119.2, 121.3, 124.0, 127.6, 127.8, 127.9, 129.5, 130.1, 132.2, 132.4, 133.3, 133.9, 139.8, 144.0, 150.3, 150.6, 156.3; MS: m/z = 453.53 (calculated), m/z = 454.0 [M + H]+ (found); elemental composition calculated for C31H23N3O: C, 82.10; H, 5.11; N, 9.27; O, 3.53; found: C, 82.13; H, 5.16; N, 9.29; O, 3.54.
[4′-(5-Dibenzo[b,f]azepin-5-ylmethyl-4,5-dihydro-isoxazol-3-yl)-biphenyl-4-ylmethyl]-phenyl-amine (8i). Yield 77%; pale brown solid; mp 173–175 °C; FT-IR (cm−1): 3444, 2251, 1661, 1053, 1005; 1H NMR (400 MHz, DMSO-d6): δ 3.26 (dd, J1–2 = 13.6 Hz, J1–1 = 6.8 Hz, 1H, C4–H isoxazoline), 3.36 (dd, J1–2 = 14.8 Hz, J1–1 = 8.4 Hz, 1H, C4–H isoxazoline), 3.76 (dd, J1–2 = 14.0 Hz, J1–1 = 6.4 Hz, 1H, N–CH), 4.01 (dd, J1–2 = 14.0 Hz, J1–1 = 6.4 Hz, 1H, N–CH), 4.30 (d, J = 5.6 Hz, 2H, N–CH2), 4.72 (m, 1H, C5–H isoxazoline), 6.22 (t, J = 6.0 Hz, 1H, ArH), 6.47 (t, J = 7.2 Hz, 1H, ArH), 6.58 (dd, J1–2 = 8.8 Hz, J1–1 = 0.8 Hz, 2H, ArH), 6.73 (d, J = 2.4 Hz, 2H, ArH), 6.99–7.02 (m, 4H, ArH), 7.06–7.26 (m, 6H, ArH), 7.33–7.40 (m, 2H, ArH), 7.51 (dd, J1–1 = 7.6 Hz, J1–2 = 1.6 Hz, 1H, ArH), 7.61 (d, J = 8.8 Hz, 2H, ArH), 6.65–7.67 (m, 3H, ArH); 13C NMR (100 MHz, DMSO-d6): δ 38.2, 46.9, 54.2, 78.6, 112.8, 116.2, 121.3, 124.0, 125.4, 125.9, 126.0, 127.3, 127.5, 128.8, 129.2, 129.4, 129.5, 132.2, 132.4, 133.9, 139.6, 141.6, 141.9, 149.0, 150.3, 150.7, 156.4; MS: m/z = 533.66 (calculated), m/z = 534.8 [M + H]+ (found); elemental composition calculated for C37H31ClN3O: C, 83.27; H, 5.86; N, 7.87; O, 3.00; found: C, 83.29; H, 5.90; N, 7.88; O, 3.01.
5-[3-(4-Thiophen-2-yl-phenyl)-4,5-dihydro-isoxazol-5-ylmethyl]-5H-dibenzo[b,f]azepine (8j). Yield 87%; pale brown solid; mp 173–175 °C; FT-IR (cm−1): 2958, 1736, 1592, 1483, 1458, 1434, 1360, 1336, 1298, 1246, 1161; 1H NMR (400 MHz, DMSO-d6): δ 3.25 (dd, J1–2 = 17.2 Hz, J1–1 = 6.8 Hz, 1H, C4–H isoxazoline), 3.37 (dd, J1–2 = 17.2 Hz, J1–1 = 10.8 Hz, 1H, C4–H isoxazoline), 3.76 (dd, J1–2 = 14.0 Hz, J1–1 = 6.4 Hz, 1H, N–CH), 3.97–4.02 (m, 1H, N–CH), 4.69–4.73 (m, 1H, C5–H isoxazoline), 6.73 (d, J = 2.8 Hz, 2H, ArH), 6.98–7.28 (m, 9H, ArH), 7.55–7.56 (m, 4H, ArH), 7.67 (d, J = 8.4 Hz, 2H, ArH); 13C NMR (100 MHz, DMSO-d6): δ 38.1, 54.2, 78.7, 121.3, 124.0, 124.9, 125.9, 126.8, 127.6, 128.7, 129.1, 129.5, 132.2, 132.4, 133.8, 135.4, 142.8, 150.37, 150.70, 156.3; MS: m/z = 434.55 (calculated) m/z = 435.2 [M + H]+ (found); elemental composition calculated for C28H22N2OS: C, 77.39; H, 5.10; N, 6.45; O, 3.68; S, 7.38; found: C, 77.41; H, 5.17; N, 6.47; O, 3.69.
5-[3-(4-Pyridin-4-yl-phenyl)-4,5-dihydro-isoxazol-5-ylmethyl]-5H-dibenzo[b,f]azepine (8k). Yield 86%; yellow solid; mp 108–110 °C; FT-IR (cm−1): 2269, 2246, 2177, 1622, 1085, 901; 1H NMR (400 MHz, DMSO-d6): δ 3.25 (dd, J1–2 = 17.2 Hz, J1–1 = 6.8 Hz, 1H, C4–H isoxazoline), 3.37 (dd, J1–2 = 17.2 Hz, J1–1 = 10.9 Hz, 1H, C4–H isoxazoline), 3.75 (dd, J1–2 = 14.0 Hz, J1–1 = 6.4 Hz, 1H, N–CH), 4.01 (dd, J1–2 = 13.6 Hz, J1–1 = 6.4 Hz, 1H, N–CH), 4.71–4.76 (m, 1H, C5–H isoxazoline), 6.73 (d, J = 2.4 Hz, 2H, ArH), 6.98–7.27 (m, 8H, ArH), 7.68 (d, J = 8.0 Hz, 2H, ArH), 7.83 (d, J = 8.4 Hz, 2H, ArH), 8.64 (s, 2H, ArH); 13C NMR (100 MHz, DMSO-d6): δ 38.1, 54.1, 78.9, 121.3, 124.0, 127.5, 127.6, 129.3, 129.5, 130.6, 132.2, 132.5, 133.9, 138.7, 146.3, 150.3, 150.7, 156.4; MS: m/z = 429.51 (calculated), m/z = 430.1 [M + H]+ (found); elemental composition calculated for C29H23N3O: C, 81.09; H, 5.40; N, 9.78; O, 3.73; found: C, 81.91; H, 5.45; N, 9.79; O, 3.74.
6-Chloro-5-[4-(5-dibenzo[b,f]azepin-5-ylmethyl-4,5-dihydro-isoxazol-3-yl)-phenyl]-1,3-dihydro-indol-2-one (8l). Yield 70%; yellow solid; mp 160–162 °C; FT-IR (cm−1): 3441, 2988, 2251, 2125, 1661, 1052, 1024, 1005; 1H NMR (400 MHz, DMSO-d6): δ 3.29 (dd, J1–2 = 16.8 Hz, J1–1 = 6.4 Hz, 1H, C4–H isoxazoline); 3.40 (dd, J1–2 = 12.0 Hz, J1–1 = 6.8 Hz, 1H, C4–H isoxazoline), 3.45 (s, 2H, CH2), 3.78 (dd, J1–2 = 13.2 Hz, J1–1 = 6.0 Hz, 1H, N–CH), 4.04 (dd, J1–2 = 14.0 Hz, J1–1 = 6.4 Hz, 1H, N–CH), 4.74–4.78 (m, 4H, C5–H isoxazoline), 6.77 (d, J = 1.6 Hz, 2H, ArH), 6.94 (s, 1H, ArH), 7.02–7.32 (m, 9H, ArH), 7.41–7.44 (m, 2H, ArH), 7.62–7.64 (m, 2H, ArH), 10.58 (s, 1H, NH); 13C NMR (100 MHz, DMSO-d6): δ 35.82, 38.25, 54.20, 78.6, 110.4, 121.3, 124.0, 126.0, 126.7, 127.3, 127.4, 128.7, 129.5, 130.1, 130.2, 132.1, 132.2, 132.4, 133.9, 140.9, 145.0, 150.3, 150.7, 156.5, 176.7; MS: m/z = 518.00 (calculated), m/z = 520.2 [M + 2H]+ (found); elemental composition calculated for C32H24ClN3O2: C, 74.20; H, 4.67; Cl, 6.84; N, 8.11; O, 6.18; found: C, 74.23; H, 4.72; N, 8.13; O, 6.19.
Assessment of antimalarial activity
P. falciparum parasites were cultured according to the method of Trager and Jensen.40 Briefly, 3D7, D6, W2 and 7G8 P. falciparum strains were cultured in RPMI 1640 medium (Gibco) supplemented with 25 mM HEPES, 29 mM sodium bicarbonate, 0.005% hypoxanthine, p-aminobenzoic acid (2 mg L−1), gentamycin sulfate (50 mg L−1) and 5% AlbuMAX II (Invitrogen) using fresh O-positive human red blood cells at 2% hematocrit. The cultures were maintained at 37 °C under 5% O2, 5% CO2, and 90% N2. The antimalarial activity of isoxazoline compounds were determined by a fluorometric method using SYBR Green I.41,42 Briefly, stock solutions of isoxazoline were prepared in DMSO (10 mM) and were serially diluted with complete culture medium to give concentrations ranging from 0.2 to 200 μM and the final concentration of DMSO was less than 0.1%. 100 μL of each testing solution was mixed with 100 μL of 0.2% parasitized red blood cells in the ring stage at 2% hematocrit in complete medium and dispensed into 96-well plates. Untreated- and DMSO (0.1%) vehicle-treated parasites were used as controls. The plates were incubated at 37 °C for 72 h and the experiment was performed in triplicate. To each well, 100 μL of lysis buffer (20 mM Tris–HCl, pH 7.5, 5 mM EDTA, 0.008% saponin, 0.08% Triton X-100) containing 0.2 μL mL−1 of SYBR Green I (Life Technology) was added. The plates were wrapped with aluminium foil and incubated in the dark at room temperature for 1 h. The fluorescence intensity was measured using multi-well plate fluorescence reader at an excitation and emission wavelengths of 485 and 535 nm, respectively. The values were expressed as relative fluorescence units. The IC50 values (the effective concentrations that inhibit parasite growth by 50%) of three independent experiments were plotted using nonlinear regression (Sigmoidal dose response) equation (GraphPad Prism version 4.01, GraphPad Software, La Jolla, CA).Cytotoxicity assay
The toxicity of 3,5-disubstituted isoxazoline derivatives against human cells was assessed by the MTS assay43,44 using human embryonic kidney cells (HEK 293 cell line) and human adenocarcinoma epithelial cells (A549 cell line). Briefly, 5 × 103 cells per well in 100 μL of DMEM medium supplemented with 10% bovine fetal serum were plated in 96-well plates. After 24 h, the stock solutions of 3,5-disubstituted isoxazoline in DMSO that were serially diluted to obtain concentrations ranging from 0.5 μM to 200 μM in 100 μL complete medium was added to each well and incubated at 37 °C for 72 h. The cells treated with DMSO (1%) alone were used as vehicle controls. To each well added 20 μL of MTS/PMS reagent (Promega, Madison, WI), incubated for 3 h at 37 °C and the absorbance measured at 490 nm. The assay was performed in triplicate and the values from three independent experiments were used to calculate the IC50 values (GraphPad Prism version 4.01).Molecular modelling methods
The software used for all computational calculations is Schrödinger Software Suite 2015-2 with hardware 2x Intel Xeon 1.9 GHz E5-2420/6C/15 MB Cache RAM 6 × 4 Gb DDR-3 1333 MHz ECC RDIMM 4 × 500 Gb Graphics Card NvidiaQuadro 600 machine.The docking studies of 3,5-disubstituted isoxazoline tethered dibenzazepine derivatives with PfA-M1 were performed by using Schrödinger Glide.45 The structures were drawn using 2D sketcher of Schrödinger maestro 10.2. The ligands were optimised for the docking using LigPrep46,47 where the structures were converted to 3D and energy optimised using OPLS2005. Ionisation states were generated for all molecules at pH range 7.0 ± 2 using Epik module, in LigPrep with all other default options. 3D structure of inhibitor bound PfA-M1 was retrieved in form from protein data bank35 (PDB ID: 4X2U). The downloaded protein structure was refined using protein preparation wizard where in missing hydrogen atoms, amino acid side chains and missing residues were added and bond order were set. The unnecessary hetero atoms and water molecules were deleted except the Zn2+ and co-crystallised inhibitor. The structure was optimized to create H-bond network and finally minimized to remove any steric clashes using OPLS2015 force field.48 The fully optimized protein structure was used for the receptor grid generation using Glide software.49 The grid was generated by choosing the centroid of bound ligand as the centroid of the grid. The prepared ligands were docked to protein grid using extra precession (XP) mode. The protein–ligand complex was further analysed and visualized using Maestro.
Molecular dynamics simulation analysis
The molecule (6k) that exhibited excellent antimalarial activity and low toxicity to human cells was assessed by molecular dynamics analysis to determine the binding stability and the binding pattern. The protein complex docked with the inhibitor was submitted for 10 ns simulation. The molecular dynamics simulation was run using Desmond.50 The orthorhombic simulation box was prepared using the system builder panel of Desmond with TIP3P explicit water model so that the minimum distance between the protein surface and the solvent surface is 10 Å. The counter ions were added to neutralise the system and 0.15 M NaCl was used to provide the iso-osmotic salt environment. The system was minimized with 2000 iterations and with convergence criteria of 1 kcal mol−1 Å−1. The minimised system was subjected to the 10 ns molecular dynamic simulation using NPT ensemble at temperature of 300 K and atmospheric pressure (1.013 bars) with the default setting of relaxation before simulation. The Nose–Hoover chain thermostat and Matrtyna–Tobias–Klein barostate was used to maintain the temperature and pressure, respectively. The time step of 2 fs was considered during the simulation and every 10 ps, the energy and structure was recorded and saved in the trajectory. From the trajectory generated, the simulation interaction diagram was generated to analyse the results.Conclusions
In summary, we have synthesized dibenzazepine tethered 3,5-disubstituted isoxazolines, and evaluated their antimalarial activity against P. falciparum drug-sensitive 3D7 strains and toxicity to human lung cancer A549 and kidney HEK 293 cells. Several compounds that showed low cytotoxic activity effectively inhibited the growth of parasite. Compounds (6j, 6k, 8c, 8k and 8l) that showed excellent antimalarial activity in initial screening were further assessed using an additional chloroquine-sensitive parasite strain (D6) and two chloroquine-resistant parasite strains (W2 and 7G8). Molecular docking studies revealed the mode of the dibenzazepine derivatives binding to PfA-M1. The molecular dynamics simulation confirmed the stability of the binding mode of the ligand 6k predicted by docking studies. The co-ordination bond formed between the isoxazoline moiety of the ligand and the enzyme-bound Zn2+ ion, π–π interaction between dibenzazepine and His496, and π–cation interactions between piperidine moiety and Tyr575, and hydrophobic interactions with residues Val493, Val523, Tyr580, Tyr575, Val459, Met462, Met1034, Ala320 stabilized the dibenzazepine binding to PfA-M1. Our experimental and theoretical finding data show that, several compounds possess excellent antimalarial activity with minimal or no cytotoxicity. Thus, overall our results suggest that the dibenzazepine tethered 3,5-disubstituted isoxazolines are promising candidates for the development of antimalarial drugs.Acknowledgements
KSV would like to thank UGC-BSR for financial support and IOE, University of Mysore, Karnataka for NMR, mass spectrometry facility. MPS thank UGC-SAP-DRS-Phase III, New Delhi and VGST-SMYSR, Bangalore, Karnataka for funding. ACV would like to thank CSIR-SRF, New Delhi for financial support. All authors acknowledge Department of Biochemistry and Molecular Biology, Penn State University College of Medicine, Hershey Medical Centre, Hershey, PA, for providing facility for assessing antimalarial activity, and Schrödinger, for providing evaluation licence.References
- World Health Organization, World Malaria Report, 2014 Search PubMed.
- J. F. Garcia-Bustos and F.-J. Gamo, Curr. Pharm. Des., 2015, 19, 270 Search PubMed.
- D. Chaturvedi, A. Goswami, P. P. Saikia, N. C. Barua and P. G. Rao, Chem. Soc. Rev., 2010, 39, 435 RSC.
- K. Banek, M. Lalani, S. G. Staedke and D. Chandramohan, Malar. J., 2014, 13, 7 CrossRef PubMed.
- A. P. Phyo, S. Nkhoma, K. Stepniewska, E. A. Ashley, S. Nair, R. McGready, C. L. Moo, S. Al-Saai, A. M. Dondorp, K. M. Lwin, P. Singhasivanon, N. P. J. Day, N. J. White, T. J. C. Anderson and F. Nosten, Lancet, 2012, 379, 1960 CrossRef.
- A. M. Dondorp, F. Nosten, P. Yi, D. Das, A. P. Phyo, J. Tarning, H. M. Lwin, F. Ariey, W. Hanpithakpong, S. J. Lee, P. Ringwald, K. Silamut, M. Imwong, K. Chotivanich, P. Lim, T. Herdman, S. S. A. S. Yeung, P. Singhasivanon, N. P. J. Day, N. Lindegardh, D. Socheat and N. J. White, N. Engl. J. Med., 2009, 361, 455 CrossRef CAS PubMed.
- A. C. Vinayaka, M. P. Sadashiva, X. Wu, S. S. Biryukov, J. A. Stoute, K. S. Rangappa and D. C. Gowda, Org. Biomol. Chem., 2014, 12, 8555 CAS.
- M. P. Sdashiva, R. Gowda, X. Wu, G. S. Inamdar, O. F. Kuzu, K. S. Rangappa, G. P. Robertson and D. Channe Gowda, Exp. Parasitol., 2015, 155, 68 CrossRef PubMed.
- L. H. Bannister, Proc. Natl. Acad. Sci. U. S. A., 2001, 98, 383 CrossRef CAS PubMed.
- M. Schlitzer, ChemMedChem, 2007, 2, 944 CrossRef CAS PubMed.
- J. H. Mckerrow, E. Sun, P. J. Rosenthal and J. Bouvier, Annu. Rev. Microbiol., 1993, 47, 821 CrossRef CAS PubMed.
- J. Liu, E. S. Istvan, I. Y. Gluzman, J. Gross and D. E. Goldberg, Proc. Natl. Acad. Sci. U. S. A., 2006, 103, 8840 CrossRef CAS PubMed.
- M. Allary, J. Schrevel and I. Florent, Parasitology, 2002, 125, 1 CrossRef CAS PubMed.
- C. S. Gavigan, J. P. Dalton and a. Bell, Mol. Biochem. Parasitol., 2001, 117, 37 CrossRef CAS PubMed.
- K. A. Kolakovich, I. Y. Gluzman, K. L. Duffin and D. E. Goldberg, Mol. Biochem. Parasitol., 1997, 87, 123 CrossRef CAS PubMed.
- M. J. Blackman, Curr. Drug Targets, 2000, 1, 59 CrossRef CAS PubMed.
- P. J. Rosenthal, Emerging Infect. Dis., 1998, 4, 49 CrossRef CAS PubMed.
- S. McGowan, C. J. Porter, J. Lowther, C. M. Stack, S. J. Golding, T. S. Skinner-Adams, K. R. Trenholme, F. Teuscher, S. M. Donnelly, J. Grembecka, A. Mucha, P. Kafarski, R. Degori, A. M. Buckle, D. L. Gardiner, J. C. Whisstock and J. P. Dalton, Proc. Natl. Acad. Sci. U. S. A., 2009, 106, 2537 CrossRef CAS PubMed.
- P. M. Jones, M. W. Robinson, J. P. Dalton and A. M. George, PLoS One, 2011, 6, 1 CrossRef.
- G. K. Rao, R. Kaur and P. N. S. Pai, J. Chem. Pharm. Res., 2010, 2, 489 CAS.
- V. K. Honnaiah, R. R. Ambati, V. Sadineni and N. Naik, J. Phys. Sci., 2010, 21, 79 CAS.
- H. V. Kumar and N. Naik, Eur. J. Med. Chem., 2010, 45, 2 CrossRef CAS PubMed.
- M. P. Sadashiva, S. Basappa, S. Nanjundaswamy, F. Li, K. A. Manu, M. Sengottuvelan, D. S. Prasanna, N. C. Anilkumar, G. Sethi, K. Sugahara and K. S. Rangappa, BMC Chem. Biol., 2012, 12, 5 CrossRef PubMed.
- M. Mazza, G. Della Marca, M. Di Nicola, G. Martinotti, G. Pozzi, L. Janiri, P. Bria and S. Mazza, Epilepsy Behav., 2007, 10, 397 CrossRef PubMed.
- World Health Organization, WHO Drug Information, 1994, vol. 8, p. 189 Search PubMed.
- A. F. Ambrósio, P. Soares-da-silva, C. M. Carvalho and A. P. Carvalho, Neurochem. Res., 2002, 27, 121 CrossRef.
- D. A. Learmonth, J. Benes, A. Parada, D. Hainzl, A. Beliaev, M. J. Bonifácio, P. M. Matias, M. A. Carrondo, J. Garrett and P. Soares-da-Silva, Eur. J. Med. Chem., 2001, 36, 227 CrossRef CAS PubMed.
- W. Sun, T. Q. Tanaka, C. T. Magle, W. Huang, N. Southall, R. Huang, S. J. Dehdashti, J. C. McKew, K. C. Williamson and W. Zheng, Sci. Rep., 2014, 4, 3743 Search PubMed.
- F. Albani, R. Riva and A. Baruzzi, Pharmacopsychiatry, 1995, 28, 235 CrossRef CAS PubMed.
- W. E. Müller, B. Siebert, G. Holoubek and C. Gentsch, Pharmacopsychiatry, 2004, 37, S189 CrossRef PubMed.
- W. Sun, T. Q. Tanaka, C. T. Magle, W. Huang, N. Southall, R. Huang, S. J. Dehdashti, J. C. McKew, K. C. Williamson and W. Zheng, Sci. Rep., 2014, 4, 3743 Search PubMed.
- J. Guan, D. E. Kyle, L. Gerena, Q. Zhang, W. K. Milhous and A. J. Lin, J. Med. Chem., 2002, 45, 2741 CrossRef CAS PubMed.
- A. Lagunin, A. Stepanchikova, D. Filimonov and V. Poroikov, Bioinformatics, 2000, 16, 747 CrossRef CAS PubMed.
- A. Lagunin, D. Filimonov and V. Poroikov, Curr. Pharm. Des., 2010, 16, 1703 CrossRef CAS PubMed.
- N. Drinkwater, R. S. Bamert, K. K. Sivaraman, A. Paiardini and S. McGowan, Proteins: Struct., Funct., Bioinf., 2015, 83, 789 CrossRef CAS PubMed.
- K. Karthikeyan, T. V. Seelan, K. G. Lalitha and P. T. Perumal, Bioorg. Med. Chem. Lett., 2009, 19, 3370 CrossRef CAS PubMed.
- T. Tsuji and M. Shirai, WO-2014/034898–A1, European Pat., 2014.
- http://www.ccdc.ac.uk/conts/retrieving.html.
- M. Kontoyianni, L. M. McClellan and G. N. Sokol, J. Med. Chem., 2004, 47, 558 CrossRef CAS PubMed.
- W. Trager and J. B. Jensen, Science, 1976, 193, 673 CAS.
- M. Smilkstein, N. Sriwilaijaroen, J. X. Kelly, P. Wilairat and M. Riscoe, Antimicrob. Agents Chemother., 2004, 48, 1803 CrossRef CAS PubMed.
- J. D. Johnson, R. A. Dennull, L. Gerena, M. Lopez-Sanchez, N. E. Roncal and N. C. Waters, Antimicrob. Agents Chemother., 2007, 51, 1926 CrossRef CAS PubMed; A. Barltrop, T. C. Owen, A. N. I. H. Cory and J. G. Cory, Bioorg. Med. Chem. Lett., 1991, 1, 611 CrossRef.
- J. A. Barltrop, T. C. Owen, A. N. I. H. Cory and J. G. Cory, Bioorg. Med. Chem. Lett., 1991, 1, 611 CrossRef CAS.
- A. H. Cory, T. C. Owen, J. A. Barltrop and J. G. Cory, Cancer Commun., 1991, 3, 207 CAS.
- Glide, version 6.7, Schrödinger, LLC, New York, NY, 2015 Search PubMed.
- LigPrep, version 2.3, Schrödinger, LLC, New York, NY, 2009 Search PubMed.
- G. Madhavi Sastry, M. Adzhigirey, T. Day, R. Annabhimoju and W. Sherman, J. Comput.-Aided Mol. Des., 2013, 27, 221–234 CrossRef CAS PubMed.
- Wizard 2015-2; Epik version 2.4, Schrödinger, LLC, New York, NY, 2015 Search PubMed; Impact version 5.9, Schrödinger, LLC, New York, NY, 2015 Search PubMed; Prime version 3.2, Schrödinger, LLC, New York, NY, 2015 Search PubMed.
- Glide, version 6.1, Schrödinger, LLC, New York, NY, 2013 Search PubMed.
- Schrödinger Release2015-1, Desmond Molecular Dynamics System, version, 4.1, D.E. Shaw Research, New York, NY, 2015 Search PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. CCDC 1049585. For ESI and crystallographic data in CIF or other electronic format see DOI: 10.1039/c5ra17926b |
| This journal is © The Royal Society of Chemistry 2015 |