Towards comprehension of complex chemical evolution and diversification of terpene and phenylpropanoid pathways in Ocimum species
Priyanka Singh
,
Raviraj M. Kalunke
and
Ashok P. Giri
*
Plant Molecular Biology Unit, Division of Biochemical Sciences, CSIR-National Chemical Laboratory, Pune 411008, Maharashtra, India. E-mail: ap.giri@ncl.res.in; Fax: +91-2025902648; Tel: +91-2025902710
First published on 1st December 2015
Abstract
Ocimum species present a wide array of diverse secondary metabolites possessing immense medicinal and economic value. The importance of this genus is undisputable and exemplified in the ancient science of Chinese and Indian (Ayurveda) traditional medicine. Unlike several other plant species of Artemisia, Salvia, Catharanthus, Taxus, Mentha, etc. that are largely exploited, detailed characterization and identification of important metabolites from Ocimum species remained unexplored. Till date, most of the analyzed Ocimum species are predominantly rich in either phenylpropanoids or terpenoids. Metabolite data suggests domination of a unique set of signature compounds in all species. However, molecular pathways leading to the production, accumulation and metabolism of these compounds are poorly understood. The past few years have witnessed an upsurge in our understanding of the complex and intricately woven secondary metabolic pathways. Such information is generated through systematic analysis and correlation of metabolite profiling with transcriptomics data sets from different Ocimum species. The present review is aimed at integrating our current knowledge to understand the active secondary metabolic pathways, the key players in flux regulation including external stimuli, differential gene expression, transcription factors, microRNAs, enzyme promiscuity, etc. Extensive analysis of available data identifies events that may have contributed to evolve Ocimum species rich with a specific set of metabolites, thus, shedding light on pathway diversification. We believe that a better understanding of the multi-level regulation of intermediates and metabolites will help us harness the inherent diversity of Ocimum species optimally.
1. Introduction
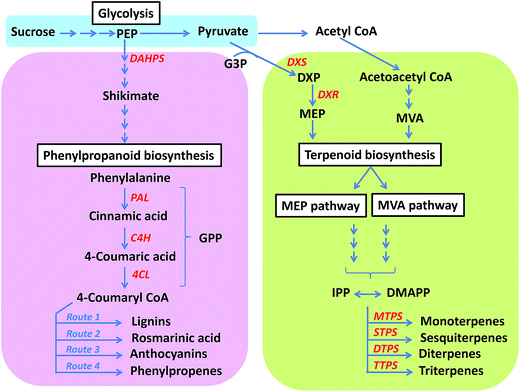
Genus Ocimum belonging to family Lamiaceae comprises between 50 to 150 species.1 The difference in the estimates of species number is partly attributed to reasons like taxonomic revisions and generic description of the genus amongst others. It was first described by Linnaeus in 1753 in the book Species Plantarum.2 The name Ocimum basilicum was derived from the Greek word Okimon (smell) and basilikon (royal), referring to its royal fragrance. While in India the Ocimum plant is considered sacred and worshipped, in other parts of the world it is hailed as the “queen of herbs” because of its strong aromatic appeal and culinary usage. With the establishment of ancient medicinal practises in India (Ayurveda) and China (Traditional Chinese Medicine), Ocimum was recognised as a medicinal herb with great healing powers.Main centres of diversity for Ocimum include tropical and subtropical regions of Africa, India and South America.3 With the exception of O. tenuiflorum and O. gratissimum that are indigenous to India, most species are native to Africa and found in wild population.4 Although Ocimum species are known to abound in medicinally important metabolites, only few species have been thoroughly profiled. Our knowledge about most other species remains limited. All species are identifiable by the presence of a large amount of signature metabolite(s) along with several other metabolites in relatively minute quantities. The diversity of metabolites produced by Ocimum plants is indeed enormous. Specific functions and/or necessity for production of such diverse and complex chemical compounds by the plant remain elusive. Interestingly, what we know is certain Ocimum species are either “terpenoid-rich” or “phenylpropanoid-rich”. However, factors determining the direction of flux are largely unknown. Terpenoids are formed from the mevalonic acid (MVA) pathway in the cytoplasm and the methylerythritol phosphate (MEP) pathway in the plastid.5 Phenylpropanoid pathway starts with the amino acid phenylalanine and eventually results in the formation of phenylpropenes such as eugenol, chavicol, anethole etc., along with intermediates for biosynthesis of lignin, rosmarinic acid, anthocyanins etc. These pathways have been well characterized in related genera including Salvia, Mentha and Lavandula6–12 but not in such details in any Ocimum species. However, with the influx of next generation sequencing data13,14 along with metabolomics, proteomics and phylogeny studies,15–19 now it seems possible to gain a deeper insight into the perplexing diversity. The present review aims at providing a comprehensive overview of the evolutionary, environmental and internal factors that may have resulted in pathway diversification and extensive chemical evolution across Ocimum species.
1.1 Importance of studying genus Ocimum
The unequivocal importance of genus Ocimum was established more than 5000 years back with the advent of ancient traditional medicinal practises in India and China. Thereafter, there have been several reports of important bioactivities of Ocimum species; tissue extracts and metabolites there in (Table 1).20–81 Although most species in this genus are associated with some or the other bioactivity, the exact compound or group of compounds, responsible for the said bioactivity remains elusive in most cases (Table 1). Basil also finds extensive application in the food, flavor, and fragrance industry, and the essential oil serves as a major source of economic wealth to the country. The plant is easy to grow and propagate, and adapts well to extreme environmental conditions including high precipitation, long dry spells and high temperature. Some species are capable of vegetative propagation through stem cuttings like O. kilimandscharicum, which makes commercial cultivation less tedious and more cost effective. Several Ocimum species grow as wild plants in various parts of the world. Since there has been no significant domestication of this wild medicinal plant, its genetic diversity has been preserved in nature, making the system more interesting to explore. Furthermore, presence of different basil types/cultivars rich in diverse metabolites provides a unique system for studying secondary metabolic pathways. In addition, glandular trichomes accord the opportunity to study the biosynthesis and regulation of these pathways at the level of a single cell. Ocimum thus presents an attractive system to explore, particularly from the point of view of secondary metabolism.| sp. | Bioactivity | Extract/compound | Dose and/or activity | Organism/cell line/assay | |
|---|---|---|---|---|---|
| a Ok (O. kilimandscharicum), Olb (O. labiatum), Ola (O. lamiifolium), Oo (O. Obovatum), Ot (O. tenuiflorum), Oa (O. americanum), Oba (O. basilicum), Og (O. gratissimum), GI50 (growth inhibition at 50%), IC50 (half maximum inhibitory concentration), EC50 (half maximal effective concentration), LC50 (median lethal concentration), p.o. (oral administration), MIC (minimum inhibitory concentration), GC-MS (gas chromatography-mass spectrometry), DCM (dichloromethane), TI (therapeutic index), HPLC (high performance liquid chromatography), HPTLC (high performance thin layer chromatography), FTIR (Fourier transform infrared spectroscopy), NMR (nuclear magnetic resonance), UPLC (ultra performance liquid chromatography). | |||||
| Ok | Free radical scavenging20 | Leaf essential oil, camphor, mixture of 1,8-cineole and limonene | Essential oil, GI50 = 8.31 μg mL−1 camphor, IC50 = 12.56 μg mL−1 limonene + 1,8-cineole, IC50 = 23.25 μg mL−1 | DPPH free-radical scavenging assay | GC-MS20 |
| Anticancer20,21 | Leaf essential oil;20 50% alcoholic aqueous leaf extract21 | Essential oil via hydrodistillation, GI50 = 31.90 mg mL−20 50% alcoholic aqueous leaf extract, dose = 200 mg kg−1 p.o.21 | Human ovarian cancer cell line;20 C(57)BL and Swiss albino mice injected intradermally with B10H16 metastatic melanoma cell line21 | GC-MS20 | |
| Anti-inflamatory20 | Leaf essential oil, camphor, mixture of 1,8-cineole and limonene | Reduction in total leucocyte migration = 82 ± 4% (30 mg kg−1 essential oil), 95 ± 4% (100 mg kg−1 of essential oil), 83 ± 9% (camphor), 80 ± 5% (1,8-cineole + limonene) | Carrageenan-induced pleurisy in mice | GC-MS20 | |
| Insecticidal22 | DCM leaf extract, camphor, limonene and β-caryophyllene | Dose = 10, 100 and 1000 ppm incorporated in artificial diet | Helicoverpa armigera (second instar larvae) | GC-MS | |
| Antidiarrhoel23 | Aqueous leaf extract | 100, 200 and 400 mg kg−1, p.o. | Castor-oil induced diarrhoea model, castor oil induced enteropooling assay in rats; charcoal meal/intestinal motility test in mice | Nil | |
| Antimicrobial24 | Essential oil, borneol, bornyl acetate, camphor, caryophyllene oxide, 1,8-cineole, limonene, linalool, α-pinene, β-pinene, spathulenol | MIC, essential oil (1.55–3.35), borneol (1.25–4.50), bornyl acetate (1.75–4.88), camphor (1.33–3.24), caryophyllene oxide (0.073 to >6.4), 1,8-cineole (2.0–9.5), limonene (>20), linalool (0.25 to >20), α-pinene (2.0–15.0), β-pinene (9.75 to >20), spathulenol (1.35 to >20) (avalues in mg mL−1) | S. aureus, S. epidermidis, S. mutans, S. viridans, E. coli, E. cloacae, K. pneumoniae, P. aeruginosa, C. albicans, C. tropicalis, C. glabrata | GC-MS | |
| Antiplasmodial25 | DCM plant extract | Extract, IC50 (CQ resistant clone) = 1.547 ± 0.226 μg mL−1 extract, IC50 (CQ sensitive clone) = 0.843 ± 0.123 μg mL−1 | SYBR green I fluorescence assay (MSF assay) against Plasmodium falciparum (CQ resistant and sensitive clone) | Nil | |
| Antioxidant26,27 | Methanolic extracts of leaves,26,27 and callus26 | Extract, dose = 1 mg mL−1 (ref. 26 and 27) | Ferric reducing antioxidant power (FRAP) assay;26 iron(III) reduction, β-carotene-linoleic acid bleaching, DPPH, superoxide anion free radical scavenging assay27 | HPLC27 | |
| Radioprotective21 | 50% alcoholic aqueous leaf extract | Extract, dose = 200 mg kg−1 p.o. | Mice irradiated by 60Co source in the cobalt therapy unit | Nil | |
| Mosquito repellent28 | Plant essential oil, dry plant material | 20% plant essential oil solution prepared in glycerine and acetone, and burning 1 kg of dry plant material; protection efficiency (PE), essential oil = 89.75% (Anopheles arabiensis) and 90.50% (Culex quinquefasciatus) | Field trials (community study) using A. arabiensis, A. gambiae and C. quinquefasciatus | Nil | |
| Olb | Antioxidant29 | Ethanolic leaf extract, labdane (isolated diterpenoid) | Extract, IC50 = 13 ± 0.8 (DPPH assay), 53.62 ± 0.57 (FRAP assay), 47.32 ± 0.76 (CUPRAC assay) and 54.86 ± 1.28 μg mL−1 (AAPH); labdane diterpenoid demonstrated minimal to no activity | DPPH, FRAP, cupric reducing antioxidant capacity (CUPRAC) and crocin bleaching assay (AAPH) | NMR |
| Anti-inflammatory29 | Ethanolic leaf extract, labdane diterpenoid | Extract, dose = 25 μg mL−1 labdane diterpenoid, dose = 50 μM (ref. 29) | Cytometric bead array (CBA) technique29 | NMR29 | |
| Ola | Antimicrobial24,30,31 | Essential oil extract; ethanolic extract of various plant parts; methanol, aqueous and n-hexane extracts | MIC = 1.75–4.90 mg mL−1;24 MIC (all extracts) <512 μg mL−1;30 MIC = 10–50 mg mL−1 (ref. 31) | S. aureus, S. epidermidis, S. mutans, S. viridans, E. coli, E. cloacae, K. pneumoniae, P. aeruginosa, C. albicans, C. tropicalis, C. glabrata;24 B. cereus, C. perfringens, L. monocytogenes, E. faecalis, S. aureus, S. pyogenes, S. epidermidis;30S. aureus, E. coli, P. aeruginosa, S. boydii | GC-MS24 |
| Mosquito-repellent32 | Volatiles from fresh, dried and smoking dried leaves | β-Ocimene (14%) strongly repelled female mosquitoes | Anopheles arabiensis, Aedes aegypti | GC-MS | |
| Antioxidant27,33 | Plant essential oils and methanolic extracts27 | Methanolic extract, dose = 1 mg mL−1;27 methanolic extract, IC50 = 8.6 ± 0.7 (DPPH assay); 12.8 ± 0.8 (linoleic acid assay);33 essential oil, IC50 = 27.5 ± 1.4 (DPPH assay); 46.1 ± 2.3 (linoleic acid assay) μg mL−1 (ref. 33) | Iron(III) reduction, β-carotene–linoleic acid bleaching, DPPH, superoxide anion free radical scavenging assay;27 DPPH and linoleic acid peroxidation assay33 | HPLC,27 GC-MS33 | |
| Anti-inflammatory34 | Aqueous and ethanolic leaf extracts | 400, 600 and 800 mg kg−1 body weight | Carrageenan, histamine, and serotonin induced mice paw edema | Nil | |
| Hepatoprotective35 | Aqueous and methanolic leaf extracts | 1 and 2 mg mL−1 | Aqueous extracts tested on CCl4-treated guinea pigs by using barbiturate induced sleep modification; methanolic extracts tested in vitro using precision cut liver slices (PCLS) against acetaminophen-induced hepatotoxicity | ||
| Analgesic36 | Aqueous and ethanolic plant extracts | 400, 600 and 800 mg kg−1 body weight | Tail-flick, hot-plate and tail-pinch assay in albino male mice | Nil | |
| Oo | Antimicrobial37 | Leaf essential oil | MIC = 50–200 μg mL−1 | Escherichia coli, Staphylococcus aureus, Klebsiella spp., Pseudomonas spp., Proteus spp. | GC-MS |
| Ot | Antidiabetic38 | 60% ethanolic leaf extract | Extract, dose = 250 and 500 mg kg−1 body weight | Male Wistar albino rats | Nil |
| Anti-hyperlipidemic38 | 60% ethanolic leaf extract | Extract, dose = 250 and 500 mg kg−1 body weight | Male Wistar albino rats | Nil | |
| Anti-oral toxicity effect38 | Hydroalcoholic leaf extract | 5–2000 mg kg−1 body weight | Male Wistar albino rats | Nil | |
| Antioxidant39 | Methanolic extracts of leaf, inflorescence, stem and callus | Extract, dose = 50–1000 μl | DPPH, hydroxyl radicals, hydrogen peroxide, chelating ferrous iron and ferric ion reducing potential assay | Reverse phase HPLC | |
| DNA damage protective40 | Anthocyanin extracts | 5, 10 and 20 μg mL−1 | Oxidative DNA damage induced via H2O2 and UV using pUC19 plasmid | UPLC | |
| Antibacterial41,42 | Essential oil | MIC (essential oil) = 25–100 μg mL−1;41 MIC (essential oil) = 0.364 mg mL−1 (S. aureus), 0.728 mg mL−1 (E. coli) | E. coli, S. enteritidis, S. typhimurium, S. typhi, S. flexneri, B. cereus, S. aureus;41E. coli and S. aureus42 | ||
| Anticancer43 | Aqueous and ethanolic leaf extracts | 50–400 μg mL | Sarcoma-180, HFS-1080 cell lines; Swiss albino Wistar mice | Nil | |
| Antiglycation44 | Methanolic and water extracts and their fractions (DCM, ethyl-acetate, n-butanol, water) | IC50 values for different fractions ranged from 21.01 ± 2.06 to 68.29 ± 1.68 μg mL−1; concentrations tested = 250 to 2000 μg mL−1 | Measuring inhibition of advanced glycation end products by fluorometry | Nil | |
| Antistress45 | OciBest (whole plant extract in gelatin capsules) | 1200 mg of actives per day | Self-evaluation by participants using symptom rating scale after 0, 2, 4, 6 week of trial period | Nil | |
| α-Amylase inhibitory46 | Isopropanol extract | IC50 = 8.9 μg mL−1 | Porcine pancreatic α-amylase (PPA) inhibition assays | GC-MS | |
| Mosquito repellent47 | Plant essential oil | EC50 = 133 ppm; EC90 = 240 ppm | Aedes aegypti | GC-MS | |
| Antiherpes48 | Methanol and DCM extracts | Therapeutic index (TI), DCM extract = 10.003 (after HSV-2G adsorption); TI for methanol extracts = 1.644, 2.473 and 29.395 before, during and after HSV-2G adsorption | African Green Monkey (GMK) cells infected with Herpes Simplex Virus (HSV) | Nil | |
| Ameliorative potential49 | Methanol extracts, saponin-rich fraction | 100 and 200 mg kg−1 p.o. | Wistar albino rats | HPTLC | |
| Oa | Free radical scavenging50 | Ethanol, butanol and ethyl-acetate extracts from leaves | 50–300 μg mL−1 | DPPH-, ABTS-, hydrogen peroxide-, nitric oxide-, hydroxyl radical-scavenging assay | |
| Anti-inflammatory Activity51 | Essential oil, linalool, 1,8-cineole | 50–300 mg kg−1 body weight | Zymosan-induced arthritis and paw edema in female balb/c mice | GC-MS | |
| Anti-herpes48 | Methanol and DCM extracts | Therapeutic index (TI) for DCM extracts = 1.865, 2.623 and 7.04 before, during and after HSV-2G adsorption; TI for methanol extracts = 2.345 and 27.357, during and after HSV-2G adsorption | African Green Monkey (GMK) cells infected with Herpes Simplex Virus (HSV) | Nil | |
| Antimicrobial52 | Plant essential oil | MIC = 0.04% v/v (for S. mutans, L. casei and C. albicans); MCC = 0.08%, 0.3% and 0.08% v/v (for S. mutans, L. casei and C. albicans resp.) | Streptococcus mutans, Lactobacillus casei, Candida albicans | Nil | |
| Oba | Antiherpes48 | Methanol and DCM extracts | Therapeutic index (TI) for DCM extracts = 1.835 and 1.817, during and after HSV-2G adsorption; TI for methanol extracts = 1.563 and 2.176, during and after HSV-2G adsorption | African Green Monkey (GMK) cells infected with Herpes Simplex Virus (HSV) | Nil |
| Anti-inflammatory53,54 | Whole plants;53 ethanol–water (25%) extract of leaves54 | Abiotic elicitors (aqueous solution), dose = 10−6 M (jasmonic acid), 10−6 M (arachidonic acid), 10−2 M (β-aminobutyric acid) sprayed on 21 day old plants;53 4 mg extract per day for five days54 | Lipoxygenase and cyclooxygenase inhibitory assay in leaves treated with abiotic elicitors;53 Swiss albino mice54 | ||
| Antiplasmodial55 | Plant ethanolic extracts (leaf, stem, root, flower) | IC50 = 43.81–78.69 μg mL−1 | Plasmodium falciparum | Nil | |
| Antioxidant and antimicrobial56–60 | Essential oil extracted via hydrodistillation;56 plant extracts prepared using ethanol, butanol, chloroform, water, ethyl acetate;58 essential oil, linalool, eugenol59 acetone and ethanol extracts60 | DPPH assay, IC50 = 0.03 to >100 μg mL−1, antimicrobial assay, MIC = 0.009–23.48 μg mL−1;56 IC50 = 124.95 μg mL−1 (DPPH assay), 25.69 (μmol Trolox/mg plant material (TEAC assay), 18.84% (HAPX assay);57 IC50 = 8.17–24.91 μg mL−1 (neutralization of DPPH radical), 6.92–25.45 μg mL−1 (neutralization of NO radical), 10.61–17.21 μg mL−1 (neutralization of superoxide radical), 17.93–71.42 μg mL−1 (neutralization of hydrogen peroxide radical);58 MIC = 60–100 μg/0.1 mL (acetone extract), 20–60 μg/0.1 mL (ethanol extract)60 | DPPH assay for antioxidation, B. cereus, M. flavus, S. aureus, E. faecalis, E. coli, P. aeruginosa, S. typhimurium, L. monocytogenes, A. fumigatus, A. niger, A. versicolor, A. ochraceus, P. funiculosum, P. ochrochloron, T. viride tested for antimicrobial activity;56 DPPH, Trolox equivalent antioxidant capacity (TEAC), hemoglobin ascorbate peroxidase activity inhibition (HAPX) and electron paramagnetic resonance (EPR);57 assay for neutralization of DPPH, NO, superoxide and hydrogen peroxide radicals;58 E. coli, E. aerogenes, E. faecalis, L. monocytogenes, P. aeruginosa, S. enterica, S. typhimurium, S. dysenteriae, S. aureus59 E. coli, K. pneumonia, S. aureus, P. aeruginosa and Proteus sp.60 | GC-MS;56,59 HPLC-MS57 | |
| Antimalarial61 | Leaf essential oil | IC50 = 21.0 ± 4.6 μg mL−1 | Plasmodium falciparum | GC-MS | |
| Anticancer62,63 | Plant methanolic extract;62 petroleum ether soluble and insoluble methanolic extracts, ursolic acid | Dose = 20–320 μg mL−1;62 ursolic acid, LC50 = 18.6 μg mL−1 | Cytotoxic activity against MCF-7 cells;62 sulforhodamine B assay using HT-144, MCF-7, NCI–H460, SF-268 cell lines, immuno-fluorescence microscopy for studying effect on cytoskeleton and nuclei of MCF-7 cells | Nil | |
| Larvicidal activity64,65 | Leaf essential oil64 | LC50 = 9.75–14.1 ppm;64 LC50 = 3.734% (first instar larvae),4.154% (second instar larvae), 4.664% (third instar larvae), 5.124% (fourth instar larvae), 5.449% (pupae)65 | Culex tritaeniorhynchus, Aedes albopictus and Anopheles subpictus;64 Aedes aegypti65 | GC-MS64 | |
| Antituberculosis66 | Methanolic extract of leaves, fruits and flowers; bacilicin | Dose = 6.25 μg mL−1, inhibition = 8–49% | Microplate Alamar blue assay (MABA) | ||
| Preventing ischemia, reperfusion-induced cerebral damage and motor dysfunctions67 | Ethyl-acetate extract of leaves | 100 and 200 mg kg−1 p.o. | Swiss albino mice | Nil | |
| Antihypertensive effects68 | Aqueous plant extract | 100, 200 and 400 mg kg−1 per day orally | Two kidney one clip Goldblatt model for renovascular hypertension in Wistar rats | Nil | |
| Vasorelaxant and anti-platelet aggregation69 | Aqueous plant extract | 0.5 g kg−1 body weight for 10 weeks | Female Wistar rats | HPLC | |
| Antigiardial activity70 | Plant essential oil, linalool, eugenol | Dose = 2 mg mL−1 (essential oil), 300 μg mL−1 (linalool), 850 μg mL−1 (eugenol) | Giardia lamblia | GC-MS | |
| Antiviral71 | Aqueous and ethanolic plant extracts, apigenin, linalool, ursolic acid | Ursolic acid, EC50 = 6.6, 4.2, 0.4, 0.5 mg L−1 (against HSV-1, ADV-8, CVB-1 and EV-71 resp.); apigenin, EC50 = 9.7, 11.1, 7.1, 12.8 mg L−1 (against HSV-2, ADV-3, hepatitis B surface antigen, hepatitis B′E′ antigen resp.); linalool, EC50 = 16.9 mg L−1 (against ADV-II) | Herpes viruses (HSV), adenoviruses (ADV), hepatitis B virus, coxsackievirus B1 (CVB1) and enterovirus 71 (EV71) | Nil | |
| Og | Protection of liver from oxidative stress and fibrosis72 | Polyphenol extract | Dose = 0 to 12 mg kg−1 body weight for 8 weeks | CCl4-induced liver fibrosis in Wistar rats | HPLC |
| Antioxidant and antimutagenic73 | Leaf aqueous extract | Antioxidant activity, IC50 = 83.0 μg mL−1 | DPPH assay for antioxidant activity; antimutagenic activity evaluated using Salmonella typhimurium (TA98 and TA100) strains using the Salmonella/microsome test | ||
| Antitrypanosomal and antiplasmodial74 | Crude ethanol extract, essential oil and pure compounds | IC50 (antitrypanosomal activity) = 1.29 to >100 μg mL−1 IC50 (antiplasmodial activity) = 41.92–76.92 μg mL−1 | Trypanosoma brucei brucei, Plasmodium falciparum | GC-MS | |
| Free radical scavenging and antioxidant75 | Aqueous extract, methanol extract and eugenol | EC50 = 242.47–254.33 μg mL−1 (DPPH assay); 10.47–46.33 μg mL−1 (hydroxyl radical scavenging activity); 14.17–37.88 μg mL−1 (nitric oxide scavenging activity) and 50.92–92.26 μg mL−1 (antioxidant activity) | DPPH assay, hydroxyl radical and nitric oxide scavenging assay, ferric thiocyanate (FTC) method, reducing power determination | HPLC, FTIR, NMR | |
| Prevention against liver fibrosis76 | Aqueous leaf extract | Doses = 0 to 40 mg kg−1 body weight) for 8 weeks | CCl4-induced liver fibrosis in Wistar rats | Nil | |
| Antimicrobial77–79 | Plant essential oil;77,78 eugenol, methyl eugenol;77 hexane and methanol extracts alone and in combination with aminoglycosides79 | MIC = 0.18–3.75 mg mL−1;77 dose, essential oil = 10, 50, 100 mg mL−1;78 | S. aureus, S. epidermidis, S. faecalis, M. flavus, M. luteus, B. subtilis, E. coli, K. pneumonia, S. marcescens, P. vulgaris, P. mirabilis, P. aeruginosa, S. typhimurium, E. aerogenes, A. niger, A. fumigatus, P. chrysogenum;77B. cereus, S. flexneri, C. albicans;78 E. coli and S. aureus (clinical and standard strains)79 | GC-MS | |
| Corrosion inhibition80 | Seed extract | Dose, extract = 4–10% (v/v) | Gravimetric methods | Nil | |
| Cerebroprotection81 | Ethanolic plant extract | 150 or 300 mg kg−1 body weight p.o. | Male Wistar rats | HPLC | |
1.2 Overview of extensive diversity within genus Ocimum
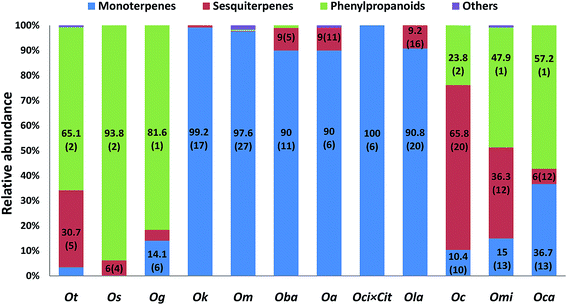
Although genus Ocimum boasts of 50–150 species, metabolite data for very few species is available. For example, O. obovatum and O. labiatum are well tested for therapeutic properties (Table 1); however, their chemical composition remains unknown. Ocimum species abound in diverse secondary metabolites including terpenoids, phenylpropanoids, rosmarinic acid, flavonoids and phenolics. Fig. 1 shows representative examples of structurally diverse classes of secondary metabolites found across genus Ocimum. These mainly include monoterpenes (example, camphor, eucalyptol, α-pinene, β-ocimene, terpinolene), sesquiterpenes (example, farnesene, β-caryophyllene, germacrene D) and phenylpropanoids (example, eugenol, eugenol methyl ether, chavicol, methyl chavicol, methyl cinnamate). Few metabolites like germacrene D and β-caryophyllene are commonly found across most species in the genus; however, others like camphor and eugenol have a specie-specific distribution (Table 2). Higher terpenes (C20 and above) and alkaloids have not been well characterized from any Ocimum species. Since most Ocimum species have not been profiled for their metabolites, the possibility that the genus represents much more diversity than what we perceive now is realistic. As mentioned previously, each species is characterized by a distinct metabolic fingerprint and presence of a signature compound(s) as the major fraction. Although metabolite profiling via conventional techniques such as gas chromatography (GC) has been routinely employed, advanced analytical techniques including liquid chromatography (LC), mass spectrometry (MS) and nuclear magnetic resonance (NMR) have not been reported, which help in gaining a better understanding of the global distribution of metabolites and pathway intermediates. Till now only 12 Ocimum species have been analysed for their essential oil composition (Fig. 2, Table 2). Overall, they can be classified as having (i) high phenylpropanoid content, (ii) high terpenoid content, and (iii) similar/comparable amounts of phenylpropanoids and terpenoids. High phenylpropanoid content group contains about 60 to 90% phenylpropanoids and includes O. gratissimum,82 O. tenuiflorum83 and O. selloi.84 High terpenoid containing group, includes O kilimandscharicum,85 O. minimum,86 O. basilicum,87 O. americanum,88 Ocimum × citriodorum89 and O. lamiifolium90 contain approximately 40 to 75% terpenoids. The third group includes O. campechianum,91 O. micranthum92 and O. canum93 which show approximately equal amount of phenylpropanoids and terpenoids. Interestingly, terpenoids unlike phenylpropanoids, show a universal presence in varying amount in all Ocimum species. Table 2 gives a comprehensive list of species-wise metabolite distribution.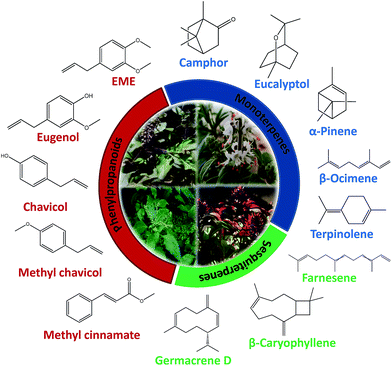 | ||
| Fig. 1 Representative examples of structurally diverse classes of secondary metabolites viz. monoterpenes, sesquiterpenes and phenylpropanoids found across genus Ocimum. | ||
| spp. | Chemical composition | ||
|---|---|---|---|
| Monoterpene (%) | Sesquiterpene (%) | Phenylpropanoids (%) | |
| a Ot (O. tenuiflorum), Os (O. selloi), Og (O. gratissimum), Ok (O. kilimandscharicum), Om (O. minimum), Oba (O. basilicum), Oa (O. americanum), Oci × cit (Ocimum × citriodorum), Ola (O. lamiifolium), Oc (O. campechianum), Omi (O. micranthum, Oca (O. canum) (parenthesis values indicate relative percentage of respective metabolite). | |||
| Ot83 | Camphene (0.79), borneol (2.74) | Germacrene (4.9), β-elemene (1.59), β-caryophyllene (8.7), farnesol (4.19), fanesene (11.27) | Methyl eugenol (62.29), eugenol (2.79) |
| Os84 | Nil | β-Caryophyllene (2.04), germacrene-D (1.3), bicyclogermacrene (1.2), pathulenol (1.30) | Methyl chavicol (93.2), methyl eugenol (0.6) |
| Og82 | Sabinene (0.31), myrcene (0.30), (Z)-ocimene (11.88), (E)-ocimene (0.77), trans-4-thujanol (0.44), terpinen-4-ol (0.44) | Copaene (0.29), bourbonene (0.43), (E)-caryophyllene (0.88), germacrene-D (2.23), cadinene (0.24), caryophyllene oxide (0.21) | Eugenol (82) |
| Ok85 | α-Pinene (1.23), camphene (7.32), β-myrcene (1.58), L-phellandrene (0.26), α-terpinene (0.33), p-cymene (0.62), limonene (13.56), 1,8-cineole (0.85), β-ocimene (2.00), γ-terpinene (0.88), trans-sabinene hydrate (0.49), α-terpinolene (1.33), linalool (1.7), cis-sabinene hydrate (0.47), camphor (56.07), 4-terpineol (3.5), myrtenol (1.24) | Trans-caryophyllene (0.33), germacrene D (0.43) | Nil |
| Om86 | α-Thujene (0.002), α-pinene (0.004), sabinene (0.01), β-pinene (0.003), myrcene (0.013), α-terpinene (0.003), limonene (0.002), β-phellandrene (0.003), eucalyptol (0.01), trans-β-ocimene (0.006), γ-terpinene (0.13), cis-linalool oxide (0.01), trans-linalool oxide (0.03), neo-allo-ocimene (0.013), plinol (0.022), terpinen-4-ol (2.352), α-terpineol (0.022), n-octyl acetate (0.007), nerol (0.034), linalyl acetate (0.194), geraniol (0.07), geranyl acetate (69.48), bornyl acetate (0.095), myrtenyl formate (0.03), carvacrol (0.043), exo-2-hydroxycineole acetate (0.018) | α-Copaene (0.028), α-cubebene (0.12) | Eugenol (0.126), chavicol (0.224) |
| Oba87 | α-Pinene (1.9), sabinene (1.9), β-pinene (3.3), β-myrcene (3.0), D-limonene (1.6), 1,8-cineole (22.6), β-phellandrene (0.1), β-cis-ocimene (0.5), β-linalool (47.6), camphor (0.8), α-terpineol (1.2) | α-Ylangene (0.76), β-cubebene (0.16), α-cis-bergamotene (0.76), α-trans-bergamotene (4.8), cis-muurola-4(14), 5-diene (2) | Eugenol (0.9) |
| Oa88 | Linalyl alcohol (2.03), β-citral (36.58), citral (47.18), nerol acetate (0.32) | Caryophyllene (1.05), α-bergamotene (0.94), humulene (0.51), germacrene D (1.4), α-selinene (0.37), bicyclogermacrene (0.92), β-elemene (0.24), α-bulnesene (0.46), cis-α-bisabolene (2.67), (−)-spathulenol (0.31), β-selinenol (0.19) | Nil |
| Oci × cit89 | Nerol (5.04), neral (33.0), geraniol (1.6), geranial (43.0) | Nil | Nil |
| Ola90 | α-Thujene (0.3), α-pinene (0.6), sabinene (33.8), β-pinene (2.2), myrcene (2.8), α-phellandrene (0.5), α-terpinene (2.0), p-cymene (2.2), β-phellandrene (4.0), limonene (1.0), (Z)-β-ocimene (17.2), (E)-β-ocimene (3.0), γ-terpinene (1.6), terpinolene (1.9), linalool (2.0), cis-p-menth-2-en-1-ol (0.5), trans-p-menth-2-en-1-ol (0.3), terpinen-4-ol (8.4), p-cymen-8-ol (0.8), α-terpineol (0.2) | α-Copaene (0.2), β-bourbonene (0.3), β-caryophyllene (5.6), α-humulene (0.1), (E)-β-farnesene (1.0), germacrene-D (1.1), γ-cadinene (0.1), δ-cadinene (0.2) | Nil |
| Oc91 | Camphene (0.4), α-pinene (0.2), sabinene (0.1), β-pinene (0.8), limonene (0.3), β-myrcene (0.2), 1.8-cineole (3.3), linalool (2.9), borneol (0.5), α-terpineol (0.3) | β-Bourbonene (9.5), α-copaene (1.9), trans-β-caryophyllene (7.8), α-guaiene (5.6), cis-β-farnesene (0.5), sesquisabinene (0.2), α-humulene (2.8), γ-muurolene (0.3), germacrene D (10.1), β-selinene (1.4), bicyclogermacrene (3.4), α-bulnesene (7.1), γ-cadinene (0.5), δ-cadinene (2.0), β-sesquiphellandrene (0.4), caryophyllene oxide (0.4), spathulenol (0.4), epi-α-muurolol (2.0), T-muurolol (0.7), 11-selinen-4-α-ol (1.1) | Eugenol (9.0), methyl eugenol (12) |
| Omi92 | R-Pinene (0.24), camphene (0.07), α-pinene (0.75), myrcene (0.26), 1,8-cineole, (5.35), cis-ocimene (2.69), trans-ocimene (0.35), linalool (1.49), allo-ocimene (2.42), borneol (0.14), mentha 1,5-dien-8-ol (0.33), R-terpineol (0.45), neral (0.06) | Elemene isomer (0.63), α-elemene (4.17), elemene isomer (0.63), α-elemene (9.06), α-caryophyllene (11.94), R-bergamotene (0.13), R-humulene (2.4), germacrene (0.13), α-selinene (0.86), bicyclogermacrene (2.9), spathulenol (1.15), caryophyllene oxide (1.23) | Eugenol (46.55) |
| Oca93 | R-Pinene (0.2), β-pinene (0.32), β-myrcene (0.18), 1,8-cineole (4.98), γ-terpinene (0.21), linalool (26.37), camphor (0.41), borneol (0.17), trans-β-terpineol (0.17), terpinen-4-ol (1.64), endo fenchyl acetate (0.21), bornyl acetate (0.64) | β-Elemene (0.29), trans-caryophyllene (0.5), trans-α-bergamotene (1.7), germacrene (0.52), bicyclogermacrene (0.6), γ-cadinene (0.38), cedrol (0.22), cadinol (1.18) | Methyl chavicol (52.71), eugenol (2.6) |
Signature compounds known in Ocimum species are as follows: camphor in O. kilimandscharicum (56%), citral in O. americanum (47%), eugenol in O. gratissimum (82%) and O. micranthum (47%), eugenol methyl ether in O. tenuiflorum (62%), linalool in O. basilicum (48%), methyl chavicol in O. selloi (93%) and O. canum (53%), geranyl acetate in O. minimum (70%), sabinene in O. lamiifolium (33%) and geranial in Ocimum × citriodorum (43%) (Fig. 2, Table 2). In plant kingdom, metabolite diversity is commonly found at the level of family or genus, but such vivid diversity at the level of species and subtypes (within species) makes genus Ocimum occupy a special niche in nature.
2. Potential evolutionary events influencing metabolite diversity via pathway modulation
Ocimum genome has evolved as a result of dramatic series of events including polyploidy, aneuploidy, chromosomal duplications/translocations/deletions etc.,16,94,95 which led to unprecedented diversification of species in Africa, India and South America. The ability to cross-pollinate and hybridize further led to the emergence of subtypes within species, which were capable of interbreeding and producing hybrids. For example, Ocimum × citriodorum is a hybrid between O. americanum and O. basilicum and has a strong lemony scent.18 O. americanum originated from O. canum and O. basilicum.96 The African blue basil subtype (O. kilimandscharicum) is evolved as a hybrid between O. kilimandscharicum and O. basilicum and abounds in camphor, linalool and eucalyptol. Interestingly, the hybrids display significantly different metabolite profile than their parents including new metabolites that are not found in the parents, indicating co-dominance, epistasis or interaction of genes.89 As reported in several other plant genera, ploidy levels also affect essential oil production, resulting in a greater accumulation of essential oils in polyploids than that in diploids.95,97–99 All these events taken together might have led to greater genetic diversity and continuous expansion of gene pool, yielding new species/subtypes/varieties over a short period.During the course of evolution, there may have been events that led to terpenoid and phenylpropanoid pathway diversification across different Ocimum species. It is interesting to note that species abounding in phenylpropanoids also have an active terpenoid pathway and vice versa. This suggests that all species evolved from an ancestor, which harbored active genes for both the pathways. However, differential expression and regulation of pathway genes determined the final chemical composition in each species.5,100 Other factors like plant habit may also have influenced the selection of one pathway over the other. For example, it has been suggested that the sanctum group has evolved to produce phenolic compounds because of its perennial woody habit, whereas the basilicum group has evolved to produce terpenoid-rich compounds owing to its annual herbaceous habit.4 Evolution of gene coding regions also had a profound impact on the diversity of Ocimum species metabolites. For example, O. basilicum fenchol synthase and myrcene synthase, and geraniol synthase and linalool synthase are 95% and 81% similar, respectively; however, they catalyse the formation of very different products. These genes most probably evolved as a result of gene duplication events and acquired mutations leading to functional differentiation,100 eventually contributing to metabolite diversity. Few pathway genes involved in the biosynthesis of selected metabolites have been reported and characterized from Ocimum and few other genera (Table 3). Genes like eugenol synthase involved in catalysing the final step of eugenol production has been well characterized (Table 3). However, most genes present upstream in the eugenol biosynthetic pathway remain functionally uncharacterized despite availability of huge transcriptomic databases. Genes from camphor biosynthesis pathway have been well characterized from related genera like Salvia and Lavandula, however, there are no reports from genus Ocimum (Table 3). Modifying enzymes like chavicol and eugenol O-methyltransferases also have been well characterized (Table 3). Information about transcription factors responsible for controlling biosynthesis of these metabolites and the transporter proteins responsible for long distance transport from source to sink tissue in Ocimum species also remains scarce. Genes reported from yet another important category of compounds, flavones and flavonoids, have been listed in Table 3. Overall, information about the biosynthesis, transport and storage of these metabolites, at the genetic level is very scarce and need to be further probed. Several other factors during species diversification and naturalization in other parts of the world have been discussed briefly, which help us in explaining the mystery behind the complex chemical evolution and pathway diversification.
| Compound | Gene | Reaction catalysed | Method of characterization |
|---|---|---|---|
| Eugenol | Eugenol synthase (EGS) (Ocimum basilicum)101 | Coniferyl acetate to eugenol | Three dimensional structure determination viz. protein X-ray crystallography and in vitro mutagenesis studies suggest that reaction proceeds via formation of quinone-methide intermediate followed by reduction; involving conserved residue Lys-132 |
| Eugenol synthase (EGS) (Fragaria ananassa)102 | Cloning, functional characterization and expression of FaEGS1a and FaEGS1b (catalysing formation of eugenol); and FaEGS2 (catalysing formation of eugenol and also isoeugenol with lower catalytic efficiency) | ||
| Coumaryl CoA ligase (4CL) (Ocimum tenuiflorum)103 | Hydroxycinnamic acids to coenzyme A (CoA) esters | Transient silencing of 4CL gene leads to reduction in eugenol accumulation, however, lignin and sinapic acid content remained unaffected, indicating involvement of a specific isoform of 4CL in eugenol biosynthesis which is different from those involved in lignin biosynthesis | |
| R2R3-MYB transcription factor (EOBII) (Fragaria ananassa)104 | Transcription factor regulating structural genes in phenylpropanoid pathway | Identification and functional characterization of FaEOBII in strawberry fruit receptacles, responsible for regulating eugenol biosynthesis by interaction with FaMYB10 | |
| Eugenol methyl ether | Eugenol O-methyl transferase (EOMT) (Ocimum basilicum)105,106 | Eugenol to eugenol methyl ether | Recombinant protein expression and characterization in E. coli, molecular modelling based on crystal structure of IOMT and site directed mutagenesis suggested single amino acid difference being responsible for substrate specificity in EOMT and CVOMT |
| Methyl chavicol | Chavicol O-methyl transferase (CVOMT) (Ocimum basilicum)105,106 | Chavicol to methyl chavicol | Linking O-methyltransferase activity with developmental timing and chemotype of O. basilicum through enzyme assays |
| Camphor | Bornyl diphosphate synthase (BPPS) (Salvia officinalis)107–110 | Geranyl diphosphate to bornyl diphosphate | Partial purification and characterization of BPPS from soluble enzyme preparations of young leaves; demonstration of GPP as preferred substrate for cyclization |
| Characterization and functional expression of recombinant BPPS | |||
| X-ray crystal structure determination using multiwave anomalous dispersion (MAD) to 2.0 Å resolution, modelling with substrates, intermediates and mechanistic implications on terpene cyclization | |||
| Molecular dynamics and multidynamic free energy simulations reveal bornyl cation to be an important enzyme induced bifurcation point; electrostatic steering by diphosphate moiety in active site guides the formation of primary product (BPP) | |||
| Borneol dehydrogenase (BDH) (Salvia officinalis)111 | Borneol to camphor | Partial purification and characterization of BDH from soluble enzyme extracts prepared using young leaves | |
| Borneol dehydrogenase (BDH) (Lavandula intermedia)112 | Cloning, functional characterization and determination of tissue-specific expression of LiBDH | ||
| Eucalyptol (1,8-cineole) | 1,8-Cineole synthetase (Salvia officinalis)113 | Neryl diphosphate to 1,8-cineole | Partial purification and characterization of cineole synthetase from soluble enzyme extracts prepared using young leaves |
| Linalool | Linalool synthase (LIS) (O. basilicum)100 | GPP to linalool | Cloning and expression of full length cDNA in E. coli followed by characterization via enzyme assays |
| Terpinolene | Terpinolene synthase (TES) (O. basilicum)100 | GPP to terpinolene (as major product) and α-pinene and limonene (as side products) | Cloning and expression of full length cDNA in E. coli followed by characterization via enzyme assays |
| Fenchol | Fenchol synthase (FES) (O. basilicum)100 | GPP to fenchol (as major product) and α-pinene and limonene (as side products) | Cloning and expression of full length cDNA in E. coli followed by characterization via enzyme assays |
| Myrcene | Myrcene synthase (MES) (O. basilicum)100 | GPP to myrcene | Cloning and expression of full length cDNA in E. coli followed by characterization via enzyme assays |
| Cadinene | Cadinene synthase (CDS) (O. basilicum)100 | FPP to γ-cadinene (as major product) and muurola 3,5-diene (as side product) | Cloning and expression of full length cDNA in E. coli followed by characterization via enzyme assays |
| Selinene | Selinene synthase (SES) (O. basilicum)100 | FPP to α & β-selinene (as major product) and β-elemene and nerolidol (as side product) | Cloning and expression of full length cDNA in E. coli followed by characterization via enzyme assays |
| Zingiberene | Zingiberene synthase (ZIS) (O. basilicum)100 | FPP to α-zingiberene (as major product) and α-bergamotene, nerolidol, β-farnesene and β-bisabolene (as side product) | Cloning and expression of full length cDNA in E. coli followed by characterization via enzyme assays |
| Germacrene-D | Germacrene D synthase (GDS) (O. basilicum)100 | FPP to germacrene D | Cloning and expression of full length cDNA in E. coli followed by characterization via enzyme assays |
| Geraniol | Geraniol synthase (GES) (O. basilicum)114 | GPP to geraniol | Cloning, expression and functional characterization of the enzyme followed by RNA gel-blot analysis revealing exclusive expression of GES in trichomes and not in leaves |
| Amyrin (triterpene) | 2,3-Oxidosqualene cyclase (AS1 and AS2) (O. basilicum)115 | 2,3-Epoxy-2,3-dihydrosqualene to α/β-amyrin | Cloning and expression of ObAS1 and ObAS2 in Saccharomyces cerevisiae strain BY4741 under GAL1 promoter; ObAS1 was identified as α-amyrin synthase, while ObAS2 produced both α-and β-amyrins |
| General phenyl propanoid pathway | Production of anthocyanin pigment 1 (PAP1) (A. thaliana)116 | Transcriptional regulator of floral scent | Introduction of PAP1 transcription factor from A. thaliana into Rosa hybrida (rose) altered the colour and scent profile of transgenic plants resulting from an increase in metabolic flux through terpenoid and phenylpropanoid pathways |
| p-Coumaroyl shikimate 3′-hydroxylase (CS3′H) (O. tenuiflorum)14 | p-Coumaroyl 5-O-shikimate to caffeoyl 5-O-shikimate | de novo sequencing of transcriptome | |
| Caffeic acid O-methyl transferase (COMT) (O. basilicum)14 | Caffeate to ferrulate | de novo sequencing of transcriptome | |
| Caffeic acid O-methyl transferase (COMT) (O. tenuiflorum)14 | Caffeate to ferrulate | de novo sequencing of transcriptome | |
| Cinnamyl alcohol dehydrogenase (CAD) (O. tenuiflorum)14 | Cinnamyl alcohol to cinnamyldehyde | de novo sequencing of transcriptome | |
| Cinnamyl alcohol dehydrogenase (CAD) (O. basilicum)14 | Cinnamyl alcohol to cinnamyldehyde | de novo sequencing of transcriptome | |
| Cinnamate-4-hydroxylase (C4H) (O. tenuiflorum)14 | Cinnamic acid to 4-coumaric acid | de novo sequencing of transcriptome | |
| Cinnamate-4-hydroxylase (C4H) (O. basilicum)14 | Cinnamic acid to 4-coumaric acid | de novo sequencing of transcriptome | |
| Flavonoid pathway | Chalcone synthase (CHS) (O. tenuiflorum)117 | Conversion of 4-coumaroyl-CoA and malonyl-CoA to naringenin chalcone | de novo sequencing of transcriptome |
| Flavone 8-hydroxylase (F8H) (O. basilicum)118,119 | Hydroxylation of salvigenin | Cloning and expression of recombinant protein ObF8H-1 in yeast and followed by characterization via enzyme assays | |
| 2-Oxoglutarate-dependent flavone demethylase (O. basilicum)120 | Accumulation of 7-O-demethylated flavone nevadensin | Enzyme assays using trichome protein extracts | |
| Flavonoid O-methyltransferase (FOMT) (Ocimum basilicum)121 | 6- and 4′-O-methylation of flavones | Cloning and expression of full length cDNA in E. coli followed by characterization via enzyme assays |
3. Factors regulating secondary metabolite flux and chemical diversity in Ocimum species
Metabolite diversity observed at the level of species in genus Ocimum is dependent on several internal and external factors (Fig. 3). Some of the known factors responsible for regulating terpenoid and phenylpropanoid pathways are discussed.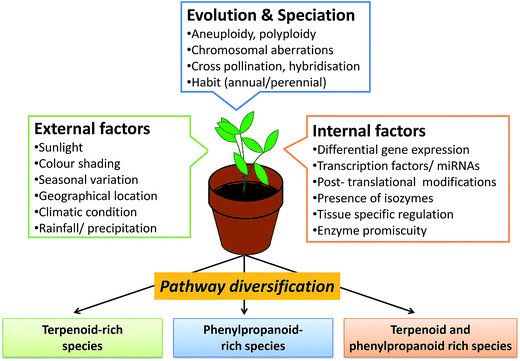 | ||
| Fig. 3 Factors responsible for chemical diversity; and terpenoid and phenylpropanoid pathway diversification in Ocimum species. | ||
3.1 Differential gene expression of enzymes in phenylpropanoid and terpenoid pathways
Gene expression plays an important role in diverting metabolic flux toward either the terpenoid or the phenylpropanoid pathway.5,13,100 In particular increased expression of terminal enzymes in the terpenoid pathway and reduced expression of phenylpropanoid entry point enzymes such as phenylalanine ammonia-lyase (PAL) has been observed in O. basilicum var. SD, rich in terpenoids. In another variety, O. basilicum var. EMX, however, the expression level of general phenylpropanoid pathway enzymes, PAL and 4-coumarate-CoA ligase (4CL) was found to be significantly higher corresponding to higher phenylpropanoid content.5 These results were supported by next generation sequencing data of O. tenuiflorum and O. basilicum.14 O. tenuiflorum rich in phenylpropanoids, shows much higher expression of general phenylpropanoid pathway enzymes including PAL, cinnamate-4-hydroxylase (C4H) and 4CL, reads per kilobase per million (RPKM) = 91.47, 34.53 and 9.52 respectively; compared to O. basilicum rich in terpenoids, RPKM = 11.3, 11.83 and 5.65 respectively. In O. basilicum, however, the entry point enzymes of the MEP pathway, representing the cytosolic pathway for terpenoid synthesis, including 1-deoxy-D-xylulose 5-phosphate reductoisomerase (DXR) was more (RPKM = 50.58) compared to O. tenuiflorum (RPKM = 15.69).14 Thus, overexpressing the entry point enzymes at major metabolic branching points also helps in directing the flux towards either phenylpropanoid or terpenoid pathway.122–124 Evidently, differential expression of enzymes strategically present at pathway branch points might play a crucial role in determining flux regulation (Fig. 4).3.2 Enzyme promiscuity
One of the major reasons for metabolite diversity observed in Ocimum species is the promiscuity of terpene synthases. These enzymes are capable of accepting a substrate and yielding a major product as well as multiple side products. For instance, Iijima et al. characterized eight terpene synthases from three cultivars of O. basilicum.100,114 In vitro recombinant protein assays using geranyl diphosphate (GPP) as substrate for putative monoterpene synthases and farnesyl diphosphate (FPP) as substrate for putative sesquiterpene synthases was performed. Terpinolene synthase gave terpinolene as the major product and α-pinene, limonene and an unidentified monoterpene as the side products. Fenchol synthase produced fenchol and limonene as major products and α-pinene and an unidentified monoterpene as the side products. Cadinene synthase produced γ-cadinene as the major product and muurola 3, 5-diene as the side product. Selinene synthase produced selinene as the major product and β-elemene and nerolidol as side products. In contrast, myrcene synthase and geraniol synthase exclusively produced myrcene and geraniol as end products.100 In another study by Major et al., using bornyl diphosphate synthase (producing camphene as the side product), it was proven that electrostatically guided dynamics determined end product formation.110 Current evidence suggests that enzyme promiscuity may play an important role in contributing to the diversity across Ocimum species.3.3 Transcription factors
Transcriptional regulation of secondary metabolism in plants for flavonoids (particularly anthocyanins), alkaloids (including nicotine, indole alkaloids and benzylisoquinolines) and terpenoids has been widely reported.125–127 Recently, PAP1 transcription factor was shown to enhance the production of both terpenoids and phenylpropanoids in rose plant.116 Deep sequencing of O. tenuiflorum and O. basilicum revealed the presence of 40 transcription factor families including MYB, WRKY, bHLH, HB, NAC, bZIP etc. which are known regulators of secondary metabolism in plants.14 A recent study performed using the red and green forma of O. tenuiflorum suggested light-mediated regulation of anthocyanin accumulation.128 It was observed that when red forma seedlings grown under natural lighting conditions, were transferred to a special greenhouse which cuts off the UV-A and UV-B radiation, the leaves turned green within 20 days. Further investigation revealed the role of transcription factors, bHLH and WD40, in downregulating the terminal enzymes of anthocyanin biosynthesis including flavonone-3′-hydroxylase, leucoanthocyanidin dioxygenase and dihydroflavonol reductase, responsible for red coloration. In another study by Misra et al., transcription factors belonging to APETALA2/ethylene responsive factor (ERF), WRKY, plant homeodomain (PHD) and zinc finger families were upregulated in methyl jasmonate (MeJa)-treated O. basilicum plants, suggesting their possible role in regulating secondary metabolism in Ocimum species.115 Thus, available data suggests transcription factors are also key regulators of terpenoid and phenylpropanoid pathway in Ocimum species and provide a more stringent control over the direction of flux.3.4 Post-translational modifications
Post-translational modifications including phosphorylation, ubiquitination and arginine monomethylation of phenylpropanoid and terpenoid pathway enzymes such as phosphoglucomutase, glucose-6-phosphate isomerase, phosphoglycerate mutase, PAL and chavicol O-methyl transferase (CVOMT) were observed in basil glandular trichomes. Post translation modifications help in explaining situations where the mRNA level does not match with the metabolite or protein level. For example, the enzyme CVOMT is responsible for methylating chavicol. O. basilicum var. SD produces negligible amount of methyl chavicol. However, the mRNA and protein levels for this enzyme were found to be very high. In contrast, very little enzyme activity and metabolites were detected. It was observed that this enzyme was ubiquitinated providing a valid explanation for the discrepancies in mRNA, protein, enzyme activity and metabolite level. Ubiquitination leads to a rapid degradation of CVOMT post translation,5 resulting in decreased formation of methyl chavicol. In another example, PAL, catalyzing the first committed step in phenylpropanoid biosynthesis, is phosphorylated in O. basilicum var. SD, rich in monoterpenes; however, other basil varieties (SW, MC, and EMX-1), rich in phenylpropanoids, lack PAL phosphorylation.5 It has been reported earlier that phosphorylation results in the reduction of PAL activity.129,130 Above examples suggest that post translation modifications provide an additional regulatory step in determining the expression of key enzyme activities in secondary metabolic pathways in Ocimum species.3.5 Presence of isozymes
Phenylpropanoid pathway produces substrates for synthesis of several important secondary metabolites. PAL, C4H and 4CL catalyse the initial few steps leading to the formation of coumaryl CoA. Latter represents a branching point, from which different end products including phenylpropenes, lignins, flavonoids and rosmarinic acid can be synthesized. Thus, 4CL represents a crucial step in pathway regulation and diversification. In recent work by Rastogi et al., it was reported that O. basilicum 4CL has 5 different isoforms.103 RNAi experiments involving the silencing of a specific isoform, Oba4CL, led to a reduced production of phenylpropanoids without affecting lignin and sinapic acid content. Thus, only one of the isoforms of 4CL was involved in the synthesis of phenylpropenes. This also represents the commitment of a specific isoform of an enzyme to a specific biosynthetic pathway at a very initial step. Presence of such pathway-committed isoforms keeps the pathway finely tuned and delicately balanced in basil.3.6 External factors
Being species native to the tropics, Ocimum plants are always subjected to severe environmental conditions including excessive heat, rainfall, humidity, dryness etc. Adaptability, thus, is the key to survival. It has been reported that external environmental factors, including the type of light, radiation, season, geographic conditions etc., influence essential oil composition. Same Ocimum specie show altered metabolic profile under different environmental factors. Red and blue shading conditions in O. selloi showed decline in level of phenylpropanoids and elevated level of in comparison with plants grown in full light.84 Plants grown under blue shading had more number of metabolites than plants subjected to full light and red shading. Decreased accumulation of methyl chavicol was observed in plants cultured under colored netting, accompanied by an increase in α-copaene, germacrene D and bicyclogermacrene content.84 This suggests a chemical defense strategy of plants against less favorable growth conditions. Similar kind of study was performed with O. basilicum cultivated in soil covered by colored mulches which demonstrated that size and aroma of leaves as well as the concentration of soluble phenols greatly improved.131 Seasonal variation of essential oil composition was observed in O. basilicum and O. tenuiflorum.83 To show the effect of geographic conditions on essential oil composition, O. gratissimum and O. campechianum were grown in Chocó Department (Columbia) and Ecuador region that resulted in different chemical composition.91 Similarly, O. basilicum and O. gratissimum grown in Benin, Cameroon, Congo and Gabon vary in chemical composition.90 O. gratissimum plants grown in Columbia showed altered metabolite profile as compared with those grown in Europe.91,132–135 This data indicates external factors including climatic conditions and geographical variations might be influencing the chemical profile of Ocimum species.3.7 Developmental and tissue specific regulation
During cinnamic acid and methylcinnamate (MC) formation from phenylalanine, activity of two enzymes, PAL and S-adenosyl-L-methionine:cinnamate carboxyl methyltransferase (SAM:CCMT) shows an important regulatory control point.136 In different developmental stages of O. basilicum, the relation between MC content, PAL and SAM:CCMT activity was examined. SAM:CCMT activity showed correlation with MC content in young leaves.136 Likewise, eugenol-O-methyltransferase (EOMT) is responsible for methylation of eugenol to form methyl eugenol in one of the final steps of phenylpropanoid pathway. The expression pattern of EOMT positively correlated with the amount of eugenol/isoeugenol and methyl eugenol in different developmental stages of all the analyzed chemotypes.137 Along with development-specific regulation of metabolite accumulation, some metabolites in Ocimum species also show tissue-specific regulation. For example, analysis of trichome, leaf, stem and root shows a strong association between eugenol content and Ob4CL expression in O. basilicum.1033.8 MicroRNA mediated regulation
Based on O. basilicum EST data set, the function of miRNAs and their targets was predicted using in silico approach.138 Four miRNA families miR164c, miR5658, miR414 and miR5021 were evaluated for their potential targets. These miRNA families showed regulatory role during stress-metabolite response. Although this study was based upon computational evaluation, further in planta experimentation is required to determine the critical role of miRNAs during secondary metabolism in Ocimum species.1384. Future perspective
Ocimum acts as a reservoir of several important secondary metabolites found in nature, thereby making it a very attractive system to explore. Although the genome of Ocimum has not yet been sequenced, the recent influx of next generation sequencing data of various tissues such as trichomes and leaves, has helped us in understanding various factors that are responsible for regulating the formation of phenylpropanoids and terpenoids in Ocimum species. Using the current information, we can genetically engineer Ocimum species to overexpress the desired metabolites by redirecting the metabolite flux.139–142 This knowledge can also be used for breeding new chemotypes producing interesting spectra of essential metabolites. Since these metabolites impart flavor and aroma, and possess medicinal properties, they can be heterologously expressed in plants, which are routinely used raw in our diet, such as tomato, thereby increasing their flavor and nutritive value. The expression of O. basilicum α-zingiberene synthase under the control of polygalacturonase promoter led to the unexpected accumulation of 15 sesquiterpenes and 10 monoterpenes, which were not present in the non-transformed fruit.143 In a separate study, the expression of O. basilicum geraniol synthase under the same promoter led to the accumulation of geraniol and its derivatives, which had profound impact on tomato flavor as well as aroma.144 Moreover, expressing terpene synthase genes from Ocimum in food crops will impart greater resistance against pathogens and pests. Till date, it is not well established whether there is a cross talk between the phenylpropanoid and terpenoid pathways. The glandular trichomes present in several Ocimum plants provide a very exciting isolated single-celled system to unravel the exchange, if any, of upstream intermediates between these two pathways. Thus, Ocimum species find useful applications in industrial, culinary, medicinal as well as scientific research areas, asserting their important position in the plant kingdom.5. Conclusions
Phenylpropanoid and terpenoid pathways in genus Ocimum have evolved as a result of various evolutionary, environmental and molecular events. Understanding the regulatory checkpoints in these pathways is a step closer towards efficiently harnessing this plant system. We believe that genes for both terpenoid and phenylpropanoid biosynthesis are present in all Ocimum species. However, several factors including genetic background, habit, ploidy levels, hybridization, differential gene expression, transcriptional and post translational modifications, isozymes etc. have played a major role in metabolic pathway diversification making Ocimum species either terpene- or phenylpropanoid-rich. The presence of many terpene synthases in single species and each one's ability to synthesize diverse metabolites from a single substrate has further complicated the chemical evolution process. Most reviews on Ocimum have concentrated only on the medicinal benefits and industrial uses of this genus. To our knowledge, this is the first review that attempts to provide cumulative information about the potential reason/s for the complex chemical evolution and discusses terpenoid and phenylpropanoid pathway diversification events across Ocimum species.Abbreviations
| MVA | Mevalonic acid |
| MEP | Methylerythritol phosphate |
| GC | Gas chromatography |
| LC | Liquid chromatography |
| MS | Mass spectrometry |
| NMR | Nuclear magnetic resonance |
| PAL | Phenylalanine ammonia-lyase |
| 4CL | 4-Coumarate-CoA ligase |
| C4H | Cinnamate-4-hydroxylase |
| RPKM | Reads per kilobase per million |
| DXR | 1-Deoxy-D-xylulose 5-phosphate reductoisomerase |
| GPP | Geranyl diphosphate |
| FPP | Farnesyl diphosphate |
| UV-A | Ultraviolet-A |
| UV-B | Ultraviolet-B |
| ERF | Ethylene responsive factor |
| PHD | Plant homeodomain |
| MeJa | Methyl jasmonate |
| CVOMT | Chavicol O-methyl transferase |
| MC | Methyl cinnamate |
| SAM | S-Adenosyl-L-methionine |
| CCMT | Cinnamate carboxyl methyltransferase |
| EOMT | Eugenol-O-methyltransferase |
| Ob4CL | Ocimum basilicum 4-coumarate-CoA ligase |
Acknowledgements
PS acknowledges support from Council of Scientific and Industrial Research (CSIR), New Delhi for research fellowship. This research work is supported by CSIR under XII five-year plan network project (BSC0124) to CSIR-National Chemical Laboratory.References
- J. E. Simon, J. Quinn and R. G. Murray, in Advances in new crops, ed. J. Janick and J. E. Simon, Timber Press, Portland, 1990, Basil: a source of essential oils, p. 484 Search PubMed.
- C. Linnaeus, in Species Plantarum, ed. Laurentii Salvii, Holmiae, Stockholm, 1753, Ocimum, p. 597 Search PubMed.
- P. D. Cantino, R. M. Harley and S. J. Wagstaff, in Advances in Labiate Science, ed. R. M. Harley and T. Reynolds, Royal Botanic Gardens, Kew, 1992, Genera of Labiatae: Status and Classification, p. 502 Search PubMed.
- M. K. Khosla, Indian J. Genet. Plant Breed., 1995, 55, 71 Search PubMed.
- Z. Xie, J. Kapteyn and D. R. Gang, Plant J., 2008, 54, 349 CrossRef CAS PubMed.
- D. Yang, X. Du, X. Liang, R. Han, Z. Liang, Y. Liu, F. Liu and J. Zhao, PLoS One, 2012, 7, e46797 CAS.
- A. Marchev, C. Haas, S. Schulz, V. Georgiev, J. Steingroewer, T. Bley and A. Pavlov, Biotechnol. Lett., 2014, 36, 211 CrossRef CAS PubMed.
- G. W. Turner and R. Croteau, Plant Physiol., 2004, 136, 4215 CrossRef CAS PubMed.
- R. B. Croteau, E. M. Davis, K. L. Ringer and M. R. Wildung, Naturwissenschaften, 2005, 92, 562 CrossRef CAS PubMed.
- R. Rios-Estepa, G. W. Turner, J. M. Lee, R. B. Croteau and B. M. Lange, Proc. Natl. Acad. Sci. U. S. A., 2008, 105, 2818 CrossRef CAS PubMed.
- A. Lane, A. Boecklemann, G. N. Woronuk, L. Sarker and S. S. Mahmoud, Planta, 2010, 231, 835 CrossRef CAS PubMed.
- G. Woronuk, Z. Demissie, M. Rheault and S. Mahmoud, Planta Med., 2011, 77, 7 CrossRef CAS PubMed.
- D. R. Gang, J. Wang, N. Dudareva, K. H. Nam, J. E. Simon, E. Lewinsohn and E. Pichersky, Plant Physiol., 2001, 125, 539 CrossRef CAS PubMed.
- S. Rastogi, S. Meena, A. Bhattacharya, S. Ghosh, R. K. Shukla, N. S. Sangwan, R. K. Lal, M. M. Gupta, U. C. Lavania, V. Gupta, D. A. Nagegowda and A. K. Shasany, BMC Genomics, 2014, 15, 588 CrossRef PubMed.
- F. Bast, P. Rani and D. Meena, Sci. World J., 2014, 2014, 847482 CrossRef PubMed.
- K. Carović-Stanko, Z. Liber, V. Besendorfer, B. Javornik, B. Bohanec, I. Kolak and Z. Satovic, Plant Syst. Evol., 2010, 285, 13 CrossRef.
- A. J. Paton, D. Springate, S. Suddee, D. Otieno, R. J. Grayer, M. M. Harley, F. Willis, M. S. J. Simmonds, M. P. Powell and V. Savolainen, Mol. Phylogenet. Evol., 2004, 31, 277 CrossRef CAS PubMed.
- A. Paton and E. Putievsky, Kew Bull., 1996, 51, 509 CrossRef.
- A. P. Singh, S. Dwivedi, S. Bharti, S. Srivastava, V. Singh and S. P. S. Khanuja, Euphytica, 2004, 136, 11 CrossRef CAS.
- V. T. de Lima, M. C. Vieira, C. A. Kassuya, C. A. Cardoso, J. M. Alves, M. A. Foglio, J. E. de Carvalho and A. S. Formagio, Phytomedicine, 2014, 21, 1298 CrossRef CAS PubMed.
- J. Monga, M. Sharma, N. Tailor and N. Ganesh, Pharm. Biol., 2011, 49, 428 CrossRef PubMed.
- P. Singh, R. H. Jayaramaiah, P. Sarate, H. V. Thulasiram, M. J. Kulkarni and A. P. Giri, PLoS One, 2014, 9, e104377 Search PubMed.
- R. V. Sarin, S. Narwal and P. A. Bafna, J. Ethnopharmacol., 2013, 148, 223 CrossRef PubMed.
- D. Runyoro, O. Ngassapa, K. Vagionas, N. Aligiannis, K. Graikou and I. Chinou, Food Chem., 2010, 119, 311 CrossRef CAS.
- B. O. Owuor, J. O. Ochanda, J. O. Kokwaro, A. C. Cheruiyot, R. A. Yeda, C. A. Okudo and H. M. Akala, J. Ethnopharmacol., 2012, 144, 779 CrossRef CAS PubMed.
- H. Song, P. Kumar, G. Arivazhagan, S. I. Lee, H. M. Yoon, I. H. Kim, H. J. Kwon, J. M. Kim and F. L. Hakkim, J. Plant Biochem. Biotechnol., 2012, 39, 146 CrossRef.
- F. L. Hakkim, G. Arivazhagan and R. Boopathy, J. Med. Plants Res., 2008, 2, 250 Search PubMed.
- E. J. Kweka, F. W. Mosha, A. Lowassa, A. M. Mahande, M. J. Mahande, C. P. Massenga, F. Tenu, E. E. Lyatuu, M. A. Mboya and E. A. Temu, Parasites Vectors, 2008, 1, 42 CrossRef PubMed.
- P. Kapewangolo, J. J. Omolo, R. Bruwer, P. Fonteh and D. Meyer, J. Inflammation, 2015, 12, 4 CrossRef PubMed.
- E. Lulekal, J. Rondevaldova, E. Bernaskova, J. Cepkova, Z. Asfaw, E. Kelbessa, L. Kokoska and P. van Damme, Pharm. Biol., 2013, 52, 614 CrossRef PubMed.
- D. Damtie and Y. Mekonnen, Afr. J. Pharm. Pharmacol., 2015, 9, 91 Search PubMed.
- F. F. Dube, K. Tadesse, G. Birgersson, E. Seyoum, H. Tekie, R. Ignell and S. R. Hill, Malar. J., 2011, 10, 375 CrossRef CAS PubMed.
- M. J. Mukazayire, J. C. Tomani, C. Stevigny, J. C. Chalchat, F. Conforti, F. Menichini and P. Duez, Food Chem., 2011, 129, 753 CrossRef CAS PubMed.
- W. Mequanint, E. Makonnen and K. Urga, J. Ethnopharmacol., 2011, 134, 32 CrossRef PubMed.
- M. J. Mukazayire, V. Allaeys, P. B. Calderon, C. Stévigny, M. J. Bigendako and P. Duez, Exp. Toxicol. Pathol., 2010, 62, 289 CrossRef PubMed.
- E. Makonnen, A. Debella, D. Abebe and F. Teka, Phytother. Res., 2003, 17, 1108 CrossRef PubMed.
- Y. Naidoo, C. T. Sadashiva, N. Kasim, A. Nicholas and G. Naidoo, J. Essent. Oil-Bear. Plants, 2014, 17, 142 CrossRef CAS.
- S. Parasuraman, S. Balamurugan, P. V. Christapher, R. R. Petchi, W. Y. Yeng, J. Sujithra and C. Vijaya, Pharmacogn. Res., 2015, 7, 156 CrossRef CAS.
- F. L. Hakkim, C. G. Shankar and S. Girija, J. Agric. Food Chem., 2007, 55, 9109 CrossRef CAS PubMed.
- B. Sarkar, D. Kumar, D. Sasmal and K. Mukhopadhyay, Nat. Prod. Res., 2014, 28, 1393 CrossRef CAS PubMed.
- P. Phanthong, P. Lomarat, M. T. Chomnawang and N. Bunyapraphatsara, ScienceAsia, 2013, 39, 472 CAS.
- M. Stefan, M. M. Zamfirache, C. Padurariu, E. Trută and I. Gostin, Open Life Sci., 2013, 8, 600 CAS.
- K. Karthikeyan, P. Gunasekaran, N. Ramamurthy and S. Govindasamy, Pharm. Biol., 1999, 37, 285 CrossRef.
- F. M. Mahomoodally, A. H. Subratty, A. Gurib-Fakim and M. I. Choudhary, BMC Complementary Altern. Med., 2012, 12, 165 CrossRef PubMed.
- R. C. Saxena, R. Singh, P. Kumar, M. P. S. Negi, V. S. Saxena, P. Geetharani, J. J. Allan and K. Venkateshwarlu, J. Evidence-Based Complementary Altern. Med., 2011, 2012, 894509 Search PubMed.
- P. Sudha, S. S. Zinjarde, S. Y. Bhargava and A. R. Kumar, BMC Complementary Altern. Med., 2011, 11, 5 CrossRef PubMed.
- O. Chokechaijaroenporn, N. Bunyapraphatsora and S. Kongchuensin, Phytomedicine, 1994, 1, 135 CrossRef CAS PubMed.
- R. Yucharoen, S. Anuchapreeda and Y. Tragoolpua, Afr. J. Biotechnol., 2013, 10, 860 Search PubMed.
- G. Kaur, A. Bali, N. Singh and A. S. Jaggi, An. Acad. Bras. Cienc., 2015, 87, 417 CrossRef PubMed.
- B. T. Aluko, O. I. Oloyede and A. J. Afolayan, Pak. J. Biol. Sci., 2013, 16, 22 CrossRef CAS PubMed.
- A. N. Yamada, R. Grespan, A. T. Yamada, E. L. Silva, S. E. Silva-Filho, M. J. Damião, M. M. de Oliveira Dalalio, C. A. Bersani-Amado and R. K. N. Cuman, Am. J. Chin. Med., 2013, 41, 913 CrossRef PubMed.
- S. Thaweboon and B. Thaweboon, Southeast Asian J. Trop. Med. Public Health, 2009, 40, 1025 Search PubMed.
- U. Szymanowska, U. Złotek, M. Karas and B. Baraniak, Food Chem., 2015, 172, 71 CrossRef CAS PubMed.
- N. P. Yadav, R. Khatri, D. U. Bawankule, A. Pal, K. Shanker, P. Srivastava, A. K. Gupta and D. Chanda, Planta Med., 75, PA72 Search PubMed.
- S. J. Inbaneson, R. Sundaram and P. Suganthi, Asian Pac. J. Trop. Med., 2012, 5, 103 CrossRef PubMed.
- D. Beatović, D. Krstić-Milošević, S. Trifunović, J. Šiljegović, J. Glamočlija, M. Ristić and S. Jelačić, Rec. Nat. Prod., 2015, 9, 62 Search PubMed.
- L. Vlase, D. Benedec, D. Hanganu, G. Damian, I. Csillag, B. Sevastre, A. C. Mot, R. Silaghi-Dumitrescu and I. Tilea, Molecules, 2014, 19, 5490 CrossRef PubMed.
- B. Kaurinovic, M. Popovic, S. Vlaisavljevic and S. Trivic, Molecules, 2011, 16, 7401 CrossRef CAS PubMed.
- I. H. N. Bassolé, A. Lamien-Meda, B. Bayala, S. Tirogo, C. Franz, J. Novak, R. C. Nebié and M. H. Dicko, Molecules, 2010, 15, 7825 CrossRef PubMed.
- K. R. Durga, S. Karthikumar and K. Jegatheesan, Afr. J. Plant Sci., 2010, 4, 163 Search PubMed.
- P. A. Ntonga, N. Baldovini, E. Mouray, L. Mambu, P. Belong and P. Grellier, Parasite, 2014, 21, 33 CrossRef PubMed.
- K. H. Al-Ali, H. A. El-Beshbishy, A. A. El-Badry and M. Alkhalaf, Pak. J. Biol. Sci., 2013, 16, 1744 CrossRef PubMed.
- A. K. Qamar, A. Dar, B. S. Siddiqui, N. Kabir, H. Aslam, S. Ahmed, S. Erum, S. Habib and S. Begum, Lett. Drug Des. Discovery, 2010, 7, 726 CrossRef.
- M. Govindarajan, R. Sivakumar, M. Rajeswary and K. Yogalakshmi, Exp. Parasitol., 2013, 134, 7 CrossRef CAS PubMed.
- K. Murugan, P. Murugan and A. Noortheen, Bioresour. Technol., 2007, 98, 198 CrossRef CAS PubMed.
- B. S. Siddiqui, H. A. Bhatti, S. Begum and S. Perwaiz, J. Ethnopharmacol., 2012, 144, 220 CrossRef CAS PubMed.
- K. S. Bora, S. Arora and R. Shri, J. Ethnopharmacol., 2011, 137, 1360 CrossRef PubMed.
- A. Umar, G. Imam, W. Yimin, P. Kerim, I. Tohti, B. Berké and N. Moore, Hypertens. Res., 2010, 33, 727 CrossRef PubMed.
- S. Amrani, H. Harnafi, D. Gadi, H. Mekhfi, A. Legssyer, M. Aziz, F. Martin-Nizard and L. Bosca, J. Ethnopharmacol., 2009, 125, 157 CrossRef PubMed.
- I. de Almeida, D. S. Alviano, D. P. Vieira, P. B. Alves, A. F. Blank, A. H. C. Lopes, C. S. Alviano and S. R. Maria do Socorro, Parasitol. Res., 2007, 101, 443 CrossRef PubMed.
- L. C. Chiang, L. T. Ng, P. W. Cheng, W. Chiang and C. C. Lin, Clin. Exp. Pharmacol. Physiol., 2005, 32, 811 CrossRef CAS PubMed.
- Y. H. Chen, Y. W. Chiu, J. C. Shyu, C. C. Tsai, H. H. Lee, C. C. Hung, J. M. Hwang, J. Y. Liu and W. H. Wang, Chin. J. Physiol., 2015, 58, 55 CrossRef CAS PubMed.
- D. C. Gontijo, L. C. Fietto and J. P. V. Leite, Rev. Bras. Plant. Med., 2014, 16, 874 CrossRef.
- G. H. Bénédicta, K. Kpoviessia, S. D. S. Kpoviessia, E. Y. Ladekan, F. Gbaguidi, M. Frédérichd, M. Moudachiroub, J. Quetin-Leclercqc, J. C. Accrombessia and J. Beroc, J. Ethnopharmacol., 2014, 155, 1417 CrossRef PubMed.
- S. K. Mahapatra and S. Roy, Asian Pac. J. Trop. Med., 2014, 7, S391 CrossRef CAS.
- Y. W. Chiu, P. Y. Chao, C. C. Tsai, H. L. Chiou, Y. C. Liu, C. C. Hung, H. C. Shih, T. J. Lai and J. Y. Liu, Am. J. Chin. Med., 2014, 42, 833 CrossRef PubMed.
- R. K. Joshi, Indian J. Pharm. Sci., 2013, 75, 457 CrossRef CAS PubMed.
- A. Katara, C. K. Pradhan, P. Singh, V. Singh and M. Ali, J. Essent. Oil-Bear. Plants, 2013, 16, 283 CrossRef CAS.
- E. F. F. Matias, F. A. V. Santos, J. M. F. Silva, C. E. S. Souza, S. R. Tintino, G. M. M. Guedes, C. R. Medeiros, M. F. B. Braga, T. S. Almeida, J. G. M. Costa, I. R. A. Menezes and H. D. M. Coutinho, Afr. J. Microbiol. Res., 2012, 6, 1902 Search PubMed.
- M. O. Nkiko and J. T. Bamgbose, Port. Electrochim. Acta, 2011, 29, 419 CrossRef CAS.
- K. S. Bora, R. Shri and J. Monga, Pharm. Biol., 2011, 49, 175 CrossRef PubMed.
- A. C. M. de Santana, G. S. Pereira, C. M. Boaventura, A. P. T. Uetenabaro, L. C. D. B. Costa and R. A. de Oliveira, Rev. Bras. Farmacogn., 2014, 24, 524 CrossRef CAS.
- S. R. Vani, S. F. Cheng and C. H. Chuah, Am. J. Appl. Sci., 2009, 6, 523 CrossRef CAS.
- L. C. Costa, J. E. Pinto, E. M. Castro, E. Alves, L. F. Rosal, S. K. Bertolucci, P. B. Alves and T. S. Evangelino, J. Essent. Oil Res., 2010, 22, 34 CrossRef CAS.
- S. K. Singh, A. Anand, S. K. Verma, M. A. Siddiqui, A. Mathur and S. Saklani, Int. J. Curr. Pharm. Res., 2011, 3, 40 CAS.
- M. Özcan and J. C. Chalchat, Czech J. Food Sci., 2002, 20, 223 Search PubMed.
- D. Ioannidis, B. Lynda and C. B. Johnson, Ann. Bot., 2002, 90, 453 CrossRef CAS PubMed.
- R. Parida, S. I. Sandeep, B. K. Sethy, S. Sahoo and S. Mohanty, Int. J. Pharm. Pharm. Sci., 2014, 6, 7 Search PubMed.
- A. S. D. Costa, M. D. F. Arrigoni-Blank, M. A. A. P. D. Silva, M. F. Alves, D. D. A. Santos, P. B. Alves and A. F. Blank, Sci. World J., 2014, 2014, 824594 Search PubMed.
- F. Tchoumbougnang, P. A. Zollo, F. Avlessi, G. A. Alitonou, D. K. Sohounhloue, J. M. Ouamba and C. Menut, J. Essent. Oil Res., 2006, 18, 194–199 CrossRef CAS.
- N. P. Benitez, E. M. M. León and E. E. Stashenko, J. Chromatogr. Sci., 2009, 47, 800 CAS.
- G. Sacchetti, A. Medici, S. Maietti, M. Radice, M. Muzzoli, S. Manfredini, E. Braccioli and R. Bruni, J. Agric. Food Chem., 2004, 52, 3486 CrossRef CAS PubMed.
- J. C. Nascimento, L. C. Barbosa, V. F. Paula, J. M. David, R. Fontana, L. A. Silva and R. S. França, An. Acad. Bras. Cienc., 2011, 83, 787 CrossRef CAS PubMed.
- O. U. Edet and P. O. Aikpokpodion, Am. J. Plant Sci., 2014, 5, 126 CrossRef.
- R. B. Bose and J. K. Choudhury, Caryologia, 1962, 15, 435 CrossRef.
- P. Pushpangadan and S. N. Sobti, Cytologia, 1982, 47, 575 CrossRef.
- T. Zderkiewicz, Acta Agrobot., 1971, 24, 121 CrossRef CAS.
- H. Dijkstra and G. J. Speckmann, Euphytica, 1980, 29, 89 CrossRef.
- E. K. Janaki Ammal and S. N. Sobti, Curr. Sci., 1962, 31, 387 Search PubMed.
- Y. Iijima, R. Davidovich-Rikanati, E. Fridman, D. R. Gang, E. Bar, E. Lewinsohn and E. Pichersky, Plant Physiol., 2004, 136, 3724 CrossRef CAS PubMed.
- G. V. Louie, T. J. Baiga, M. E. Bowman, T. Koeduka, J. H. Taylor, S. M. Spassova, E. Pichersky and J. P. Noel, PLoS One, 2007, 2, e993 Search PubMed.
- I. Aragüez, S. Osorio, T. Hoffmann, J. L. Rambla, N. Medina-Escobar, A. Granell, M. A. Botella, W. Schwab and V. Valpuesta, Plant Physiol., 2013, 163, 946 CrossRef PubMed.
- S. Rastogi, R. Kumar, C. S. Chanotiya, K. Shanker, M. M. Gupta, D. A. Nagegowda and A. K. Shasany, Plant Cell Physiol., 2013, 54, 1238 CrossRef CAS PubMed.
- L. Medina-Puche, F. J. Molina-Hidalgo, M. Boersma, R. C. Schuurink, I. López-Vidriero, R. Solano, J. M. Franco-Zorrilla, J. L. Caballero, R. Blanco-Portales and J. Muñoz-Blanco, Plant Physiol., 2015, 168, 598 CrossRef CAS PubMed.
- D. R. Gang, N. Lavid, C. Zubieta, F. Chen, T. Beuerle, E. Lewinsohn, J. P. Noel and E. Pichersky, Plant Cell, 2002, 14, 505 CrossRef CAS.
- E. Lewinsohn, I. Ziv-Raz, N. Dudai, Y. Tadmor, E. Lastochkin, O. Larkov, D. Chaimovitsh, U. Ravid, E. Putievsky, E. Pichersky and Y. Shoham, Plant Sci., 2000, 160, 27 CrossRef CAS PubMed.
- R. Croteau and F. Karp, Arch. Biochem. Biophys., 1979, 198, 512 CrossRef CAS PubMed.
- M. L. Wise, T. J. Savage, E. Katahira and R. Croteau, J. Biol. Chem., 1998, 273, 14891 CrossRef CAS PubMed.
- D. A. Whittington, M. L. Wise, M. Urbansky, R. M. Coates, R. B. Croteau and D. W. Christianson, Proc. Natl. Acad. Sci. U. S. A., 2002, 99, 15375 CrossRef CAS PubMed.
- D. T. Major and M. Weitman, J. Am. Chem. Soc., 2012, 134, 19454 CrossRef CAS PubMed.
- R. Croteau, C. L. Hooper and M. Felton, Arch. Biochem. Biophys., 1978, 188, 182 CrossRef CAS PubMed.
- L. S. Sarker, M. Galata, Z. A. Demissie and S. S. Mahmoud, Arch. Biochem. Biophys., 2012, 528, 163 CrossRef CAS PubMed.
- R. Croteau and F. Karp, Arch. Biochem. Biophys., 1977, 179, 257 CrossRef CAS PubMed.
- Y. Iijima, D. R. Gang, E. Fridman, E. Lewinsohn and E. Pichersky, Plant Physiol., 2004, 134, 370 CrossRef CAS PubMed.
- R. C. Misra, P. Maiti, C. S. Chanotiya, K. Shanker and S. Ghosh, Plant Physiol., 2014, 164, 1028 CrossRef CAS PubMed.
- M. M. B. Zvi, E. Shklarman, T. Masci, H. Kalev, T. Debener, S. Shafir, M. Ovadis and A. Vainstein, New Phytol., 2012, 195, 335 CrossRef PubMed.
- A. K. Upadhyay, A. R. Chacko, A. Gandhimathi, P. Ghosh, K. Harini and A. P. Joseph, et al., BMC Plant Biol., 2015, 15, 212 CrossRef PubMed.
- A. Berim, J. Park and D. R. Gang, Plant J., 2014, 80, 385 CrossRef CAS PubMed.
- A. Berim and D. R. Gang, Phytochemistry, 2013, 92, 33 CrossRef CAS PubMed.
- A. Berim and D. R. Gang, J. Biol. Chem., 2013, 288, 1795 CrossRef CAS PubMed.
- A. Berim, D. C. Hyatt and D. R. Gang, Plant Physiol., 2012, 160, 1052 CrossRef CAS PubMed.
- N. J. Bate, J. Orr, W. T. Ni, A. Meromi, T. Nadlerhassar, P. W. Doerner, R. A. Dixon, C. J. Lamb and Y. Elkind, Proc. Natl. Acad. Sci. U. S. A., 1994, 91, 7608 CrossRef CAS.
- L. Carretero-Paulet, I. Ahumada, N. Cunillera, M. Rodriguez-Concepcion, A. Ferrer, A. Boronat and N. Campos, Plant Physiol., 2002, 129, 1581 CrossRef CAS PubMed.
- M. Rodriguez-Concepcion, Phytochem. Rev., 2006, 5, 1 CrossRef CAS.
- Z. X. Yu, J. X. Li, C. Q. Yang, W. L. Hu, L. J. Wang and X. Y. Chen, Mol. Plant, 2012, 5, 353 CrossRef CAS PubMed.
- S. Mishra, V. Triptahi, S. Singh, U. J. Phukan, M. M. Gupta, K. Shanker and R. K. Shukla, PLoS One, 2013, 8, e52784 CAS.
- M. A. Naoumkina, X. He and R. A. Dixon, BMC Plant Biol., 2008, 8, 132 CrossRef PubMed.
- P. Vyas, I. Haque, M. Kumar and M. Mukhopadhyay, Acta Physiol. Plant., 2014, 36, 2091 CrossRef CAS.
- E. G. Allwood, D. R. Davies, C. Gerrish, B. E. Ellis and G. P. Bolwell, FEBS Lett., 1999, 457, 47 CrossRef CAS PubMed.
- G. P. Bolwell, Phytochemistry, 1992, 31, 4081 CrossRef CAS.
- J. H. Loughrin and M. J. Kasperbauer, J. Agric. Food Chem., 2001, 49, 1331 CrossRef CAS PubMed.
- M. G. de Vasconcelos, A. A. Craveiro, F. J. Abreu, M. I. L. Machado and J. W. Alencar, Fitoterapia, 1999, 70, 32 CrossRef.
- C. M. Murbach, M. Ortiz and M. Costa, J. Ethnopharmacol., 2006, 105, 161 CrossRef PubMed.
- E. Guenther, in The Essential oils, van Nostrand Co. Inc., New York, USA, 1995, vol. III, V, pp. 427–428 Search PubMed.
- R. Hiltunen and Y. Holm, Basil the genus Ocimum, Harwood Academic Publishers, 1999 Search PubMed.
- Z. Hao, Biosynthesis of methylcinnamate in Ocimum basilicum L., Purdue University, 1998 Search PubMed.
- I. K. Renu, I. Haque, M. Kumar, R. Poddar, R. Bandopadhyay, A. Rai and K. Mukhopadhyay, Mol. Biol. Rep., 2014, 41, 1857 CrossRef CAS PubMed.
- N. Singh and A. Sharma, Gene, 2014, 552, 277 CrossRef CAS PubMed.
- B. M. Lange and A. Ahkami, Plant Biotechnol. J., 2013, 11, 169 CrossRef CAS PubMed.
- N. Dudareva, A. Klempien, J. K. Muhlemann and I. Kaplan, New Phytol., 2013, 198, 16 CrossRef CAS PubMed.
- M. J. Fischer, S. Meyer, P. Claudel, M. Bergdol and F. Karst, Biotechnol. Bioeng., 2011, 108, 1883 CrossRef CAS PubMed.
- N. Misawa, Curr. Opin. Biotechnol., 2011, 22, 627 CrossRef CAS PubMed.
- R. Davidovich-Rikanati, E. Lewinsohn, E. Bar, Y. Iijima, E. Pichersky and Y. Sitrit, Plant J., 2008, 56, 228 CrossRef CAS PubMed.
- R. Davidovich-Rikanati, Y. Sitrit, Y. Tadmor, Y. Iijima, N. Bilenko, E. Bar, B. Carmona, E. Fallik, N. Dudai, J. E. Simon, E. Pichersky and E. Lewinsohn, Nat. Biotechnol., 2007, 25, 899 CrossRef CAS PubMed.
| This journal is © The Royal Society of Chemistry 2015 |