Selecting analytical tools for characterization of polymersomes in aqueous solution†
Joachim Habelab,
Anayo Ogbonnab,
Nanna Larsenc,
Solène Cherréd,
Søren Kyndee,
Søren Roi Midtgaarde,
Koji Kinoshitaf,
Simon Krabbeg,
Grethe Vestergaard Jensene,
Jesper Søndergaard Hansenh,
Kristoffer Almdald and
Claus Hèlix-Nielsen*abi
aTechnical University of Denmark, Department of Environmental Engineering, Miljøvej, building 113, 2800 Kgs. Lyngby, Denmark. E-mail: clhe@env.dtu.dk; Tel: +45 27 10 20 76
bAquaporin A/S, Ole Maaløes Vej 3, 2200 Copenhagen, Denmark
cUniversity of Copenhagen, Copenhagen Biocenter, Ole Maaløes Vej 5, 2200 Copenhagen, Denmark
dTechnical University of Denmark, Department of Micro- and Nanotechnology, Produktionstorvet, building 423, 2800 Kgs. Lyngby, Denmark
eUniversity of Copenhagen, Niels Bohr Institute, Hans Christian Ørsted building D, Universitetsparken 5, 2100 Copenhagen, Denmark
fUniversity of Southern Denmark, Department of Physics, Chemistry and Pharmacy, Campusvej 55, 5230 Odense, Denmark
gUniversity of Copenhagen, Department of Biology, August Keogh Building, Universitetsparken 13, 2100 Copenhagen, Denmark
hLund University, Department of Experimental Medical Science, Box 117, 22100 Lund, Sweden
iUniversity of Maribor, Laboratory for Water Biophysics and Membrane Processes, Faculty of Chemistry and Chemical Engineering, Smetanova ulica 17, 2000 Maribor, Slovenia
First published on 9th September 2015
Abstract
Selecting the appropriate analytical methods for characterizing the assembly and morphology of polymer-based vesicles, or polymersomes are required to reach their full potential in biotechnology. This work presents and compares 17 different techniques for their ability to adequately report size, lamellarity, elastic properties, bilayer surface charge, thickness and polarity of polybutadiene–polyethylene oxide (PB–PEO) based polymersomes. The techniques used in this study are broadly divided into scattering techniques, visualization methods, physical and electromagnetical manipulation and sorting/purification. Of the analytical methods tested, Cryo-transmission electron microscopy and atomic force microscopy (AFM) turned out to be advantageous for polymersomes with smaller diameter than 200 nm, whereas confocal microscopy is ideal for diameters >400 nm. Polymersomes in the intermediate diameter range can be characterized using freeze fracture Cryo-scanning electron microscopy (FF-Cryo-SEM) and nanoparticle tracking analysis (NTA). Small angle X-ray scattering (SAXS) provides reliable data on bilayer thickness and internal structure, Cryo-TEM on multilamellarity. Taken together, these tools are valuable for characterizing polymersomes per se but the comparative overview is also intended to serve as a starting point for selecting methods for characterizing polymersomes with encapsulated compounds or polymersomes with incorporated biomolecules (e.g. membrane proteins).
1 Introduction
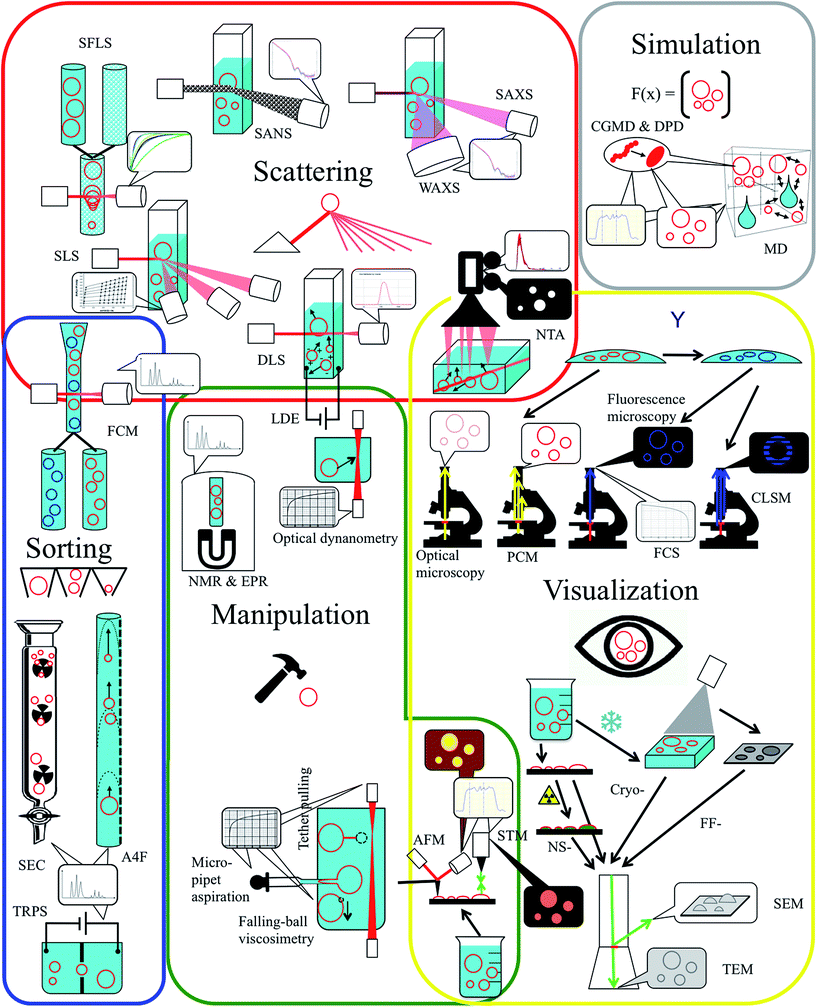
Polymersomes are hollow spheres arising from spontaneous self-assembly of amphiphilic block copolymers in solution.1–15 They have the potential to replace liposomes in biomolecule encapsulation for drug delivery applications,16–20 for incorporating proteins in their bilayer to create artificial cells14,21–24 and as design elements in ion-25,26 and water selective biomimetic membranes.27–31In order to use these versatile nanoscopic tools, a reliable and reproducible characterization is crucial. The characterization of polymersomes is however often a compromise between the convenience of instrumentation and preparation on one side and precision of the measurements on the other. More precise techniques typically implicate more invasive sample preparation. Here we summarize analysis techniques in five thematic groups with overlapping borders as shown in Fig. 1 and summarized in Table 1: scattering techniques, visualization methods, physical and electromagnetical manipulation, sorting/purification, and simulation tools.
| Characterization method | Subgroup | SDL [nm] | Parameters for analysis | Advantages | Disadvantages | References |
|---|---|---|---|---|---|---|
| Scattering methods | ||||||
| Dynamic light scattering | 2 | Size | Simple | Only for monodisperse samples | 39–41 | |
| Quasi-elastic light scattering | Minimal sample volume | Difficult comparison between instruments | ||||
| Photon correlated spectroscopy | Sensitivity to few large vesicles | Misinterpretation of aggregates | ||||
| Static light scattering | Multi-angle light scattering | 10 | Molecular weight, radius of gyration | Simple | RI and concentration required | 42 and 43 |
| X-ray scattering | Small-angle X-ray scattering | 0.5 | Bilayer thickness, lamellarity, encapsulation | Single polymer residue information | Elaborated setup | 33, 44–47 |
| X-ray scattering | Wide-angle X-ray scattering | 0.05 | Structural information | Atomic resolution | Elaborated setup | 43 and 48 |
| Neutron scattering | Small-angle neutron scattering | 0.5 | Bilayer thickness, lamellarity, encapsulation | Single polymer residue information | Elaborated setup & sample preparation | 47, 49–51 |
| Stopped-flow light scattering | Permeability | Simple | Only for monodisperse samples | 52 | ||
| Nanoparticle tracking analysis | 70 | Size, concentration | Simple | Underestimation of small vesicles | 37, 53 and 54 | |
| High accuracy | Misinterpretation of vesicles in z-plane | |||||
| Visualization methods via photons | Native environment | Resolution limit | ||||
| Optical microscopy | 200 | Size, concentration | Widely available | Low contrast | 55 | |
| Optical microscopy | Phase-contrast microscopy | 200 | Size, concentration | Better contrast | Additional staining | 55–57 |
| Fluorescence microscopy | Generalized polarization microscopy | Size, lamellarity, concentration | Specific labeling | Photobleaching | 43 and 58 | |
| Multiple staining | Only for labelled vesicles | |||||
| Exquisite sensitivity | Phototoxic effects | |||||
| Fluorescence microscopy | Fluorescence correlation spectroscopy | Size, lamellarity, concentration | Can distinguish free & incorporated dye | Additional staining | 59 | |
| Fluorescence microscopy | Confocal laser scanning microscopy | 200 | Size, lamellarity, concentration | High sensitivity | Difficult quantitative analysis | 54, 60 and 61 |
| Improved signal-to-noise-ratio | ||||||
| Fluorescence microscopy | Stimulated emission depletion microscopy | 20 | Size, lamellarity, concentration | Resolution | Elaborated setup | 62 |
| Visualization methods via electrons | Resolution | In vacuum | ||||
| Contrast | Elaborative sample preparation | |||||
| Transmission electron microscopy | Negative-staining TEM | 0.5 | Size, morphology, lamellarity | Resolution | Staining artifacts | 37, 49 and 52 |
| Vesicle shrinkage | ||||||
| Transmission electron microscopy | Freeze fracture TEM | 0.1 | Size, morphology, lamellarity | Structure preservation | Freezing artifacts | 49, 63 and 64 |
| Uncertainty in true size | ||||||
| Transmission electron microscopy | Cryo-TEM | 0.1 | Size, lamellarity, morphology | Structure preservation | Freezing artifacts | 49 and 65 |
| Uncertainty in bilayer thickness | ||||||
| Scanning electron microscopy | Freeze fracture Cryo-SEM | 2 | Size, lamellarity, morphology | 3D appearance | Freezing artifacts | 66 and 67 |
| Scanning electron microscopy | Environmental SEM | 30 | Size, lamellarity, morphology, concentration | Native environment | Poor resolution | 40, 60 and 68 |
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) |
||||||
| Electromagnetic manipulation methods | ||||||
| Scanning probe microscopy | 3D information | High sensitivity to vibration | ||||
| Scanning probe microscopy | Atomic force microscopy | 1 | Size, topology, elastic properties | Sensitivity | Shape alteration upon attachment | 37, 40, 54, 60 and 69 |
| Scanning force microscopy | Amphiphile adsorption on cantilever | |||||
| Scanning probe microscopy | Scanning tunneling microscopy | 0.1 | Size, topology | No mechanical contact to sample | Cantilever tip condition crucial | 70 and 71 |
| Nuclear magnetic resonance | P31-nuclear magnetic resonance | Lamellarity | High accuracy | Signal decrease due to convenient buffer | 72–74 | |
| Electron paramagnetic resonance | Encapsulation, bilayer flexibility, | Specific to unpaired electrons | Signal decrease due to water | 40, 75 and 76 | ||
| Electron spin resonance | Bilayer polarity, lamellarity | |||||
| Laser Doppler electrophoresis | Zeta potential, surface charge potential | Fast | Calibration required frequently | 77 and 78 | ||
| Optical tweezer | Elastic properties | High sensitivity | Elaborated alignment | 79 | ||
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) |
||||||
| Mechanical manipulation methods | ||||||
| Tether pulling | Elastic properties | Vesicle directly accessible | Only for large vesicles | 79 | ||
| Micropipette aspiration | 2500 | Elastic properties | Vesicle directly accessible | Only for large vesicles | 79 and 80 | |
| Falling-ball viscosimetry | Elastic properties | Vesicle directly accessible | Only for large vesicles | 79 | ||
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) |
||||||
| Sorting methods | ||||||
| Flow cytometry | Fluorescence-activated cell sorting | 270 | Size, concentration | Obtaining multiple properties | RI required | 39, 53, 81 and 82 |
| Widely available | Size restriction | |||||
| Field flow fractionation | Asymmetric flow field-flow fractionation | 1 | Size, concentration | Wide range of vesicle sizes | Elaborated setup | 39 and 49 |
| Calibration standard necessary | ||||||
| Sample loss through adsorption | ||||||
| Size exclusion chromatography | 17 | Size, concentration | Well-established | Not for large vesicles | 39, 83–85 | |
| Amphiphile adsorption on column material | ||||||
| Calibration standard necessary | ||||||
| Slow amphiphile diffusion | ||||||
| Size exclusion chromatography | High performance liquid chromatography | 1 | Size, concentration | More selective & rapid than SEC | Elaborated setup | 84 |
| Tunable resistive pulse sensing | 70 | Size, concentration | Accurate for concentration measurement | Potential pore clogging | 53 and 54 | |
| Coulter counting | Only for monodisperse samples | |||||
| Tunable resistive pulse sensing | Scanning ion occlusion sensing | 50 | Size, concentration | High sensitivity | Calibration required | 54 and 86 |
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) |
||||||
| Simulation methods | ||||||
| Molecular dynamics | Structural information | Highest resolution | Elaborated calculation | 87 | ||
| Molecular dynamics | Coarse grain molecular dynamics | Structural information | Simpler than MD | Less accurate than MD | 88 | |
| Molecular dynamics | Dissipative particle dynamics | Structural information | Simpler than coarse grain | Less accurate than coarse grain | 89 | |
Among all methods, dynamic light scattering (DLS) is the most convenient technique for routine supplementary size and morphology measurements (resolution limit: 2 nm). DLS is based on the scattering of laser light at different intensity induced by the brownian motion of particles in solution. Velocity and therewith particle size is calculated from these intensity fluctuations, using the Stokes–Einstein relation.32 DLS is simple and fast, however care has to be taken when interpreting polydisperse samples, which are the case with most polymers and most preparation methods. Small-angle X-ray33 or neutron scattering (SAXS or SANS; resolution limit of both: 0.5 nm)34 provide detailed information about the polymersome bilayer,35 but due to the need for access to large scale radiation facilities, their use for routine measurements for their information is somewhat limited. With SAXS and SANS, particle shape and size information are collected by monitoring the elastic X-ray respectively neutron scattering at low angles (0.1–10°). X-ray interact with the electron clouds of molecules or elements, where neutrons interact with the nuclei.33,34 Another scattering method for polymersome permeability measurements is stopped-flow light scattering (SFLS). The mechanism behind SFLS is a rapid mixing of the polymersome solution with an osmotically active substance, called osmotic agent (usually sucrose or NaCl). The osmotic shock causes the polymersomes to change volume, resulting in changed light scattering, which is monitored by a photomultiplier tube collecting 90° angle scatter from the mixing chamber. Thus, osmotically induced polymersome shrinking leads to increased light scattering. Nanoparticle tracking analysis (NTA), a novel analysis method,36–38 combines scattering and visualization. This method assesses the hydrodynamic diameter of single particles in a bulk solution without influence of density or refractive index (RI) in contrast to DLS. Particle-induced scattered laser light is captured by a CCD camera, where each particle is tracked separately. Their size is again calculated by the Stokes–Einstein relation.36 To our knowledge this is the first publication, where polymersomes have been analyzed using the NTA technique.
Generally, electron microscopy (EM), especially transmission electron microscopy (TEM, resolution limit: 0.1–0.5 nm) is the most frequently used in-depth analysis technique for polymersome size, morphology, lamellarity or bilayer thickness. Almost all studies on polymersomes and other self-assembly morphologies are based on TEM. Image formation in TEM is based on sample interaction of electrons transmitting through a thin sample slice. Regions of high electron density in the sample (strong interaction) appear as black, whereas regions of low electron density (low interaction) are white.90 The same holds for scanning electron microscopy (SEM), but here the electrons are not passing the sample but interact and excite sample atoms that emits so-called secondary electrons giving information about the sample electron density dependent on their energy.91 To improve contrast, samples are stained with electron-rich heavy metal atoms, gathering around particles and emphasizing their shape in the image (negative staining, NS). The great drawback is that EM (with exception of environmental scanning electron microscope, ESEM92) only works under vacuum conditions. To overcome vacuum-induced sample deformation, the sample is quick-frozen in liquid alkanes in order to capture them in original shape in liquid solution and observed at −180 °C (Cryo-SEM or -TEM). Optionally the sample can be fractured to reveal the particle interior (freeze fracture, FF-SEM or -TEM).93 Optical microscopy can visualize polymersomes in their native environment however the main size dimension of interest (nm range) is below the diffraction limit of photons. There are two fluorescence-mediated optical microscope techniques used in this study: confocal laser scanning microscopy (CLSM) and generalized polarization microscopy (GPM). CLSM can maximize image resolution within the diffraction limit by optical sectioning, where only focal plane ”sample slices” are taken sequentially and in this case visualized by fluorophore laser excitement.94 GPM is based on the emission of the polarity sensitive fluorophore 6-lauroyl-2-(dimethylamino)-naphthalene (Laurdan) that exhibits a red shift with increasing environment polarity.95 To overcome the diffraction limit, super-resolution microscopes, mainly based on fluorescence signaling, has been developed.62 However, the required use of fluorophores comes with additional mixing and purification steps, limiting the use for routine measurements.
Besides EM, atomic force microscopy (AFM, resolution limit: 1 nm) has become a versatile tool for routine measurements, especially on size and topographic information. AFM utilizes an elastic lifting arm (cantilever) with a microscopic tip scanning the sample at small distance using a piezoelectric device. Generally, when the tip interacts with the sample, the cantilever bends and this is monitored by a change of the laser reflection on the cantilever surface, resulting in topographical information about the sample.96 There are various operation modes. The greatest advantage and disadvantage at the same time is the sensitivity of the cantilever tip. It enables atomic resolution imaging, but is prone to vibration noise and undesired sample interactions.80,97 All mechanical-based manipulation techniques (tether pulling,98 micropipette aspiration99 and falling-ball viscosimetry100) can be used complementary to AFM but are usually limited to micrometer-sized polymersomes.79 Micropipette aspiration, used in this study, provides information about the elastic properties of particles by micropipette suction. Here, the particle surface is aspirated into a micrometer-sized glass tube while the leading edge of its surface is monitored.99 Other bilayer properties such as lamellarity, polarity and zeta potential can be obtained as well by utilization of the electron or nuclear spin after applying an electrical field (electron paramagnetic resonance (EPR)),72,76,101 nuclear magnetic resonance (NMR)72,76,102 or laser Doppler electrophoresis (LDE). The latter method, employed in this study, is based on particle movement due to particle charge interaction with an applied electric field. The Doppler shift of particle scattered laser light is used to calculate particle velocity (equally to DLS) and zeta potential using the Smulochowski model.78
Sorting analysis tools such as flow cytometry (FCM), size exclusion chromatography (SEC) and asymmetric flow field-flow fractionation (A4F) are usually combined with light scattering. FCM103 is used mainly for cell analysis (as well as the most fluorescence microscopes). The main drawback of FCM is the limited detection level which makes detection challenging for polymersome diameter (dP) less than 300 nm. SEC104 is the most well-established sorting technique, but suffers from polymer adsorption on the column material. A4F105 has the advantage of separating a wide range of particle size but requires an expensive setup.
Here we attempt to provide a comparative and representative overview of a total of 17 analyzing techniques for polymersomes of a conventional chemistry for polymersome formation (polybutadiene polyethylene oxide, PB–PEO, chemical structure in Fig. 2)6,106–108 prepared with a conventional formation method (film rehydration).8,109,110
So far there has not been an experimental comparative study on polymersome analysis methods to the extent presented here. Another comparative study with six methods of all families was done by Till et al.,49 however more with a focus on comparing A4F to the other methods than on comparison among them. For liposomes several analytical tools have been investigated,53,60 where CLSM stands out as a particularly popular method. There have also been extensive studies related to specific types of analysis such as EM,64,111 fluorescence microscopy,112 mechanical manipulation,79,80 single particle techniques like NTA,37 AFM40,113 and scanning tunneling microscopy (STM).70 Comparative analysis reviews have been presented for liposomes,39,54 whereas for some polymersome reviews, characterization methods generally only constitute a smaller part of the review.8,55
This work is a step towards integrating all these studies in a more broad perspective. Precision and reliability of a single method can be differentiated better when compared to other methods. A comparison of 17 methods should provide a facilitated insight and also help in deciding when it may be desirable to switch from a fast technique to an in-depth technique. Results here are based upon polymersomes, however in terms of dP analysis, it also pertains to liposome analysis.
 | ||
| Fig. 2 Chemical structure of polybutadiene–polyethylene oxide (PB–PEO). PB, the hydrophobic has a blue background, PEO, the hydrophilic polymer has red background. | ||
We first analyze methods for determining dP of PB–PEO polymersomes where DLS, NTA, TEM, NS-TEM, Cryo-TEM, FF-TEM, SEM, FF-Cryo-SEM, CLSM and AFM will be compared. This discussion will be followed by a polymersome lamellarity respectively multivesicularity and bilayer or hydrophobic core thickness (tP) analysis using Cryo-TEM, AFM, SAXS and SANS. Multilamellar polymersomes are concentric polymersome, where multivesicular polymersomes are randomly encapsulated polymersomes that do not share a common centre. Finally, elastic properties and permeability of the bilayer will be compared using micropipette aspiration and SFLS, whereafter surface charge potential analyzed by LDE and polarity by GPM with Laurdan-labeling will be briefly introduced. Polarity measurements can help to obtain information about the hydrophobic barrier properties, which are essential in drug delivery applications.16 All measurements were performed on polymersomes formed from PB33–PEO18 except polarity experiments (PB12–PEO9, PB22–PEO23, PB46–PEO23, PB46–PEO30). All measurements on dP, lamellarity, tP, zeta potential and polarity were repeated with three independent samples and for all direct measurements (All EM, CLSM and AFM) 100 polymersomes per sample were measured. Micropipette aspiration and SFLS were performed with two independent samples.
2 Material & methods
Polymer synthesis
PB33–PEO18 was synthesized using anionic polymerization.106 Butadiene (Bd) and ethylene oxide (EO) monomers were purified and afterwards dried using liquid nitrogen and distilled over n-dibutylmagnesium (n-Bu2Mg) and n-butyl lithium (n-BuLi) to remove any traces of water. Tetrahydrofuran (THF) as the synthesis solvent was purified and dried afterwards via reflux and stirring over sodium in paraffin and benzophenone.Bd was polymerized first. THF was first introduced in a pre-dried reactor and cooled to −40 °C. Afterwards, Bd and n-BuLi were added and the reaction was allowed to run for 4 h at −20 °C. The polybutadienyl lithium appeared in a orange or yellow colour. The mixture was afterwards cooled again to −40 °C and approximately 1 ml of EO was added to end-cap the polymerization, visible by the disappearance of the yellow colour. Afterwards, the reaction was left for 1.5 h at −40 °C, where a precursor was drawn and analyzed via SEC on a SIL-10AD from Shimadzu, Kyoto, Japan. The SEC system consisted of a Shimadzu LC-10AD high performance liquid chromatography (HPLC) pump together with a Viscotek Differential Refractometer model 200 and the following columns: 5 cm Polymer Laboratories, Guard column 3 μm, 30 × 7.8 cm Waters Styragel HMW 6E, and 30 × 7.5 cm Polymer Laboratories, PLgel 5 μm Mixed-D. The columns were thermostatted at room temperature (RT) during measurement. Non-stabilized THF was used as eluent, and the system was calibrated with polystyrene standard samples having very narrow molar mass distributions. For EO polymerization, the remaining EO was added and 1-tert-butyl-4,4,4-tris(dimethylamino)-2,2-bis[tris(dimethylamino)-phosphor-anylidenamino]-2λ5,4λ5-catenadi(phosphazene) (tBuP4) was injected afterwards in a molar n-BuLi![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) tBuP4 ratio of 1
tBuP4 ratio of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 to ensure exact stoichiometry.106 The reaction was heated afterwards to 40 °C to start EO chain propagation. Finally, the reaction was quenched with acetic acid after two days. The polymer was precipitated in cold acetone and vacuum dried. Bd-EO stoichiometry was subsequently analyzed by NMR at 300 or 400 MHz, whereafter polydispersity index PDIM was analyzed via SEC. The polymer was stored at −20 °C until use. PB12–PEO9, PB22–PEO23, PB46–PEO23, PB46–PEO30 (only used for the polarity measurement using GPM) were purchased from Polymer source, Montreal, Canada. All polymers are listed in Table 2.
1 to ensure exact stoichiometry.106 The reaction was heated afterwards to 40 °C to start EO chain propagation. Finally, the reaction was quenched with acetic acid after two days. The polymer was precipitated in cold acetone and vacuum dried. Bd-EO stoichiometry was subsequently analyzed by NMR at 300 or 400 MHz, whereafter polydispersity index PDIM was analyzed via SEC. The polymer was stored at −20 °C until use. PB12–PEO9, PB22–PEO23, PB46–PEO23, PB46–PEO30 (only used for the polarity measurement using GPM) were purchased from Polymer source, Montreal, Canada. All polymers are listed in Table 2.
| Polymer | Mn [g mol−1] | PDIM | f |
|---|---|---|---|
| PB12–PEO9 | 1050 | 1.09 | 0.319 |
| PB33–PEO18 | 2561 | 1.087 | 0.251 |
| PB22–PEO23 | 2200 | 1.09 | 0.388 |
| PB46–PEO23 | 3500 | 1.09 | 0.233 |
| PB46–PEO30 | 3800 | 1.04 | 0.284 |
Polymersome formation via film rehydration
PB–PEO and 1,2-dioleoyl-sn-glycero-3-phosphocholine (DOPC, Avanti Lipids, Alabaster, USA) was dissolved in CHCl3 to create a 10 mg ml−1 polymer suspension. Afterwards, it was sonicated for 5 min and stored at −20 °C until use. 2.5 ml of the stock solution was injected in a 5 ml round flask and put afterwards on a rotary evaporator for at least 2 h at room temperature and 2 mbar at a rotation speed of 120 rpm to evaporate CHCl3. The polymer was then present as a smooth film on the flask wall. Afterwards, the sample was rehydrated with 200 μl of tris buffer (10 mM tris pH 8.0, 50 mM NaCl) with 13 mg ml−1 n-octyl-β-D-glucopyranoside (OG) and left stirring at least overnight at RT. The sample was diluted with 800 μl tris buffer (polymersome concentration 25 mg ml−1), whereafter 20 mg SM2 biobeads (Bio-Rad, Hercules, USA) were added to remove the OG between the polymersome bilayer.52 Afterwards, the sample was left on a shaker with 200 rpm for 3 h at RT, whereafter another 20 mg of biobeads were added. The sample was then left overnight shaking with 200 rpm at 4 °C. Biobeads were removed using a squeezed syringe. Polymersomes prepared by this procedure were used for all analysis techniques except GPM analysis, they were used though for CLSM.DLS
A Nano Zetasizer (Malvern, Worcestershire, UK) was used for DLS experiments. PB33–PEO18 polymersomes in tris buffer was injected (1 ml, 0.025 mg ml−1) in a disposable cuvette and subsequently measured three times with 6 runs of 10 s per measurement at RT.NTA
For NTA analysis, 0.025 mg ml−1 PB33–PEO18 polymersomes in tris buffer were introduced manually in the analysis chamber of the NTA analysis instrument LM10 (Nanosight, Wiltshire, UK) equipped with a laser of wavelength 638 nm and the NTA software version 3.0. For each samples, three videos of 60 s were recorded with a camera level of 11 and a frame rate of 30 frames per s. The videos were analyzed with a detection threshold of 4 (blur size and minimum track length: auto).SAXS
Prior to SAXS measurement, 20 mg ml−1 PB33–PEO18 polymersomes in tris buffer were dialyzed 1.5 days with a 300 kDa 1 ml Float-a-lyzer (VWR, Herlev, Denmark) and subsequently extruded 20 times through track-etched polycarbonate membranes with 200 nm pore size. SAXS measurements were performed at the BioSAXS beamline BM29 at the European Synchrotron Radiation Facility (ESRF) in Grenoble, France, and again using the U-SAXS beamline ID02 also at ESRF two days later. Scattering intensity was measured as a function of the magnitude q of the scattering vector given as q = 4π![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) sin(θ)/λ where 2θ is the scattering angle and λ is the wavelength of the incoming radiation. The setting at BM29 covered a q-range from 0.04 to 5 nm−1 and the ultra small-angle setup at ID02 covered the range from 0.002 to 0.25 nm−1 giving a combined q-range from 0.002 to 5 reciprocal nm with a substantial overlap. Data were background subtracted and radially averaged using the standard software at the beamlines. Absolute calibration was done using water as a known scattering standard.
sin(θ)/λ where 2θ is the scattering angle and λ is the wavelength of the incoming radiation. The setting at BM29 covered a q-range from 0.04 to 5 nm−1 and the ultra small-angle setup at ID02 covered the range from 0.002 to 0.25 nm−1 giving a combined q-range from 0.002 to 5 reciprocal nm with a substantial overlap. Data were background subtracted and radially averaged using the standard software at the beamlines. Absolute calibration was done using water as a known scattering standard.
SANS
Prior to SANS measurement, 20 mg ml−1 PB33–PEO18 polymersomes in deuterated tris buffer were dialyzed 1.5 days with a 300 kDa 1 ml Float-a-lyzer (VWR, Herlev, Denmark), and subsequently extruded 20 times through track-etched polycarbonate membranes with 200 nm pore size. SANS measurements were performed at the Forschungs-Neutronenquelle Heinz Maier-Leibnitz (FRM II), in Munich, Germany. Measurements were performed at three different settings to obtain a large q-range. Sample-detector distances were 1.27, 4 and 8 meter with corresponding collimation lengths of 4, 4, and 8 meter. These settings covered the q-ranges [0.35![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 4.5] nm−1, [0.12
4.5] nm−1, [0.12![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1.6] nm−1 and [0.057
1.6] nm−1 and [0.057![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0.77] nm−1. Data were background subtracted and radially averaged using the standard software at the beamline. Absolute calibration was performed using a plexiglass standard.
0.77] nm−1. Data were background subtracted and radially averaged using the standard software at the beamline. Absolute calibration was performed using a plexiglass standard.
SFLS
A SFM-300 (BioLogic, Claix, France) with a Xe–Hg lamp was used to measure shrinking and swelling of polymersomes. The measured data was fitted to an exponential rise equation to calculate the water permeability of the bilayer Pf, using the following expression:114
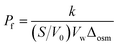 | (1) |
TEM
Eight micro liters MilliQ water were placed on a glow-discharge 400 mesh holey carbon copper grid (Agar scientific, Essex, UK) and blotted off, followed by 3.5 μl of 2.5 mg ml−1 PB33–PEO18 polymersomes in tris buffer that was allowed to adsorb for 2 min. The grid was loaded with another 8 μl MilliQ water to wash out the remaining salt. The grid was placed in CM100 TEM (Philips, Amsterdam, Netherlands) the same day. This TEM has an installed Veleta 2k CCD camera (Olympus, Shinjuku, Japan). The applied voltage on a tungsten source was 80 kV with a 100 μm objective lens aperture.NS-TEM
Eight microliters MilliQ water were dropped on a glow-discharge 400 mesh holey carbon copper grid (Agar scientific, Essex, UK), blotted off and rehydrated twice with 8 μl phosphor tungsten acid. This procedure was followed by injecting 3.5 μl of 2.5 mg ml−1 PB33–PEO18 polymersomes in tris buffer with adsorption time of 2 min, followed by washing with 8 μl MilliQ water. Finally, 8 μl phosphor tungsten acid were adsorbed for 0.5 min and blotted off afterwards. The grid was placed directly afterwards in the same TEM with the same parameters as for the TEM analysis. Size analysis of the TEM pictures was performed by manual measurement using the image analysis software Gimp 2.8.Cryo-TEM
Three micrometers of 25 mg ml−1 PB33–PEO18 polymersomes in tris buffer was admitted to a glow-discharged 300 mesh holey carbon formvar copper grid (Agar scientific, Essex, UK), mounted on a Vitrobot mark 5 (FEI, Hillsboro, USA). After removal of excessive sample by automated blotting, the sample was rapidly frozen by being plunched into liquid ethane and subsequently cooled down further to approximately −174 °C. The grid was then moved to the Cryo transporter system and mounted in the Gatan cryoholder (FEI, Hillsboro, USA). The sample was observed with a Tecnai G2 200 kV with a 4 × 4k CCD eagle camera, both (FEI, Hillsboro, USA). The applied voltage on applied voltage on a LaB6 source was 200 kV. Size analysis of the TEM pictures was performed by manual measurement over Gimp. Lamellarity measurements was done by manual counting uni- and multilamellar polymersomes at several images per sample with a reasonable amount of polymersomes.FF-TEM
FF was performed on a MED020 with EM VCT100 shuttle attached (Leica, Wetzlar, Germany). 1.2 μl of 25 mg ml−1 PB33–PEO18 polymersomes in tris buffer was injected into a 3 mm aluminium sample carrier at the side with 300 μm depth. Afterwards, another one was placed on top with the 200 μm depth side, but care had to be taken to avoid air bubbles in between. This sandwich was then plunged into liquid ethane for 20 s and then immediately in liquid N2. The sample carrier was afterwards fixed at the sample holder and introduced in a high vacuum chamber (2 × 10−6 mbar) at −140 °C. After the lower sample carrier had been removed, the sample was coated at the same temperature with 2 nm carbon, afterwards by 4 nm platinum with 45° tilted and finally with a 19 nm carbon protection layer without tilt. Outside of the chamber, the carrier was thawed for 5 min at RT, whereafter it was carefully placed at 45° angle into a 200 μl bath of tris buffer with 100 mg ml−1 OG for 5 min for solubilizing the polymersomes. Finally, the removed replica or single pieces of it were placed on uncoated 400 mesh copper grid that were as well carefully placed in the bath at 45°.The grid was observed in the same way, as described in the TEM analysis section. Size analysis of the TEM pictures was performed by manual measurement over Gimp, using a correction factor of 4/π to balance out the diameter error when the fracturing is not in equatorial plane.64
SEM
PB33–PEO18 polymersomes in tris buffer with concentrations 25 mg ml−1, 2.5 mg ml−1 and 0.25 mg ml−1, respectively, were dropped on a SEM holder with carbon tape on and left overnight for air drying were dropped on a SEM holder with carbon tape on and left overnight for air drying. The holder was then placed in a Quanta FEG 3D SEM (Philips, Amsterdam, Netherlands) with 5 kV electrons, spot size 1 and 30 μm objective lens aperture. Size analysis of the SEM pictures was performed by manual measurement over Gimp.FF-Cryo-SEM
FF was performed on a MED020 with EM VCT100 shuttle attached (Leica, Wetzlar, Germany). 1.2 μl of 25 mg ml−1 PB33–PEO18 polymersome in tris buffer was injected into a 3 mm aluminium sample carrier at the side with 300 μm depth. Another one was placed on top with the 200 μm depth side. This sandwich was then plunged into liquid ethane for 20 s and then immediately in liquid N2. The sample carrier was afterwards fixed at the sample holder and introduced in a high vacuum chamber (2 × 10−6 mbar) at −110 °C. After the lower sample carrier had been removed, the sample was left for sublimation at −110 °C for 1 min. The sample was subsequently coated at the same temperature with 8 nm platinum with 45° tilted and finally with a 19 nm carbon protection layer without tilting. The transfer chamber was then mounted at the cooling stage of the Quanta FEG 3D SEM (Philips, Amsterdam, Netherlands) and introduced in the vacuum chamber. The SEM was operated as described in the SEM analysis section. Size analysis of the SEM pictures was performed by manual measurement via Gimp, using a correction factor of 4/π.64 Lamellarity measurements were done by manual counting uni- and multilamellar polymersomes on at least three images per sample with a reasonable amount of polymersomes.CLSM
Five micrometers of 25 mg ml−1 PB33–PEO18 polymersomes in tris buffer, labelled with Coumarin 6 (Sigma-Aldrich, Brøndby, Denmark), were placed inside a Zeiss LSM 710 confocal laser scanning microscope (Carl-Zeiss, Jena, Germany) with a 63× 1.4NA oil objective. Sections of different focus area were taken, with 10 images per section. dP was measured automatically from the LSM 710 software.GPM
Vesicles of PB12–PEO9, PB22–PEO23, PB46–PEO23, PB46–PEO30 and DOPC were prepared either via gentle film rehydration58 or electroformation.115 In gentle film rehydration, 10 mg ml−1 amphiphiles in CHCl3 and glucose from a 1 mg ml−1 stock in methanol at a molar ratio of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 as well as 16 μl Laurdan (Invitrogen, Carlsbad, USA) were injected in glass vial. The sample was dehydrated using blow-drying with nitrogen flow that a film appeared on the glass wall and dried on vacuum for 3–12 h. MilliQ water was added carefully to the sample without shaking the sample. The solution was left for 12 h in the dark at RT. Electroformation was done in a VesiclePrepPro chamber (Nanoion, Munich, Germany). Ten mg ml−1 amphiphiles in CHCl3 and Laurdan were dehydrated on a indium tin oxide (ITO) coated glass slide for 1 h at RT. A greased o-ring was put around the dehydrated spot. MilliQ water was injected into the space in order to give a final amphiphile concentration of 0.1 mg ml−1. Another ITO coated slide was placed onto the ring with care not to produce air bubbles. The VesiclePrepPro chamber was then closed and connected to the station. Vesicles were formed at 3 V, 5 Hz over a period of 2 h at 36° C. The formed vesicle were put in a Eppendorf tube without exposure to light.
1 as well as 16 μl Laurdan (Invitrogen, Carlsbad, USA) were injected in glass vial. The sample was dehydrated using blow-drying with nitrogen flow that a film appeared on the glass wall and dried on vacuum for 3–12 h. MilliQ water was added carefully to the sample without shaking the sample. The solution was left for 12 h in the dark at RT. Electroformation was done in a VesiclePrepPro chamber (Nanoion, Munich, Germany). Ten mg ml−1 amphiphiles in CHCl3 and Laurdan were dehydrated on a indium tin oxide (ITO) coated glass slide for 1 h at RT. A greased o-ring was put around the dehydrated spot. MilliQ water was injected into the space in order to give a final amphiphile concentration of 0.1 mg ml−1. Another ITO coated slide was placed onto the ring with care not to produce air bubbles. The VesiclePrepPro chamber was then closed and connected to the station. Vesicles were formed at 3 V, 5 Hz over a period of 2 h at 36° C. The formed vesicle were put in a Eppendorf tube without exposure to light.
Vesicles were observed with a Varian Cary eclipse fluorescence spectrometer (Varian, Palo Alto, USA) with excitation wavelength of 380 nm and emission recorded in the range of 400–700 nm. Pictures were taken at 40× magnification with spectrometer-supporting software AxioVision. Picture processing and GP analysis of Laurdan were performed with ImageJ (NHI, Bethesda, USA).58
AFM
Ten microliters of 0.25 mg ml−1 PB33–PEO18 polymersomes in tris buffer were dropped on a piece of silicon wafer (Topsil, Poland, 1 × 1 cm2, p-doped, single sided polish) pre-coated with N-(6-aminohexyl)aminopropyltrimethoxysilane (92%, AB110807, abcr GmbH, Karlsruhe, Germany). After 1 min of absorption of the vesicle on the support, about 500 μl of MilliQ water was dropped using a pasteur pipette. Using a tissue, the liquid was absorbed. The procedure was repeated three times. After the last washing step, the silicon wafer was blown dried with air and directly placed in a XE-150 AFM (Park Systems, Suwon, South Korea). Polymersomes were imaged in non-contact tapping mode using the AFM system. The acquisition program was XEP 1.7.70 (Park Systems, Suwon, South Korea). The image size was 5 × 5 μm2 with a resolution of 256 × 256 pixel. The AFM probe (Tap300Al-G, BudgetSensors, Sofia, Bulgaria) had a force constant of 40 N m−1, a tip radius of 10 nm and a resonance frequency of 300 kHz. The images were leveled in x- and y-direction and dP was obtained using the threshold method available in the analysis software XEI 1.8.0 (Park Systems, Suwon, South Korea).Micropipette aspiration
Some drops of approximately 0.25 mg ml−1 PB33–PEO18 polymersomes in tris buffer were injected in a glass specimen chamber. Observation was performed using an Axioconvert 100 (Carl Zeiss, Jena, Germany) with 100× magnification of oil immersion objective. A micropipette with approximately 1.9 μm inner diameter was introduced in the specimen chamber, whereafter a polymersome (dP 2.5–3 μm) was soaked by the suction pressure controlled pipette. During the micropipette manipulation experiment, live-images with CCD camera (DAGE-MIT, Michigan City, USA) and pressure with pressure transducer (Validyne Engineering, Northridge, Canada) were monitored & recorded on a computer by using a LabVIEW program. The elastic modulus properties, elastic-area compressibility modulus Ka and the bending elasticity modulus kc were calculated from the geometrical shape of the micropipette inner diameter, diameter of the spherical segment of the polymersomes exterior to the pipette and projection length.80,116,117LDE
Zeta potential measurements were performed using the Zetasizer (Malvern, Worcestershire, UK). One milliliter of 25 mg ml−1 PB33–PEO18 polymersomes in tris buffer were injected in a zeta potential cuvette carefully to avoid air bubbles. The number of runs, depending on the quality of the results, varied between 20 and 100. All measurements were done at 25 °C.3 Results & discussion
Size
Size analysis was performed with DLS, NTA, TEM, NS-TEM, Cryo-TEM, FF-TEM, SEM, FF-Cryo-SEM, CLSM and AFM. Distributions of dP from EM-related techniques are shown in Fig. 3a, while results from the other techniques are depicted in Fig. 3b. Most dP measurements are in the range of 0 to 300 nm, whereas there are two peaks between 400 and 500 nm for the CLSM measurements. Small peaks in the 100–300 nm dP range are observed with AFM, Cryo-TEM and FF-TEM. With NTA, DLS and FF-Cryo-SEM the highest peaks are in the range between 150–250 nm and for NS-TEM the highest peak was below 50 nm.The size distribution for DLS was performed with the focus on minimizing the known drawback of DLS size analysis i.e. the bias towards few larger aggregates or polymersomes as these scatter significantly more. The original intensity size distribution curve based of the Rayleigh scattering intensity was converted to volume size distribution. The intensity of Rayleigh scattering I has a relationship to the particle diameter d of I ∼ d6, meaning that a particle of 1000 nm diameter will scatter one million times more than a particle of 100 nm diameter. Volume size distribution takes Mie scattering into account, where the relation between diameter and scattering intensity is more realistic.118 A further drawback of DLS is related to how the algorithm used to extract size (diameter) information, deals with polydisperse samples. The usual algorithm for obtaining the intensity-weighted hydrodynamic dP (ZD) and the polymersome polydispersity index ( , where σ is the distribution width of dP119), is cumulant analysis. However for PDIDLS above 0.25, the non-negatively constrained least squares (NNLS) should be applied according the manufacturers recommendations. This algorithm provides higher correction, stabilizing and weighting factors that, when combined, can extract more information out of the scattering intensity than the cumulant algorithm.120 Both algorithms are visualized in Fig. 4a with ZD (in the figure referred to as Z-average) and PDIDLS (in the figure referred to as PdI), derived from cumulant analysis, at the left side and the peak statistics, derived from NNLS on the right side and plotted in the diagram below. Since PDIDLS values larger than 0.25 can be expected for PB–PEO polymersomes prepared with film rehydration, we used volume size distribution values, derived from NNLS. Even though the first distribution peak at about 80 nm dP was in accord with several other analysis techniques like Cryo-TEM or AFM, two more broad peaks at 1500 nm and 5000 nm were observed (not shown). Although we used an algorithm which should minimize the bias towards larger polymersomes, it seemed that DLS was still not capable of giving a realistic size distribution of polydisperse samples.
, where σ is the distribution width of dP119), is cumulant analysis. However for PDIDLS above 0.25, the non-negatively constrained least squares (NNLS) should be applied according the manufacturers recommendations. This algorithm provides higher correction, stabilizing and weighting factors that, when combined, can extract more information out of the scattering intensity than the cumulant algorithm.120 Both algorithms are visualized in Fig. 4a with ZD (in the figure referred to as Z-average) and PDIDLS (in the figure referred to as PdI), derived from cumulant analysis, at the left side and the peak statistics, derived from NNLS on the right side and plotted in the diagram below. Since PDIDLS values larger than 0.25 can be expected for PB–PEO polymersomes prepared with film rehydration, we used volume size distribution values, derived from NNLS. Even though the first distribution peak at about 80 nm dP was in accord with several other analysis techniques like Cryo-TEM or AFM, two more broad peaks at 1500 nm and 5000 nm were observed (not shown). Although we used an algorithm which should minimize the bias towards larger polymersomes, it seemed that DLS was still not capable of giving a realistic size distribution of polydisperse samples.
 | ||
| Fig. 4 DLS & NTA analysis: (a) representative scheme of DLS measurement. ZD (here referred as Z-average) and PDIDLS on the left (here referred as PdI) are calculated using cumulant analysis, where peak statistics on the right and in the diagram below are based on NNLS. (b) Screenshot of video record of NTA analysis with PB33–PEO18 polymersomes with a concentration of 0.025 mg ml−1. It is crucial to obtain a balance in the camera exposure time, between too high, producing diffraction rings as seen on the right side38 and too low that smaller polymersomes will be left out. Scale bar is 200 μm. (c) Size vs. concentration of 0.025 mg ml−1 PB33–PEO18 polymersomes analyzed with NTA. The main dPs were in the range from 150 to 300 nm, where there were small peaks also between 400 and 600 nm. The differences between the measurements mirrors the polydispersity of the sample. The sharp peak of the first measurement (blue) could be due to automatic finite track length adjustment (FTLA). Inset: concentration distribution over diameter. | ||
NTA revealed one single broad peak with a maximum around 150 nm. This diameter is larger compared to the prevalent diameters coming out of the other techniques with the exception of CLSM. In contrast to DLS, NTA tracks single particles and therefore has a number-averaged size distribution, based on a significantly higher number of polymersomes than with any other of the studied techniques. According to the NTA experiment report, approximately a mean number of 126 polymersomes per frame were measured. With an assumed mean polymersome presence of 0.5 s in the laser spot (which is a reasonable assumption when watching the observation video, which can be found in the ESI†), 1800 frames per video and three videos per sample, a total of 136![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 polymersomes were identified and analyzed. This is 12 times larger than for the CLSM analysis and 1360 times larger than the manual measurements.
000 polymersomes were identified and analyzed. This is 12 times larger than for the CLSM analysis and 1360 times larger than the manual measurements.
However, several error sources are hidden in the interplay between the scattered light projections and the camera aperture. On the one hand, a larger aperture, integrates scattered light projections of polymersomes of all dP but could lead to oversaturation of the CCD camera due to the scattered light projection of the larger polymersomes. On the other hand, when using a lower aperture there is an increased risk of overseeing poorly scattered light projections from small polymersomes. Oversaturation of the camera leads to blurry projection and tracking errors (see Fig. 4b dotted circle and ESI†). This can result in an underestimation of the number of smaller polymersomes, as noted by van der Pol.53 Additionally, oversaturated scattered light projections of polymersome aggregates can erroneously be classified as one large polymersome. Another error source is the minimum track length, meaning the minimum number of frames, in which a polymersome is present. If the threshold for this length is set too high, small polymersomes are excluded, if it set too low, the sizing accuracy of individual polymersomes is reduced.38 Automatic adjustment of the track length (finite track length adjustment, FTLA) can result in reliable diameter determination,38 but for vesicles it can lead to undesirable narrowing of peak sizes.53 Therefore, care has to be taken when analyzing sharp peaks after using FTLA (Fig. 4c measurement 1). A third reason of a bias towards larger polymersomes could be that Brownian motion is in x, y and z-direction, however the camera only detects movement in the x- and y-plane. The z-directed movement of polymersomes could therefore be observed as artificially slow, leading to artificially greater dP.54 DLS that also measures over time, has the same error source. In the higher dP range between 400 and 600 nm, a small peak could be observed in the reports of the single samples Fig. 4c, which is consistent with the results from the CLSM measurements. In the Fig. 3b overview scheme, these peaks are not visible anymore due to their minimal contribution to the whole distribution compared to the smaller polymersomes. This did not occur in the case of the CLSM analysis, due to the diffraction limit-related cut-off of polymersomes with smaller dP than 200 nm.
Simple TEM without staining or freezing did not result in visible PB33–PEO18 polymersomes, even though this process was successfully applied, using polymersomes of other chemistries like polystyrene polyacrylic acid (PS-PAA).121 In previous experiments with PB12–PEO9, prepared by detergent-mediated film rehydration, polymersomes could be visualized directly with TEM. However for these vesicles, only empty grids were observed.
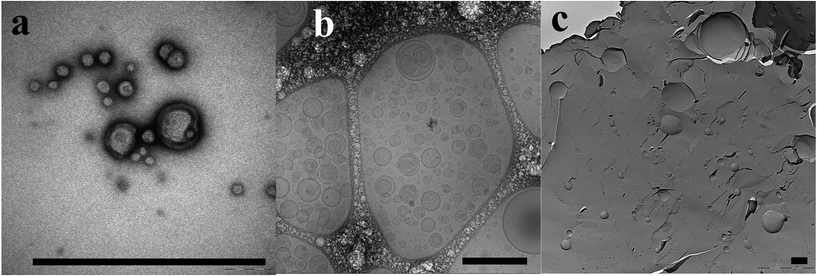
When observed with NS-TEM, polymersomes were visible on the grids. The majority was intact, as can be seen in Fig. 5a however some of them appeared ruptured and deflated. In contrast, PB12–PEO9 vesicles appeared mainly in collapsed form.52 This could be due to the smaller polymer Mw or the staining agent (phosphotungstic acid versus uranyl formate used in52). There were less polymersomes observed in the NS-TEM images compared to FF-TEM, Cryo-TEM or FF-Cryo-SEM due to the higher dilution, chosen since higher concentrations would break the holey carbon film of the TEM grid. In the size overview in Fig. 3a and b, NS-TEM analyzed polymersomes had dP values about 20 nm less than for polymersomes analyzed with AFM, Cryo-TEM and FF-TEM. The main reason behind this decrease was most probably osmotic shrinkage due to phosphotungstic acid, which will have a higher osmotic activity than the tris buffer in the polymersome lumen due to higher ion concentration. Large polymersomes (>300 nm) were not observed. This could be due to chance, as far as only regions of the whole grid were observed. However, different regions were observed per sample, so the chance of missing out larger polymersomes (which could be observed with other EM techniques and CLSM) was low. On the other hand, larger polymersomes would be less stable towards osmotic shrinkage due to staining and drying involved in the NS-TEM sample preparation. This argument is consistent with the occasional observation of polymer bilayer membrane fractions that were found on some images (see ESI†).
Cryo-TEM revealed polymersome dPs in the range from below 10 nm up to almost microns, including highly multilamellar ones (see Fig. 5b and ESI†). The samples were highly heterogeneous, even within individual samples local accumulations of small and large polymersomes could be observed (see ESI†). As shown in Fig. 3a, the majority of polymersome dPs were around 50 nm, though some polymersomes of dP between 200 and 400 nm could be observed.
The plunge-freezing of the polymersomes can be seen as taking a ”snapshot” of the polymersomes in their native state. Due to the fact that the freezing rate of liquid ethane is quite high (12![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 K s−1),122 freezing artifacts can be assumed to be negligibly small. For small and monodisperse samples, this technique can enable an undistorted direct imaging and quantification of the particle diameter, leading to realistic number-weighted size distributions, as stated by Egelhaaf.111 For larger particles such as polymersomes, deformation effects could occur due to thickness fluctuations in the vitrified water layer. This layer is thinner towards the middle of a hole in a carbon holey grid. These thickness fluctuations could be of the same order of magnitude as the polymersome dP, resulting in overestimation or underestimation of dP.111 For mainly larger polymersomes (dP > 500 nm), photon-mediated techniques like CLSM would therefore be the more suitable analysis technique. In contrast to NS-TEM, no staining or drying steps are involved in the Cryo-TEM preparation that could destabilize larger polymersomes. For this reason and due to the higher concentration compared to NS-TEM discussed before, larger polymersomes could be observed, as can be seen in the ESI.†
000 K s−1),122 freezing artifacts can be assumed to be negligibly small. For small and monodisperse samples, this technique can enable an undistorted direct imaging and quantification of the particle diameter, leading to realistic number-weighted size distributions, as stated by Egelhaaf.111 For larger particles such as polymersomes, deformation effects could occur due to thickness fluctuations in the vitrified water layer. This layer is thinner towards the middle of a hole in a carbon holey grid. These thickness fluctuations could be of the same order of magnitude as the polymersome dP, resulting in overestimation or underestimation of dP.111 For mainly larger polymersomes (dP > 500 nm), photon-mediated techniques like CLSM would therefore be the more suitable analysis technique. In contrast to NS-TEM, no staining or drying steps are involved in the Cryo-TEM preparation that could destabilize larger polymersomes. For this reason and due to the higher concentration compared to NS-TEM discussed before, larger polymersomes could be observed, as can be seen in the ESI.†
During FF-TEM, the freeze-captured polymersomes were fractured and coated, where the replica from this coating is observed, separated from the original polymersomes. The great advantage of FF-TEM over Cryo-TEM is the three-dimensional appearance of the micrographs, making it desirable for polymersome surface structure research with higher resolution than what can be obtained with FF-Cryo-SEM. The sample can also be observed at any time after preparation, whereas for Cryo-TEM samples should be observed directly afterwards and always be maintained at cryogenic temperatures. Even so there is still a risk of melting the samples under the electron beam, thus observation time can be limited. A disadvantage with FF-TEM is the need for separation of the thawed polymersomes from their carbon-platinum copy. Even with minimal agitation and highest possible lateral stability the replica is prone to damage during solubilization of the polymersomes in detergent. Additional forces are applied when the replica is removed from the detergent solution and dried on a copper grid. The weakest point of the replica is the polymersome fracture plane,123 thus if replicas break, they usually break at the edge of larger polymersomes, as can be seen in Fig. 5c or in areas of high polymersome accumulation (see ESI†). This could lead to an underestimation of the number of larger polymersomes, compared to FF-Cryo-SEM that showed significantly more polymersomes above dP of 400 nm, even though the preparation in terms of freeze fracturing was the same. An additional drawback for size analysis with both methods is that the fracture plane is not going through the equatorial plane of the polymersome. Two studies from Coldren et al. and Egelhaaf et al. have addressed this problem and made several suggestions for how to align FF-TEM and Cryo-TEM size distribution analyses.64,111 Coldren et al. found a correction factor of 4/π, which we applied here.64 However, both studies were performed on monodisperse liposome or surfactant vesicles and Egelhaaf et al. reported an effect on the observed dP depending on polydispersity.111 They furthermore found an underrepresentation of small liposomes due to the higher probability of larger liposomes to get fractured and due to invisibly small caps or cavities from small liposomes. This could explain the lower number of polymersome with dP 30–50 nm compared to NS-TEM and Cryo-TEM in Fig. 3a. For polydisperse samples Egelhaaf recommends a larger amount of analyzed vesicles.111 Thus, if more than the 300 polymersomes had been analyzed, the size distribution may have become more similar to the measured size distribution of Cryo-TEM. Automatic imaging software (e.g. ImageJ or similar programs) would be challenged by the low contrast between polymersomes and background, and manual analysis may be required to get good statistics.
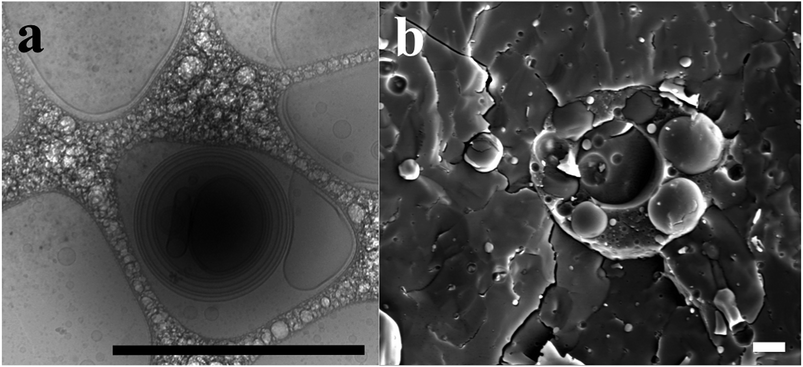
Regarding SEM, we analyzed two preparation approaches: one based on simple dropping of polymersomes onto a SEM holder with air-drying and subsequent observation, and another using freeze-fracturing as in FF-TEM but with observation of the coated sample directly instead of observing the separated replica (FF-Cryo-SEM). The first approach gave surprisingly good agreement for polymersomes with dP > 400 nm, as shown in Fig. 3a. However for dP < 400 nm, the majority of polymersomes had dPs where no other size measurement technique had any significant dP peaks. Upon inspection dehydrated cavities of polymersome shells were observed (Fig. 6a) and for large polymersomes there was enough material to deform the underlying carbon tape on the SEM holder, whereas for smaller polymersomes this was not possible, thus they remained invisible.
The size distribution of FF-Cryo-SEM was significantly shifted towards larger polymersomes compared to Cryo-TEM and AFM, less shifted compared to FF-TEM and in good agreement with NTA analysis. This could be related to the resolution limit, which was not 2 nm in all images for analysis.67 Magnifications >25 000× resulted in a higher electron density per area and the samples started to melt. At 50![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000× magnification, it was difficult to focus due to the rapid melting and cracks appearing on the surface (see the ESI†). Polymersomes with dP < 200 nm were hardly visible at lower non-invasive magnification, similar to the observations with NTA, when the scattering intensity was too low. NTA and FF-Cryo-SEM could therefore be ideal size analysis tools for filling the dP gap between Cryo-TEM, FF-TEM or AFM on the one side (where size limitation for micron-sized polymersomes could occur due to deformation effects at Cryo-TEM or AFM and replica fragilities at FF-TEM) and photon-mediated techniques on the other side, where size limitation for polymersomes with dP < 200 nm could occur due to the diffraction limit.
000× magnification, it was difficult to focus due to the rapid melting and cracks appearing on the surface (see the ESI†). Polymersomes with dP < 200 nm were hardly visible at lower non-invasive magnification, similar to the observations with NTA, when the scattering intensity was too low. NTA and FF-Cryo-SEM could therefore be ideal size analysis tools for filling the dP gap between Cryo-TEM, FF-TEM or AFM on the one side (where size limitation for micron-sized polymersomes could occur due to deformation effects at Cryo-TEM or AFM and replica fragilities at FF-TEM) and photon-mediated techniques on the other side, where size limitation for polymersomes with dP < 200 nm could occur due to the diffraction limit.
Images of FF-Cryo-SEM revealed high amounts of multivesicular and a few multilamellar polymersomes, as can be seen in Fig. 6b. The same correction factor used for FF-TEM was also applied at FF-Cryo-SEM, because both have the same fracture plane issue, discussed earlier. The size distribution of FF-Cryo-SEM was shifted towards higher dP due to the resolution limit mentioned before but also because there was no sample fragility as in the case of FF-TEM. Especially in regions with multivesicular polymersomes as in Fig. 6b there were plenty of cavities where no stabilizing carbon was present at the surface.123 Consequently, these regions broke apart during FF-TEM preparation and could not be visualized in the TEM anymore. Regions with lower accumulations of polymersomes that usually contained fewer large, multilamellar or -vesicular polymersomes (see ESI†) provided a more even surface and thus a higher lateral stability of the replica. Those regions were dominant at FF-TEM, whereas at FF-Cryo-SEM, regions with high and low polymersome accumulation were equally distributed.
Of the methods tested, CLSM was the only direct visualization method that allowed observation in completely undisturbed native environment. CLSM revealed Coumarin 6-labelled polymersomes, where the Coumarin 6 mainly filled the surface completely at the smaller polymersomes, however the lumen could be seen for a few larger polymersomes or tubular structures, visible in Fig. 7a (further images and a video can be found in the ESI†). There were few polymersomes with approximately dP of 330 nm, two peaks between dP of 400 and 500 nm, and a few polymersomes with dP between 500 and 2000 nm. The ones above dP 700 nm were not shown in Fig. 3b for clarity purposes. The two peaks between dP of 400 and 500 nm were in good agreement with the results of the SEM analysis and in reasonable agreement with the FF-TEM and FF-Cryo-SEM analyses. This suggests that the polymersomes did not alter shape during rapid freezing or even air drying above dP of 400 nm, and that the π/4 correction factor could be applied for polymersomes with dP > 400 nm. The size histogram analysis was with a bin width of 10 nm (the same that was used in Fig. 3). As the total number of analyzed polymersomes was larger than 11![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000, the size distribution can be seen as representative. No multilamellar polymersomes could be observed using CLSM, even though other techniques revealed their presence and studies reported that this would be possible.60 The size restriction remains a challenge for this method, as for most optical measurement techniques. A novel approach by Kunding et al. used the fluorescence intensity of immobilized labelled vesicles to obtain vesicle diameter below 200 nm with good agreement with Cryo-TEM.61 However the immobilisation procedure may lead to potential polymersome shape changes. Other methods like stimulated emission depletion microscopy (STED) can provide significantly sharper images below the diffraction limit, as shown in various studies on tissues62 and polymersomes.112
000, the size distribution can be seen as representative. No multilamellar polymersomes could be observed using CLSM, even though other techniques revealed their presence and studies reported that this would be possible.60 The size restriction remains a challenge for this method, as for most optical measurement techniques. A novel approach by Kunding et al. used the fluorescence intensity of immobilized labelled vesicles to obtain vesicle diameter below 200 nm with good agreement with Cryo-TEM.61 However the immobilisation procedure may lead to potential polymersome shape changes. Other methods like stimulated emission depletion microscopy (STED) can provide significantly sharper images below the diffraction limit, as shown in various studies on tissues62 and polymersomes.112
The size distribution of AFM analysis (Fig. 3b) correlated well with Cryo-TEM (Fig. 3a). Both analysis techniques have the highest peak at dP 50 nm with the same peak width, but the AFM peak was significantly smaller compared to the peak obtained from Cryo-TEM analysis. For polymersomes with dP between 100 and 150 nm, AFM analysis revealed slightly higher peaks. These latter peaks were in good agreement with results from the FF-TEM analysis. This could be due to the immobilization procedure since larger polymersomes could become deflated which result in capping structures similar to the ones obtained by FF-TEM or FF-Cryo-SEM, whereas smaller (and stiffer) polymersomes would not alter their shape upon immobilization. Force measurements could reveal more information here and this will be discussed in the next section. Each AFM measurement was performed within one hour, where the sample was thought to remain unchanged.80 Also no sticking of amphiphiles to the cantilever was observed during the measurements in tapping-mode. Compared to FF-TEM, Ruozi et al. reported better surface information for AFM analysis with liposomes.40 On large polymersomes, some holes in the surface were observed (see ESI†), however for the most polymersomes, surfaces appeared smooth as visualized with FF-TEM, see Fig. 7b. The holes could reflect deflated interiors of some polymersomes, or be due to cantilever-induced artifacts. AFM analysis did not reveal any information about polymersome interior (multilamellarity or multivesicularity) in contrast to what has been achieved with liposomes.113 The contrast and contour of FF-TEM visualized polymersomes was higher than the ones visualized using AFM, which is in agreement with other studies.40 Another possibility of AFM, which was not performed in this study, would be to analyze polymersomes in liquid mode. Here they would have been captured in natural conditions. However they need to be immobilized, which would most likely give a difference in their properties compared to when they are diffusing freely.
Bilayer thickness
The only methods, where information about tP could be obtained were Cryo-TEM, SAXS and SANS. The tP values of PB33–PEO18 polymersomes obtained using these three techniques, are summarized in Table 3. They agree with tP values found for PB–PEO from other studies.44,124 Cryo-TEM has shown to be a powerful tool for obtaining tP.125 The reliability of tP measurement by Cryo-TEM however depends on dP. tP in Cryo-TEM images is visualized as the difference in electron scattering intensity of the bilayer compared to the aqueous background.64 Near the equatorial plane of the polymersome, the electron beam enters the bilayer in a very small angle. Consequently, the electrons have to pass a long distance through the bilayer, lowering the contrast and the certainty of tP. At the interior polymersome edge, this effect is more pronounced than at the exterior edge. Furthermore, this effect has a higher influence with smaller polymersomes, where the tP is larger in comparison to dP.64 Thus, tP analysis with Cryo-TEM has to be seen in the light of these limitations.| Method | tP [nm] | Multilamellar/vesicular per 100 polymersomes |
|---|---|---|
| Cryo-TEM | 6.74 ± 1.28 | 10.71 ± 4.5 |
| FF-Cryo-SEM | 3.32 ± 0.36 | |
| SAXS & SANS | 7.6 ± 0.2 |
SAXS and SANS on the other hand usually can provide more precise information.126 The SAXS and SANS scattering patterns, arising from particles of simple geometrical shapes, can be calculated analytically. The structural parameters of this model can then be optimized by χ2 fitting of the calculated curve to the measured data (shown in Fig. 8a). Assuming the polymersomes to be spherical and polydisperse, we analyzed the data using a multi contrast shell model with three layers. The cross-section of the model is shown in Fig. 8b. The inner and outer shell, representing the PEO-chains, whereas the middle shell represents the hydrophobic core of the bilayer, containing the PB-chains. The same model is used to fit SAXS and SANS data simultaneously.
It is clear from Fig. 8a that the model does not fit the data well at very low q. This is owing to the presence of large vesicles of dP of more than 100 nm. In the q-range from 0.2 to 2 nm−1, on the other hand the fit quality is very good. In this regime length scales from 30 to 3 nm are probed, corresponding to tP and cross-section.
The fitted parameters of the model are shown in Table 4. The thickness of a PEO monolayer could not be unambiguously determined and was fixed at the reasonable value of 1.8 nm corresponding to two radii of gyration of a free PEO18-chain in solution. Further details of the model can be found in the ESI.†
| Fit parameter | Value |
|---|---|
| Mean polymersomes radius RP | 29.6 ± 0.5 nm |
| Gaussian width of RP distribution | 12.0 ± 0.2 nm |
| Thickness of PB33 bilayer, tP | 7.6 ± 0.2 nm |
| Thickness of PEO18 monolayer | 1.81 nm (fixed) |
| Width of polymersome interface s | 0.73 ± 0.03 nm |
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) |
|
| Deduced parameters | |
| Volume fraction of water in PEO shells | 0.34 |
| Interface area per block copolymer | 0.87 nm2 |
tP values determined from SAXS/SANS modeling is in good agreement with theoretical estimates.127 However it has to be taken into account that the preparation method of the samples for SAXS/SANS involved an additional dialysis step compared to the samples for Cryo-TEM. Thus, the bilayer could be stabilized52 and therewith become thicker during dialysis as pretreatment to SAXS/SANS analysis. In some of our previous SAXS measurements (not published), we found that the polymersome bilayer smoothes out with dialysis treatment. This could potentially result in a change in tP due to molecular rearrangement and stretching of the block copolymers in the bilayer. We observed the same during NS-TEM observations.52
Lamellarity
Cryo-TEM analysis gave a mean value of 10.71 ± 4.5 multilamellar or -vesicular polymersomes per 100 polymersomes, which was three times higher than the amount observed by FF-Cryo-SEM (see Table 3 and Fig. 9a). Interestingly, the amount of observed multivesicular polymersomes was higher in FF-Cryo-SEM (Fig. 9b and ESI†). Cryo-TEM is more likely to show the real distribution, because the interior of the polymersome is always revealed, allowing for identification of both structures. The higher number of multilamellar structures could be due to the film rehydration procedure, where their formation is more likely.128 The higher appearance of multivesicular polymersomes at FF-Cryo-SEM is consistent with these structures being more prone to fracturing than multilamellar polymersomes. In FF-Cryo-SEM structures that can be fractured easier are more likely to be observed. The interior forces are also more homogeneously distributed in multilamellar polymersomes, whereas in multivesicular polymersomes the uneven distribution of polydisperse polymersomes result in heterogeneous force distributions, all weakening the outer bilayer. SAXS and SANS showed an unilamellar behaviour due to prior extrusion preparation. They would otherwise be powerful tools to show multilamellar polymersomes.Elastic properties
The two elastic moduli, kc and Ka were analyzed by micropipette aspiration (Fig. 10a and video in the ESI†) following a procedure after Evans.116,117,129 The fractional surface area change (α = ΔA/A0) polymersomes with increasing suction pressure was monitored by camera. The isotropic tension change of the polymersome surface τ was calculated from the geometrical shape of the polymersomes under applied pressure, as described elsewhere.80 The linear slope from α against τ plot yielded a Ka of 60–170 mN m−1, see Fig. 10b. No kc could be obtained due to the small dP. Ka was significantly different from studies for PB–PEO polymersomes by Dimova (Ka 470 ± 15 mN m−1).79 However, Dimova et al. used a different preparation procedure, including sucrose in the buffer. Sucrose has a significant effect on the self-assembly behavior of polymersomes and thereby also on the elastic properties of their bilayers. PB12–PEO19 polymersomes are only forming if sucrose is present in the solution, otherwise they assemble to worm- or sperm like structures.52 They obtained polymersomes with a mean dP of 15 μm, whereas polymersomes of this work did seldom exceed 1 μm dP. In general, kc is reflecting thermal undulations and the soft regime of area compliance. The initial soft-exponential rise with area expansion in Fig. 10b reveals smoothing of thermal shape fluctuations.117 Ka had a large variety most likely due to the differences in preparation compared to the work of Dimova et al.79 A video showing the micropipette aspiration of a single polymersome is given in the ESI.†Permeability
For shrinking/swelling studies polymersomes must be extruded and we analyzed PB33–PEO18 polymersomes based on extrusion to a nominal dP of 200 nm. SFLS revealed a polymersome permeability of 5.87 ± 0.31 μm s−1, which is in the range of permeabilities, obtained by other studies on PB–PEO polymersomes (3.1 ± 1.6 μm s−1,130) and lower than what has been obtained with phosphatidylcholine liposomes (10–150 μm s−1,131), see Fig. 10c. In previous experiments, we measured PB12–PEO9, PB22–PEO23, PB35–PEO14, PB46–PEO23, PB46–PEO30 that revealed permeabilities between 7 and 80 μm s−1 with an increase with increasing hydrophobic block length, except for PB22–PEO23. This was due to the significantly smaller dP of the PB22–PEO23 polymersomes. PB12–PEO9 had a five times higher permeability than second highest permeable PB35–PEO14 (80 μm s−1 versus 14.5 μm s−1). On a different SFLS instrument we even measured 190 μm s−1 for PB12–PEO9 polymersomes.52 It seems that from a certain number of hydrophobic blocks the membrane permeability is exponentially increasing. Regarding the hydrophobic block length, PB33–PEO18 should have a permeability close to that of PB35–PEO14, however it is only one third. In the polymersomes from previous experiments of our group and Kumar et al.52 we used sucrose in the buffer as well, which has a significant influence on the bilayer as discussed before. Interestingly, for liposomes, there is no relationship between permeability and Ka, kc or tP.132 However permeability correlates to the area per lipid. Only when cholesterol was added, a linear relationship between Ka and permeability was found, probably because Ka becomes linear with area per lipid ratio.132Polarity
Polarity measurements are used to determine the hydrophobicity of a polymersome bilayer. This can help to deduct water or ion permeability or incorporation capability for membrane proteins. For polarity experiments, using GPM, we compared polymersomes of four different polymers (PB12–PEO9, PB22–PEO23, PB46–PEO23, PB46–PEO30) and DOPC liposomes. Laurdan labeled giant polymersomes and liposomes were produced using GUV formation and electroformation as described in Hansen et al.58,133 Further details are given in the Materials & methods chapter. Laurdan is a polarity sensitive fluorophore located in the bilayer with its functional group polarized parallel to the hydrophobic chains of the bilayer.95 Laurdan exhibits a red shift in spectral wavelength emission when set into polar environment or elevated temperatures.95,109 This shift can be quantified as values ranging between −1 and 1 using the GP function.134 All bilayers of the polymersomes appeared bluish indicating that they are significantly more hydrophobic than DOPC liposomes which appeared green,95 see Fig. 11. GPM and RGB processed images of vesicle bilayers of all polymersomes compared to DOPC are shown in Fig. 11. Laurdan labeling was distributed equally in PB12–PEO9, PB46–PEO23 and PB46–PEO30, whereas it varied in PB22–PEO23 vesicles. From the GP histogram in Fig. 12 of polymersomes and DOPC liposomes, one can recognize the red shift for DOPC liposomes and the blue shift for the polymersomes. Among the polymersomes, the blue shift increased linearly with increasing hydrophobic block length. The spectral emission shift from 22 hydrophobic units to 46 was quite small when compared to the one between 12 and 22. This could arise from the small dP values for both PB46–PEO23 and PB46–PEO30 polymersomes.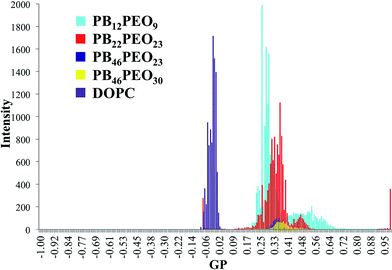 | ||
| Fig. 12 Generalized polarization (GP) histogram of Laurdan spectral emission shift for all vesicles, shown in values between −1 and 1 by integrating the intensities in a Laurdan-specific GP function.134 The spectral emission shift of Laurdan depends on the polarity of its environment. An unlinear blue shift with an increase of hydrophobic units was observed. | ||
Zeta potential
Zeta potential was measured using LDE on a Nano Zetasizer (Malvern, Worcestershire, UK), the same instrument that was used for DLS measurements. The measured zeta potential for PB33–PEO18 polymersomes was 3.32 ± 0.35 mV which indicated that polymersomes of this chemistry seemed to exhibit minimal surface charge. This is confirmed in other studies.3,135,136 The zeta potential is a measure of the electrostatic repulsion between particles. Particles with a zeta potential >30 mV or <−30 mV will repel each other and thereby avoid aggregation, where they will aggregate in the intermediate range.137 Thus, PB–PEO polymersomes are more prone to aggregation than polymersomes with a higher surface charge. As charges may be screened by the presence of ions (here due to 50 mM NaCl concentration in the buffer), the zeta potential (and potential aggregation) of polymersomes may be controlled to some extent.1384 Conclusion
Here we have presented 17 techniques to analyze (PB–PEO)-polymersomes and discussed them in terms of dP, tP, lamellarity, elastic properties, polarity and zeta potential. Advantages as well as drawbacks of each method were discussed and exemplified using specific PB–PEO polymersome samples. Although our review focus on polymersome analysis, it also pertains to liposome analysis. In summarizing the results we arrive at the following conclusions:DLS has the advantage of being simple, fast and well-established. For highly polydisperse samples, as used in this study, it is though not suited, even though the parameters and algorithms for calculating the size distribution was optimized towards minimizing the bias towards larger polymersomes.
NTA can be seen as a superior fast technique compared to DLS, due to size-distributions based on number-averages, in contrast to algorithm-based averages with DLS measurements. NTA has the same easy preparation as DLS, however it faces as well the same problems as DLS, when it comes to polydisperse samples and the large difference in scattering intensity between larger and smaller polymersomes.37 Care has to be taken with the interpretation of NTA size analysis especially for polymersomes that generally have a low RI.36
For analysis focusing on vesicle morphology and size measurements, NS-TEM is the method of choice for polymersomes with dP < 200 nm, due to its simple preparation procedures compared to FF-TEM, Cryo-TEM and FF-Cryo-SEM. For encapsulation, NS-TEM has advantages over Cryo-TEM due to the possibility of differential staining.113 However, results from NS-TEM analysis needs to be substantiated by data from other methods, due to osmotic shrinkage caused by the staining agent.
Cryo-TEM remains the method of choice for obtaining a reliable size distribution of polymersomes below 400 nm. For dP and lamellarity measurements, it is the most realistic imaging of all methods analyzed here, due to the “capture” of polymersomes in native environments. There are no size limitations or additional labeling steps necessary as the case with CLSM or super resolution microscopy techniques such as STED. However, for obtaining the tP values, care has to be taken with smaller polymersomes, which may be artificially thicker due to the contrast fluctuations near the equatorial plane of the polymersomes.
FF-TEM has good practical feasibility in the sense that the samples can be analyzed and stored over long time periods, in contrast to sample preparation for Cryo-TEM analyses. Also FF-TEM is well suited for analysis of membrane proteins incorporation.139 However, if exact dP determination are required, Cryo-TEM is superior for getting a reliable size distribution.
FF-Cryo-SEM is the method of choice for morphology and topology analysis of multivesicular (and to a less extent) multilamellar polymersomes. Due to good resolution and three dimensional projection between 50 nm and micrometer scale, it can fill the “analysis-gap” between TEM and photon-mediated visualization techniques. Also information obtained with FF-Cryo-SEM can be enhanced within NTA analysis. However, FF-Cryo-SEM has a poorer resolution compared to TEM and more rapid damaging of the sample when observing at higher magnifications.
CLSM provides reliable dP measurements for polymersomes with dP > 400 nm. For polymersomes with smaller dPs (i.e. <200 nm), the technique requires novel enhancements from the super resolution family such as STED. Establishing comparative dP studies with CLSM and EM-based methods, scattering and mechanical manipulation approaches will be a major future challenge.
AFM is a versatile fast technique for smaller polymersomes. AFM size distributions closely matches distributions achieved with Cryo-TEM. Deflation or deformation effects for polymersomes below dP of 400 nm seems to have only little influence on the size distribution, in contrast to measurements on liposome samples. AFM gives results comparable to what can be achieved with FF-TEM. However, AFM is not well suited for multilamellarity and multivesicularity analysis of polymersomes.
SAXS and SANS provides reliable information on tP, size and polydispersity of the polymersomes, where tP can be measured with higher accuracy compared to TEM. However their use will always be limited by complex analytical data analysis and the need for access to large-scale radiation facilities.
Micropipette aspiration is a great technique to obtain elastic properties of polymersomes. The downside is that it is quite time consuming in terms of manually aspirating single polymersomes. A big advantage compared to AFM is the visualization of the deformation of the entire polymersome during surface tension measurement with applied pressure that will reduce errors of measurement. AFM could use a spot of something else than the polymersome for obtaining the elastic moduli.
SFLS is an easy and quick tool for measuring permeabilities through polymersome bilayers. Depending on the instrument and the algorithm used, calculated permeabilities can be significantly different. Furthermore, monodisperse and unilamellar polymersomes are required to obtain reliable values.
Finally, LDE in combination with DLS can provide reliable information on the zeta potential of polymersomes. For PB–PEO polymersomes, the zeta potential was minimal, thus this system is prone to aggregation. Care has to be taken to the buffer solution, as ions can greatly influence the measurements.
The work presented here on polymersomes can be seen as a preparative step prior towards further processing and use of polymersomes. Thus, all the analysis methods discussed here are directly applicable in polymersome-based applications such as drug delivery systems,16 artificial cells21 or biomimetic membrane technology.27 In this development, the methods presented here will have to be supplemented by methods focusing on characterizing biomolecule encapsulation and protein incorporation characterization. Integration of these characterization methods constitutes an interesting challenge to be addressed in future polymersome research and associated technological developments.
Abbreviations/nomenclature
| A0 | Initial polymersome surface area |
| A4F | Asymmetrical flow field-flow fractionation |
| AFM | Atomic force microscopy |
| Bd | 1,3-Butadiene |
| n-BuLi | n-Butyl lithium |
| n-Bu2Mg | n-Dibutylmagnesium |
| CGMD | Coarse grain molecular dynamics |
| CLSM | Confocal laser scanning microscopy |
| d | Particle diameter |
| dP | Polymersome diameter |
| DLS | Dynamic light scattering |
| DOPC | 1,2-Dioleoyl-sn-glycero-3-phosphocholine |
| DPD | Dissipative particle dynamics |
| EM | Electron microscopy |
| EO | Ethylene oxide |
| EPR | Electron paramagnetic resonance |
| ESEM | Environmental scanning electron microscopy |
| ESRF | European Synchrotron Radiation Facility |
| f | Hydrophilic volume ratio |
| FACS | Fluorescence-activated cell-sorting |
| FCM | Flow cytometry |
| FCS | Fluorescence correlation microscopy |
| FF | Freeze fracture |
| FRM II | Forschungs-Neutronenquelle Heinz Maier-Leibnitz |
| FTLA | Finite track length adjustment |
| GPM | Generalized polarization microscopy |
| I | Light scattering intensity |
| ITO | Indium tin oxide |
| HPLC | High performance liquid chromatography |
| k | Rate constant of initial rise in stopped-flow light scattering curve |
| Ka | Elastic-area compressibility modulus |
| kc | Bending elasticity modulus |
| Laurdan | 6-Lauroyl-2-(dimethylamino)-naphthalene |
| LDE | Laser Doppler electrophoresis |
| Mn | Number-averaged molecular weight |
| Mw | Weight-averaged molecular weight |
| MD | Molecular dynamics |
| NMR | Nuclear magnetic resonance |
| NNLS | Non-negatively constrained least squares |
| NS | Negative staining |
| NTA | Nanoparticle tracking analysis |
| OG | n-Octyl-β-D-glucopyranoside |
| Pf | Bilayer permeability |
| PAA | Polyacrylic acid |
| PB | 1,2-Polybutadiene |
| PCM | Phase contrast microscopy |
| PDIM | Polydispersity index of the polymer length, defined as Mw/Mn |
| PDIDLS | Polydispersity index of DLS analysis |
| PEO | Polyethylene oxide |
| PS | Polystyrene |
| q | X-ray and neutron scattering vector |
| RP | Outer polymersome radius |
| RI | Refractive index |
| RT | Room temperature |
| S | Vesicle surface area |
| SANS | Small-angle neutron scattering |
| SAXS | Small-angle X-ray scattering |
| SEC | Size exclusion chromatography |
| SDL | Size detection limit |
| SEM | scanning electron microscopy |
| SFLS | Stopped-flow light scattering |
| SLS | Static light scattering |
| STED | Stimulated emission depletion microscopy |
| STM | Scanning tunneling microscopy |
| tP | Thickness of the hydrophobic core of polymersomes |
| tBuP4 | 1-tert-Butyl-4,4,4-tris(dimethylamino)-2,2-bis[tris (dimethyl-amino)-phosphoranylidenamino]-2λ5,4λ5-catenadi(phosphazene) |
| TEM | Transmission electron microscopy |
| THF | Tetrahydrofuran |
| TRPS | Tunable resistive pulse sensing |
| V0 | Vesicle volume before osmotic shock at stopped-flow light scattering |
| Vw | Molar volume of water (18 cm3 mol−1) |
| WAXS | Wide-angle X-ray scattering |
| ZD | Intensity-weighted hydrodynamic diameter |
| α | Fractional surface area change of polymersomes |
| δ | Distribution width of polymersome diameter at DLS analysis |
| Δosm | Difference in osmolarity |
| ΔA | Polymersomes surface area change |
| θ | X-ray or neutron scattering angle |
| λ | Wavelength |
| τ | Isotropic tension change of the polymersome surface |
Acknowledgements
We thank Lars Schulte and Simon Levinsen, DTU Nanotech for assistance with the polymer synthesis; Fadoua Sbai, Aquaporin A/S, for assistance with SFLS analysis; Klaus Qvortrup, Ramon Liebrechts and Thomas Braunstein, University of Copenhagen, for providing assistance with freeze fracture and CLSM and access to the freeze fracture unit, the TEM and the CLSM and Noemi Rozlosnik, DTU Nanotech for providing access to the AFM. We thank NXUS for the collection and analysis of the SAXS-data. NXUS is a pilot project enabling industrial use of large scale research facilities in collaboration with university experts in the field of neutron and X-ray science. NXUS is funded by the University of Copenhagen and the capital region of Denmark. JH was supported by an industrial PhD grant from Innovation Fund Denmark. CHN was supported by the IBISS – Industrial Biomimetic Sensing and separation platform (http://www.ibiss.dtu.dk) funded by the Danish Innovation Fund Grant no. 097-2012-4.References
- L. Zhang and A. Eisenberg, Science, 1995, 268, 1728–1731 CrossRef CAS PubMed.
- B. M. Discher, Y.-Y. Won, D. Ege, J. Lee, F. S. Bates, D. E. Discher and D. Hammer, Science, 1999, 284, 1143–1146 CrossRef CAS PubMed.
- B. M. Discher, D. Hammer, F. S. Bates and D. E. Discher, Curr. Opin. Colloid Interface Sci., 2000, 5, 125–131 CrossRef CAS.
- C. Nardin, T. Hirt, J. Leukel and W. P. Meier, Langmuir, 2000, 16, 1035–1041 CrossRef CAS.
- C. Nardin, J. Widmer, M. Winterhalter and W. P. Meier, Eur. Phys. J. E, 2001, 4, 403–410 CrossRef CAS.
- D. E. Discher and A. Eisenberg, Science, 2002, 297, 967–973 CrossRef CAS PubMed.
- M. Antonietti and S. Förster, Adv. Mater., 2003, 15, 1323–1333 CrossRef CAS.
- K. Kita-Tokarczyk, J. Grumelard, T. Haefele and W. P. Meier, Polymer, 2005, 46, 3540–3563 CrossRef CAS.
- D. E. Discher and F. Ahmed, Annu. Rev. Biomed. Eng., 2006, 8, 323–341 CrossRef CAS PubMed.
- D. E. Discher, V. Ortiz, G. Srinivas, M. L. Klein, Y. Kim, D. Christian, S. Cai, P. Photos and F. Ahmed, Prog. Polym. Sci., 2007, 32, 838–857 CrossRef CAS PubMed.
- D. H. Levine, P. P. Ghoroghchian, J. Freudenberg, G. Zhang, M. J. Therien, M. I. Greene, D. A. Hammer and R. Murali, Methods, 2008, 46, 25–32 CrossRef CAS PubMed.
- C. Lopresti, H. Lomas, M. Massignani, T. Smart and G. Battaglia, J. Mater. Chem., 2009, 19, 3557–3776 RSC.
- M. Massignani, H. Lomas and G. Battaglia, Advances in Polymer Sciences, Springer, Berlin, Germany, 2010, pp. 115–154 Search PubMed.
- P. Tanner, P. Baumann, R. Enea, O. Onaca, C. G. Palivan and W. P. Meier, Acc. Chem. Res., 2011, 44, 1039–1049 CrossRef CAS PubMed.
- J. Liao, C. Wang, Y. Wang, F. Luo and Z. Qian, Curr. Pharm. Des., 2012, 18, 3432–3441 CrossRef CAS PubMed.
- J. Nicolas, S. Mura, D. Brambilla, N. Mackiewicz and P. Couvreur, Chem. Soc. Rev., 2013, 42, 1147–1235 RSC.
- J. Yang, H. Liu and X. Zhang, Biotechnol. Adv., 2014, 32, 804–817 CrossRef CAS PubMed.
- K. Langowska, C. G. Palivan and W. P. Meier, Chem. Commun., 2013, 49, 128–130 RSC.
- P. V. Pawar, S. V. Gohil, J. P. Jain and N. Kumar, Polym. Chem., 2013, 4, 3160–3176 RSC.
- R. Langer, Science, 1990, 249, 1527–1533 CrossRef CAS PubMed.
- S. Egli, M. Nussbaumer, V. Balasubramanian, M. Chami, N. Bruns, C. G. Palivan and W. P. Meier, J. Am. Chem. Soc., 2011, 133, 4476–4483 CrossRef CAS PubMed.
- L. Kuang, D. A. Fernandes, M. O'Halloran, W. Zheng, Y. Jiang, V. Ladizhansky, L. S. Brown and H. Liang, ACS Nano, 2014, 8, 537–545 CrossRef CAS PubMed.
- H.-J. Choi and C. D. Montemagno, Nano Lett., 2005, 5, 2538–2542 CrossRef CAS PubMed.
- V. Malinova, M. Nallani, W. P. Meier and E. K. Sinner, FEBS Lett., 2012, 586, 2146–2156 CrossRef CAS PubMed.
- A. González-Pérez, K. Stibius, T. Vissing, C. Hélix-Nielsen and O. G. Mouritsen, Langmuir, 2009, 25, 10447–10450 CrossRef PubMed.
- M. Sauer, T. Haefele, A. Graff, C. Nardin and W. P. Meier, Chem. Commun., 2001, 23, 2452–2453 RSC.
- C. Hélix-Nielsen, Anal. Bioanal. Chem., 2009, 395, 697–718 CrossRef PubMed.
- A. S. Aquaporin, C. Y. Tang, C. Qiu, Y. Zhao, W. Shen, A. Vararattanavech, R. Wang, X. Hu, J. Torres, A. G. Fane and C. Hélix-Nielsen, Aquaporin based thin film composite membranes, Nanyang Technological University, WO 2013/043118, 2013.
- C. Y. Tang, Y. Zhao, R. Wang, C. Hélix-Nielsen and A. G. Fane, Desalination, 2013, 308, 34–40 CrossRef CAS.
- P. S. Zhong, T.-S. Chung, K. Jeyaseelan and A. Armugam, J. Membr. Sci., 2012, 407–408, 27–33 CrossRef CAS.
- H. L. Wang, T.-S. Chung, Y. W. Tong, K. Jeyaseelan, A. Armugam, H. H. P. Duong, F. Fu, H. Seah, J. Yang and M. Hong, J. Membr. Sci., 2013, 434, 130–136 CrossRef CAS.
- B. J. Berne and R. Pecora, Dynamic light scattering, Courier Dover Publications, Mineola, NY, USA, 2000 Search PubMed.
- O. Glatter and O. Kratky, Small Angle X-ray Scattering, Academic Press, London, UK, 1982 Search PubMed.
- I. Grillo, Small-Angle Neutron Scattering and Applications in Soft Condensed Matter, Springer, Heidelberg, Germany, 2008 Search PubMed.
- J. S. Pedersen, Adv. Colloid Interface Sci., 1997, 70, 171–210 CrossRef CAS.
- B. Carr and M. Wright, Nanoparticle tracking analysis – A review of Applications and Usage 2010–2012, Nanosight, 2013, pp. 1–193 Search PubMed.
- R. A. Dragovic, C. Gardiner, A. S. Brooks, D. S. Tanetta, D. J. Ferguson, P. Hole, B. Carr, C. W. G. Redman, A. L. Harris, P. J. Dobson, P. Harrison and I. L. Sargent, Nanomedicine, 2011, 7, 780–788 CrossRef CAS PubMed.
- C. Gardiner, Y. J. Ferreira, R. A. Dragovic, C. W. G. Redman and I. L. Sargent, J. Extracell. Vesicles, 2013, 2, 19671 CrossRef PubMed.
- K. Edwards and A. Baeumner, Talanta, 2006, 68, 1432–1441 CrossRef CAS PubMed.
- B. Ruozi, G. Tosi, E. Leo and M. Vandelli, Talanta, 2007, 73, 12–22 CrossRef CAS PubMed.
- E. Tomaszewska, K. Soliwoda, K. Kadziola, B. Tkacz-Szczesna, G. Celichowski, M. Cichomski, W. Szmaja and J. Grobelny, J. Nanomater., 2013, 2013, 313081 Search PubMed.
- S. Hupfeld, A. M. Holsæter, M. Skar, C. B. Frantzen and M. Brandl, J. Nanosci. Nanotechnol., 2006, 6, 3025–3031 CrossRef CAS PubMed.
- X. Zhang, P. Tanner, A. Graff, C. G. Palivan and W. P. Meier, J. Polym. Sci., Part A: Polym. Chem., 2012, 50, 2293–2318 CrossRef CAS.
- Y. Won, A. K. Brannan, H. Davis and F. S. Bates, J. Phys. Chem. B, 2002, 106, 3354–3364 CrossRef CAS.
- S.-H. Choi, F. S. Bates and T. P. Lodge, Macromolecules, 2014, 47, 7978–7986 CrossRef CAS.
- J. Braun, N. Bruns, T. Pfohl and W. P. Meier, Macromol. Chem. Phys., 2011, 212, 1245–1254 CrossRef CAS.
- M. Hirai, R. Kimura, K. Takeuchi, Y. Hagiwara, R. Kawai-Hirai, N. Ohta, N. Igarashi and N. Shimuzu, J. Synchrotron Radiat., 2013, 20, 869–874 CrossRef CAS PubMed.
- A. I. Norman, J. T. Cabral, A. Karim and E. J. Amis, Macromol. Rapid Commun., 2004, 25, 307–311 CrossRef CAS.
- U. Till, M. Gaucher-Delmas, P. Saint-Aguet, G. Hamon, J.-D. Marty, C. Chassenieux, B. Payré, D. Goudounèche, A.-F. Mingotaud and F. Violleau, Anal. Bioanal. Chem., 2014, 406, 7841–7853 CrossRef CAS PubMed.
- Y. Won, H. Davis and F. S. Bates, Macromolecules, 2003, 36, 953–955 CrossRef CAS.
- P. S. Ho, J. Leu and W. W. Lee, Low Dielectric Constant Materials for IC Applications, Springer, Berlin, Germany, 2003 Search PubMed.
- M. Kumar, J. Habel, Y.-x. Shen, W. P. Meier and T. Walz, J. Am. Chem. Soc., 2012, 134, 18631–18637 CrossRef CAS PubMed.
- E. van der Pol, F. A. W. Coumans, A. E. Grootemaat, C. Gardiner, I. L. Sargent, P. Harrison, A. Sturk, T. G. van Leeuwen and R. Nieuwland, J. Thromb. Haemostasis, 2014, 12, 1182–1192 CrossRef CAS PubMed.
- C. Chen, S. Zhu, T. Huang, S. Wang and X. Yan, Anal. Methods, 2013, 5, 2150–2157 RSC.
- J. P. Jain, W. Y. Ayen and N. Kumar, Curr. Pharm. Des., 2011, 17, 65–79 CrossRef CAS PubMed.
- A. A. Reinecke and H.-G. Döbereiner, Langmuir, 2003, 19, 605–608 CrossRef CAS.
- http://www.microbehunter.com/phase-contrast-vs-bright-field%20microscopy/.
- J. S. Hansen and C. Hélix-Nielsen, Biochem. Biophys. Res. Commun., 2011, 415, 686–690 CrossRef CAS PubMed.
- M. Erbakan, Y.-X. Shen, M. Grzelakowski, P. J. Butler, M. Kumar and W. R. Curtis, PLoS One, 2014, 9, e86830 CrossRef PubMed.
- B. Ruozi, D. Belletti, A. Tombesi, G. Tosi, L. Bondioli, F. Forni and M. A. Vandelli, Int. J. Nanomed., 2011, 6, 557–563 CrossRef CAS PubMed.
- A. H. Kunding, M. W. Mortensen, S. M. Christensen and D. Stamou, Biophys. J., 2008, 95, 1176–1188 CrossRef CAS PubMed.
- S. W. Hell, Science, 2007, 316, 1153–1158 CrossRef CAS PubMed.
- J. Habel, A. Ogbonna, N. Larsen, L. Schulte, K. Almdal and C. Hèlix-Nielsen, J. Polym. Sci., Part B: Polym. Phys. Search PubMed , submitted..
- B. Coldren, R. van Zanten, M. J. Mackel, J. A. Zasadzinski and H.-T. Jung, Langmuir, 2003, 19, 5632–5639 CrossRef CAS.
- Y.-Y. Won, Korean J. Chem. Eng., 2004, 21, 296–302 CrossRef CAS.
- J. K. Harris, G. D. Rose and M. L. Bruening, Langmuir, 2002, 18, 5337–5342 CrossRef CAS.
- D. C. Joy, J. Microsc., 1984, 136, 241–258 CrossRef CAS.
- F. J. Doucet, J. R. Lead, L. Maguire, E. P. Achterberg and G. E. Millward, J. Environ. Monit., 2005, 7, 115–121 RSC.
- S. Li and A. F. Palmer, Macromolecules, 2005, 38, 5686–5698 CrossRef CAS.
- M. R. Mozafari, M. H. Zareie, E. Piskin and V. Hasirci, Drug Delivery, 1998, 5, 135–141 CrossRef CAS PubMed.
- C. Bai, Scanning Tunneling Microscopy and Its Application, Springer, Heidelberg, Germany, 2nd edn, 2000, vol. 32 Search PubMed.
- B. Baruah and A. Surin, JBIC, J. Biol. Inorg. Chem., 2012, 17, 899–910 CrossRef CAS PubMed.
- Y. P. Patil and S. Jadhav, Chem. Phys. Lipids, 2014, 177, 8–18 CrossRef CAS PubMed.
- A. Laouini, C. Jaafar-Maalej, I. Limayem-Blouza, S. Sfar, C. Charcosset and H. Fessi, J. Colloid Sci. Biotechnol., 2012, 1, 147–168 CrossRef CAS.
- K. Takeshita and T. Ozawa, J. Radiat. Res., 2004, 45, 373–384 CrossRef CAS PubMed.
- D. Wu, M. Spulber, F. Itel, M. Chami, T. Pfohl, C. G. Palivan and W. P. Meier, Macromolecules, 2014, 47, 5060–5069 CrossRef CAS.
- C. Matos, B. de Castro, P. Gameiro, J. L. F. C. Lima and S. Reis, Langmuir, 2004, 20, 369–377 CrossRef CAS PubMed.
- J. A. Schwarz and C. I. Contescu, Surfaces of Nanoparticles and Porous Materials, CRC Press, New York, NY, USA, 1st edn, 1999, vol. 78 Search PubMed.
- R. Dimova, U. Seifert, B. Pouligny, S. Förster and H.-G. Döbereiner, Eur. Phys. J. E, 2002, 7, 241–250 CrossRef CAS.
- S. Dieluweit, A. Csiszár, W. Rubner, J. Fleischhauer, S. Houben and R. Merkel, Langmuir, 2010, 26, 11041–11049 CrossRef CAS PubMed.
- R. R. Jahan-Tigh, C. Ryan, G. Obermoser and K. Schwarzenberger, J. Invest. Dermatol., 2012, 132, e1 CrossRef CAS PubMed.
- D. Burger, S. Schock, C. S. Thompson, A. C. Montezano, A. M. Hakim and R. M. Touyz, Clin. Sci., 2013, 124, 423–441 CrossRef CAS PubMed.
- M. Holzer, S. Barnert, J. Momm and R. Schubert, J. Chromatogr. A, 2009, 1216, 5838–5848 CrossRef CAS PubMed.
- C. Grabielle-Madelmont, S. Lesieur and M. Ollivon, J. Biochem. Biophys. Methods, 2003, 56, 189–217 CrossRef CAS PubMed.
- Z. Zhong, Q. Ji and J. A. Zhang, J. Pharm. Biomed. Anal., 2010, 51, 947–951 CrossRef CAS PubMed.
- L. Yang, M. F. Broom and I. G. Tucker, Pharm. Res., 2012, 29, 2578–2586 CrossRef CAS PubMed.
- J. N. Sachs, P. S. Crozier and T. B. Woolf, J. Chem. Phys., 2004, 121, 10847 CrossRef CAS PubMed.
- G. Srinivas, D. E. Discher and M. Klein, Nano Lett., 2005, 5, 2343–2349 CrossRef CAS PubMed.
- M. Xiao, J. Liu, J. Yang, R. Wang and D. Xie, Soft Matter, 2013, 9, 2434–2442 RSC.
- L. Reimer, Transmission Electron Microscopy: Physics of Image Formation, Springer, New York, NY, USA, 5th edn, 2008 Search PubMed.
- D. McMullan, Scanning, 1995, 17, 175–185 CrossRef CAS.
- G. D. Danilatos, Advances in Electronics and Electron Physics, Academic Press, San Diego, CA, USA, 1988, pp. 109–250 Search PubMed.
- A. Cavalier, D. Spehner and B. M. Humbel, Handbook of cryo-preparation methods for electron microscopy, CRC Press, Boca Raton, FL, USA, 2008 Search PubMed.
- J. Pawley, Handbook of Biological Confocal Microscopy, Springer, Berlin, Germany, 3rd edn, 2006 Search PubMed.
- L. A. Bagatolli, Biochim. Biophys. Acta, Biomembr., 2006, 1758, 1541–1556 CrossRef CAS PubMed.
- F. J. Giessibl, Rev. Mod. Phys., 2003, 75, 949–983 CrossRef CAS.
- C. Rein, K. Pszon-Bartosz, K. B. Stibius, T. Bjørnholm and C. Hélix-Nielsen, Langmuir, 2011, 27, 499–503 CrossRef CAS PubMed.
- Y. Chen, PhD thesis, Washington University, St. Louis, MO, USA, 2008.
- R. M. Hochmuth, J. Biomech., 2000, 33, 15–22 CrossRef CAS PubMed.
- R. Dimova, C. Dietrich, A. Hadjiisky, K. Danov and B. Pouligny, Eur. Phys. J. B, 1999, 12, 589–598 CrossRef CAS.
- V. Chechik and D. M. Murphy, Electron Paramagnetic Resonance, RSC, London, UK, 2014, vol. 24 Search PubMed.
- J. Keeler, Understanding NMR Spectroscopy, Jon Wiley & Sons, Hoboken, NJ, USA, 2nd edn, 2010 Search PubMed.
- M. G. Omerod, Flow Cytometry – A practical approach, Oxford University Press, Oxford, UK, 3rd edn, 2000 Search PubMed.
- A. M. Striegel, J. J. Kirkland, W. W. Yau and D. D. Bly, Modern Size Exclusion Chromatography: Practice of Gel Permeation and Gel Filtration Chromatography, Jon Wiley & Sons, Hoboken, NJ, USA, 2nd edn, 2009 Search PubMed.
- C. Bria, F. Violleau and S. K. R. Williams, LC·GC Eur., 2013, 26, 660–671 CAS.
- S. Förster and E. Krämer, Macromolecules, 1999, 32, 2783–2785 CrossRef.
- S. Jain, M. H. E. Dyrdahl, X. Gong, L. E. Scriven and F. S. Bates, Macromolecules, 2008, 41, 3305–3316 CrossRef CAS.
- J. A. Zupancich, F. S. Bates and M. A. Hillmyer, Biomacromolecules, 2009, 10, 1554–1563 CrossRef CAS PubMed.
- J. Lee, H. Bermudez, B. M. Discher, M. Sheehan, Y.-Y. Won, F. S. Bates and D. E. Discher, Biotechnol. Bioeng., 2001, 73, 135–145 CrossRef CAS PubMed.
- J. R. Howse, R. A. L. Jones, G. Battaglia, R. E. Ducker, G. J. Leggett and A. J. Ryan, Nat. Mater., 2009, 8, 507–511 CrossRef CAS PubMed.
- S. U. Egelhaaf, E. Wehrli, M. Muller, M. Adrian and P. Schurtenberger, J. Microsc., 1996, 184, 214–228 CrossRef CAS.
- G. Battaglia, C. Lopresti, M. Massignani, N. J. Warren, J. Madsen, S. Forster, C. Vasilev, J. K. Hobbs, S. P. Armes, S. Chirasatitsin and A. J. Engler, Small, 2011, 7, 2010–2015 CrossRef CAS PubMed.
- O. Stauch, T. Uhlmann, M. Fröhlich, R. Thomann, M. El-Badry, Y.-K. Kim and R. Schubert, Biomacromolecules, 2002, 3, 324–332 CrossRef CAS PubMed.
- M. Kumar, M. Grzelakowski, J. Zilles, M. M. Clark and W. P. Meier, Proc. Natl. Acad. Sci. U. S. A., 2007, 104, 20719–20724 CrossRef CAS PubMed.
- M. I. Angelova and D. S. Dimitrov, Faraday Discuss. Chem. Soc., 1986, 81, 303–311 RSC.
- E. Evans, W. Rawicz and B. A. Smith, Faraday Discuss., 2013, 161, 591–611 RSC.
- W. Rawicz, K. C. Olbrich, T. McIntosh, D. Needham and E. Evans, Biophys. J., 2000, 79, 328–339 CrossRef CAS PubMed.
- G. Mie, Ann. Phys., 1908, 330, 377–445 CrossRef.
- P. A. Hassan and S. K. Kulshreshtha, J. Colloid Interface Sci., 2006, 300, 744–748 CrossRef CAS PubMed.
- C. F. Bohren, The Scattering of light & other electromagnetic radiation, Jon Wiley & Sons, New York, NY, USA, 1983 Search PubMed.
- L. Ma and A. Eisenberg, Langmuir, 2009, 25, 13730–13736 CrossRef CAS PubMed.
- K. P. Ryan, Scanning Microsc., 1992, 6, 715–743 CAS.
- L. P. Aggerbeck and T. Gulik-Krzywicki, Methods Enzymol., 1986, 128, 457–472 CrossRef CAS PubMed.
- H. Bermudez, A. K. Brannan, D. A. Hammer, F. S. Bates and D. E. Discher, Macromolecules, 2002, 35, 8203–8208 CrossRef CAS.
- F. Itel, M. Chami, A. Najer, S. Lörcher, D. Wu, I. A. Dinu and W. P. Meier, Macromolecules, 2014, 47, 7588–7596 CrossRef CAS.
- R. Hite, S. Raunser and T. Walz, Curr. Opin. Struct. Biol., 2007, 17, 389–395 CrossRef CAS PubMed.
- L. Fetters, W. Graessley, R. Krishnamoorti and D. Lohse, Macromolecules, 1997, 30, 4973–4977 CrossRef CAS.
- P. Walde and S. Ichikawa, Biomol. Eng., 2001, 18, 143–177 CrossRef CAS PubMed.
- E. Evans and D. Needham, J. Phys. Chem., 1987, 91, 4219–4228 CrossRef CAS.
- A. Carlsen, N. Glaser, J.-F. Le Meins and S. Lecommandoux, Langmuir, 2011, 27, 4884–4890 CrossRef CAS PubMed.
- J.-L. Rigaud and D. Levy, Methods Enzymol., 2003, 372, 65–86 CAS.
- J. C. Mathai, S. Tristram-Nagle, J. F. Nagle and M. L. Zeidel, J. Gen. Physiol., 2008, 131, 69–76 CrossRef CAS PubMed.
- F. M. Menger and M. I. Angelova, Acc. Chem. Res., 1998, 31, 789–797 CrossRef CAS.
- S. A. Sanchez, M. A. Tricerri, G. Gunter and E. Gratton, Modern Research and Educational Topics in Microscopy, Formatex, Extremadura, Spain, 2007, pp. 1007–1014 Search PubMed.
- H. Aranda-Espinoza, H. Bermudez, F. S. Bates and D. E. Discher, Phys. Rev. Lett., 2001, 87, 208301 CrossRef CAS PubMed.
- J. Habel, A. Ogbonna, N. Larsen, S. Krabbe, K. Almdal and C. Hèlix-Nielsen, Phys. Chem. Chem. Phys. Search PubMed , submitted..
- R. W. O'Brien, B. R. Midmore, A. Lamb and R. J. Hunter, Faraday Discuss. Chem. Soc., 1990, 90, 301–312 RSC.
- B. D. Coday, T. Luxbacher, A. E. Childress, N. Almaraz, P. Xu and T. Y. Cath, J. Membr. Sci., 2015, 478, 58–64 CrossRef CAS.
- F. Itel, S. Al-Samir, F. Oberg, M. Chami, M. Kumar, C. T. Supuran, P. M. T. Deen, W. P. Meier, K. Hedfalk, G. Gros and V. Endeward, FASEB J., 2012, 26, 5182–5191 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available: Polymersome images of NTA, NS-TEM, Cryo-TEM, FF-TEM, Cryo-FF-SEM, CLSM, AFM, modelling of small-angle scattering, videos of NTA, CLSM and micropipette aspiration analysis. See DOI: 10.1039/c5ra16403f |
| This journal is © The Royal Society of Chemistry 2015 |