Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
The urea cycle of rat white adipose tissue
Sofía Arriarána,
Silvia Agnellia,
Xavier Remesarabc,
José-Antonio Fernández-López abc and
Marià Alemany
abc and
Marià Alemany *abc
*abc
aDepartment of Nutrition and Food Science, Faculty of Biology, University of Barcelona, Av. Diagonal 643, 08028 Barcelona, Spain. E-mail: malemany@ub.edu
bInstitute of Biomedicine, University of Barcelona, 08028 Barcelona, Spain
cCIBER-OBN Research Web, Barcelona, Spain
First published on 19th October 2015
Abstract
White adipose tissue (WAT) contains a powerful metabolic machinery related to energy metabolism and partition. The adaptive, regulatory, size and structural variety of WAT agrees with this control role. However, its nitrogen metabolism has been sparsely studied and we know close to nothing about its implication on N-related processes and homoeostasis. We have studied, and found a complete urea cycle (liver type) in four WAT sites (gene expressions and enzyme activities). We postulated a possible function of the cycle (under basal conditions) in the control of arginine handling through citrulline synthesis. In our opinion, the metabolic and control potential of WAT on energy metabolism may be second only to liver. This impression should be extended to amino acid metabolism too, WAT providing an extra-intestinal source of citrulline to maintain the body availability of arginine independently of the operation of intestine–liver urea cycle for N disposal.
Introduction
Adipose tissue is one of the most versatile, adaptable and resilient tissues of the mammal's body. A number of additional functions complement the well-known energy-storage role of adipose tissue, such as protection, both physical (filling, packing, barrier action against trauma, thermal insulation) and defensive (immune organ, tissue regeneration site). White adipose tissue (WAT) plays an important role in metabolic regulation, as a paracrine and endocrine organ, but also as a key energy partition system. WAT also shows (at least in part), a thermogenic function, which is used for the elimination of unwanted excess energy as heat, and the maintenance of body temperature.Our appreciation of adipose tissue is growing in physiological transcendence in parallel to the discovery of the full extent of its functions. A turning point has been the acknowledgment that different adipose tissue sites may carry out different functions,1 sharing some but not all the possibilities such malleable tissue/organ possesses. Extreme differentiation resulted in the practical division of adipose tissue in at least three different types according to specialized function, cell structure, main active metabolic pathways, and anatomical placement. White (WAT), made up of large cells, usually with a single fat-filled vacuole, largely glycolytic, with low oxygen consumption and a limited number of mitochondria;2 this is the most abundant adipose tissue type. Brown (BAT), formed by plurivacuolar cells; highly oxidative, with a large number of mitochondria, specialized thermogenic function, and highly vascularized and innervated. In addition, beige or brite, sharing characteristics of brown and white, but not a transition stage between both, of independent origin, and functions also related mainly to thermogenesis.
This largely functional and morphologic division of adipose tissue affects, essentially, the functions of energy storage and thermogenesis, but there are also marked differences in location, functional adaptability and regulatory ability of WAT in different body masses and sites.3,4 These differences are in part a consequence of their different developmental lineage,5 but also of their specific function, with a critical influence of the cell environment.
This considerable variability is enhanced under conditions of metabolic derangement, as in metabolic syndrome, when most WAT sites are massively infiltrated by immune cells,6 to avert a possible metabolic aggression, eliciting an alarm response,7 which tends to become chronic, inducing generalized alterations of adipose tissue cell composition and functions.8
In addition to glycolysis, production of acetyl-CoA, lipogenesis, synthesis, storage and hydrolysis of triacylglycerols, WAT produces lactate9 and alanine10 as part of Cori and glucose–alanine cycles. WAT may use amino acids as energy (or lipogenesis) substrates,11 contains active glycogen depots,12 and converts glucose to lactate and glycerol to lower its glycemic load.13 BAT is capable of using almost any circulating substrate (including amino acids)14 to sustain thermogenesis.
WAT is a significant producer of nitric oxide,15 and its blood flow and metabolic activity are regulated by catecholamines and insulin.16,17 In sum, adipose tissues, and WAT in particular because of its size and distribution, have an enormous metabolic potential, which probably is activated on demand, depending on the needs of the particular niche/function the tissue covers. In this context, the regulatory capabilities of WAT have been explored extensively in the last years, resulting in the discovery of a sizeable number of adipokines and other regulatory agents and mechanisms.18
The study of metabolic pathways in WAT has been, so far, limited; with only a few publications dealing with issues other than glucose metabolism and lipogenesis; amino acid metabolism remains a considerably neglected field, limited to few papers from last century.11,19 The large relative mass of WAT and the key role that arginine plays in the control of WAT hemodynamics (via synthesis of nitric oxide), especially in metabolic syndrome,20 made us think on the possibility that WAT implication in the control of arginine availability and metabolism20 could be deeper than currently assumed. In the present work, we analyzed whether WAT can synthesize arginine through the urea cycle. We are aware that this cycle is fully complete and functional only in liver, and (at least partially) in kidney and intestine. In doing the study, we tested for a significant (and largely ignored) importance of WAT in amino acid metabolism.
Experimental
Ethics statement
All animal handling procedures and the experimental setup were in accordance with the animal handling guidelines of the corresponding European and Catalan Authorities. The Committee on Animal Experimentation of the University of Barcelona specifically authorized the procedures used in the present study.Experimental design and animal handling
Nine week old male Wistar rats (Harlan Laboratory Models, Sant Feliu de Codines, Spain) were used. The rats (N = 6) were housed in two-rat cages with wood shards for bedding. They had free access to water and ate normal rat chow (type 2014, Harlan). They were kept in a controlled environment (lights on from 08:00 to 20:00; 21.5–22.5 °C; 50–60% humidity) for at least one month.The rats were killed by exsanguination (by aortic puncture), under isoflurane anesthesia, at the beginning of a light cycle; then, were rapidly dissected, taking large samples of liver and WAT: mesenteric, epididymal, retroperitoneal and subcutaneous (inguinal fat pads). All samples were blotted and frozen in liquid nitrogen. To minimize the problems of tissue sampling for analyses, the frozen samples were weighed, and then were ground with mortar and pestle under liquid nitrogen. The coarse powder was stored at −80 °C until processed. Later, the dissection of the rats continued, extracting the remaining liver and WAT in ME, EP and RP sites; the rats were skinned, and the whole subcutaneous WAT was dissected. The weights of the recovered WAT were added to those of the frozen samples in order to determine the precise mass of the four WAT sites liver weight was determined in the same way.
Tissue homogenate preparation
Frozen tissue samples were homogenized, using a tissue disruptor (Ultraturrax IKA-T10, Ika Werke, Staufen, Germany), in 5 volumes of chilled 70 mM hepes buffer pH 7.4 containing 1 mM dithiothreitol (Sigma, St Louis MO USA), 50 mM KCl, 1 g L−1 Triton X-100 (Sigma) and 1 g L−1 lipid-free bovine serum albumin (Sigma). In homogenates to be used for carbamoyl-P synthase 2 estimation, the concentration of Triton X-100 was halved to prevent foaming. The homogenates were centrifuged for 10 min at 5000 × g; the floating fat layer and gross debris precipitate were discarded. The homogenates were kept on ice, and were used for the estimation of protein and for enzymatic analyses within 2 h after their preparation. Liver homogenates were obtained using 10 volumes of chilled buffer. They were further diluted with buffer, as needed, in the reaction mixtures for the estimation of enzyme activities.Tissue protein content was estimated with the Lowry method.21 After development of color, turbidity was eliminated with small amounts of finely powdered solid MgO, which adsorbed the remaining suspended fat, and centrifuging the tubes before reading the absorbance. In the measurements of homogenate protein content, homogenization buffer (containing 1 g per L albumin) was used as blank.
Enzyme activities were expressed in nkat per g protein.
Enzyme activity analyses
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) methanol
methanol![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) acetic acid (1
acetic acid (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2
2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2 by volume). After drying, the developed chromatograms, were sprayed (only the lane with non-labelled standards) with ninhydrin, and heated with a hair dryer. The ornithine and citrulline spots were marked and the rest of the plate penciled in horizontal 1 cm zones, which were cut and counted with the liquid scintillation system described above. The labeled areas were identified and quantified. To limit interference by size of sample and contamination, the label in the citrulline spot was expressed as a percentage of the total label counted in each TLC lane. These data allowed the calculation of newly formed citrulline at each time. The Vi value for each sample was plotted, and considered an expression of Vmax under the conditions tested.
2 by volume). After drying, the developed chromatograms, were sprayed (only the lane with non-labelled standards) with ninhydrin, and heated with a hair dryer. The ornithine and citrulline spots were marked and the rest of the plate penciled in horizontal 1 cm zones, which were cut and counted with the liquid scintillation system described above. The labeled areas were identified and quantified. To limit interference by size of sample and contamination, the label in the citrulline spot was expressed as a percentage of the total label counted in each TLC lane. These data allowed the calculation of newly formed citrulline at each time. The Vi value for each sample was plotted, and considered an expression of Vmax under the conditions tested.The chemical reaction using 1-phenyl-2-oxime-1,2-propanodione to measure the urea released from arginine by arginase had, in our case, the advantage of reacting also with arginine and citrulline. Thus, the values obtained from the perchloric acid supernatants at time 0 represent the sum of urea, arginine and citrulline (all reacting with the oxime23) present in the tissue before incubation to measure the lyase activity. These values were later used for calculation of their molal concentrations in tissues (WAT sites and liver) as a composite value of its content in urea + arginine + citrulline.
Analysis of plasma parameters
Blood plasma was deproteinized with acetone24 and used for the analysis of individual amino acids using an amino acid analyzer (Biochrom 30+, Biochrom, Cambridge, UK). Plasma urea was estimated with kit #11537 (Spinreac, Sant Esteve de Bas, Spain). Plasma and tissue water contents were calculated as previously described13 for the estimation of molal concentrations of arginine, citrulline and urea.Gene expression analysis
Total tissue RNA was extracted from the frozen tissue samples using the Tripure reagent (Roche Applied Science, Indianapolis IN USA), and was quantified in a ND-100 spectrophotometer (Nanodrop Technologies, Wilmington DE USA). These data were also used to determine the total RNA content of the tissue (per g of weight or g of protein) in order to establish comparisons between the quantitative importance of gene expressions. RNA samples were reverse transcribed using the MMLV reverse transcriptase (Promega, Madison, WI USA) system and oligo-dT primers.Real-time PCR (RT-PCR) amplification was carried out using 10 μL amplification mixtures containing Power SYBR Green PCR Master Mix (Applied Biosystems, Foster City, CA USA), 4 ng of reverse-transcribed RNA and 150 nM of primers. Reactions were run on an ABI PRISM 7900 HT detection system (Applied Biosystems) using a fluorescent threshold manually set to 0.15 for all runs.
A semi-quantitative approach for the estimation of the concentration of specific gene mRNAs per unit of tissue/RNA or protein weight was used.25 Rplp0 was the charge control gene.26 We expressed the data as the number of transcript copies per gram of protein in order to obtain comparable data between the groups. The genes analyzed and a list of primers used is presented in Table 1.
| Enzyme | A | EC number | Gene | 5′ > 3′ | 3′ > 5′ | BP |
|---|---|---|---|---|---|---|
| a A = activity measured in addition to the analysis of gene expression. | ||||||
| Carbamoyl-phosphate synthase [ammonia], mitochondrial type 1 | 6.3.4.16 | Cps1 | ACCCATCATCCCCTCTGACT | ACACGCCACCTCTCCAGTAG | 118 | |
| Glutamine-dependent carbamoyl-phosphate synthase, type 2 | X | 6.3.5.5 | Cad | AGTTGGAGGAGGAGGCTGAG | ATTGATGGACAGGTGCTGGT | 90 |
| Ornithine carbamoyltransferase | X | 2.1.3.3 | Otc | CTTGGGCGTGAATGAAAGTC | ATTGGGATGGTTGCTTCCT | 126 |
| Arginino-succinate synthase 1 | X | 6.3.4.5 | Ass1 | CAAAGATGGCACTACCCACA | GTTCTCCACGATGTCAATGC | 100 |
| Arginino-succinate lyase | X | 4.3.2.1 | Asl | CCGACCTTGCCTACTACCTG | GAGAGCCACCCCTTTCATCT | 104 |
| Arginase, liver (type 1) | X | 3.5.3.1 | Arg1 | GCAGAGACCCAGAAGAATGG | GTGAGCATCCACCCAAATG | 126 |
| Arginase-2 | 3.5.3.1 | Arg2 | GCAGCCTCTTTCCTTTCTCA | CCACATCTCGTAAGCCAATG | 122 | |
| N-Acetyl-glutamate synthase | 2.3.1.1 | Nags | GCAGCCCACCAAAATCAT | CAGGTTCACATTGCTCAGGA | 82 | |
| Glutaminase, kidney isoform | 3.5.1.2 | Gls | CCGAAGGTTTGCTCTGTCA | AGGGCTGTTCTGGAGTCGTA | 63 | |
| Glutamate-ammonia ligase [glutamine synthetase] | 6.3.1.2 | Glul | AACCCTCACGCCAGCATA | CTGCGATGTTTTCCTCTCG | 148 | |
| Adenosine monophosphate deaminase 2 | 3.5.4.6 | Ampd2 | CGGCTTCTCTCACAAGGTG | CGGATGTCGTTACCCTCAG | 78 | |
| Glutamate dehydrogenase 1 (NADH) | 1.4.1.3 | Glud1 | GGACAGAATATCGGGTGCAT | TCAGGTCCAATCCCAGGTTA | 122 | |
| Nitric oxide synthase 3, endothelial cell type | 1.14.13.39 | Nos3 | CAAGTCCTCACCGCCTTTT | GACATCACCGCAGACAAACA | 138 | |
| 60S acidic ribosomal protein 0 (housekeeping gene) | — | Rplp0 | GAGCCAGCGAAGCCACACT | GATCAGCCCGAAGGAGAAGG | 62 | |
The possible contamination of RNA with DNA was checked before PCR cycling by charging known RNA on a number of samples of each batch. No spurious signals were observed. All the primers used for measurement of urea-cycle enzyme gene expressions were checked by Northern blots of the PCR-synthesized cDNA. In all cases, the cDNA obtained had the expected molecular weights.
Statistics
One-way ANOVA comparisons between groups, correlations and curve fitting (including Vi estimations) were analyzed with the Prism 5 program (GraphPad Software, San Diego CA USA).Results
Tissue distribution and analysis
The rats used weighed 373 ± 15 g. Table 2 shows the absolute and relative weights of the four WAT sites analyzed. The sum of the four WAT sites represent about 8% of in vivo body weight. The largest store was that of subcutaneous (SC) followed by epididymal (EP), retroperitoneal (RP) and mesenteric (ME) fat pads. Their protein content practically reversed this order, with the exception of the relatively high protein content in subcutaneous fat. The distribution of total RNA was different from that of protein, with marked differences between sites, mesenteric WAT showing 18-fold more RNA per g of tissue than retroperitoneal WAT; subcutaneous WAT RNA was only about 1/4th of mesenteric, and epididymal WAT dropped to 1/9th.| Units | SC WAT | ME WAT | EP WAT | RP WAT | ∑WAT | P | |
|---|---|---|---|---|---|---|---|
| a All values are the mean ± SEM of 6 different animals. Statistical significance of the differences between WAT sites were calculated using a one-way ANOVA analysis; post hoc Tuckey test: in each row, different superscript letters represent different (p < 0.05) site values. | |||||||
| Weight | g | 12.2 ± 0.20A | 4.94 ± 0.49B | 7.34 ± 0.64C | 6.29 ± 0.79BC | 30.8 ± 1.7 | <0.0001 |
| % BW | 3.27 ± 0.07A | 1.33 ± 0.18B | 1.97 ± 0.13C | 1.69 ± 0.22BC | 8.26 ± 0.47 | <0.0001 | |
| Protein | mg g−1 | 63.1 ± 11.6 | 74.2 ± 7.4 | 44.3 ± 1.6 | 65.1 ± 6.3 | NS | |
| RNA | μg g−1 | 248 ± 51.1A | 880 ± 84.3B | 94.3 ± 6.0AC | 48.8 ± 4.11C | <0.0001 | |
Urea-cycle enzyme activities and expressions
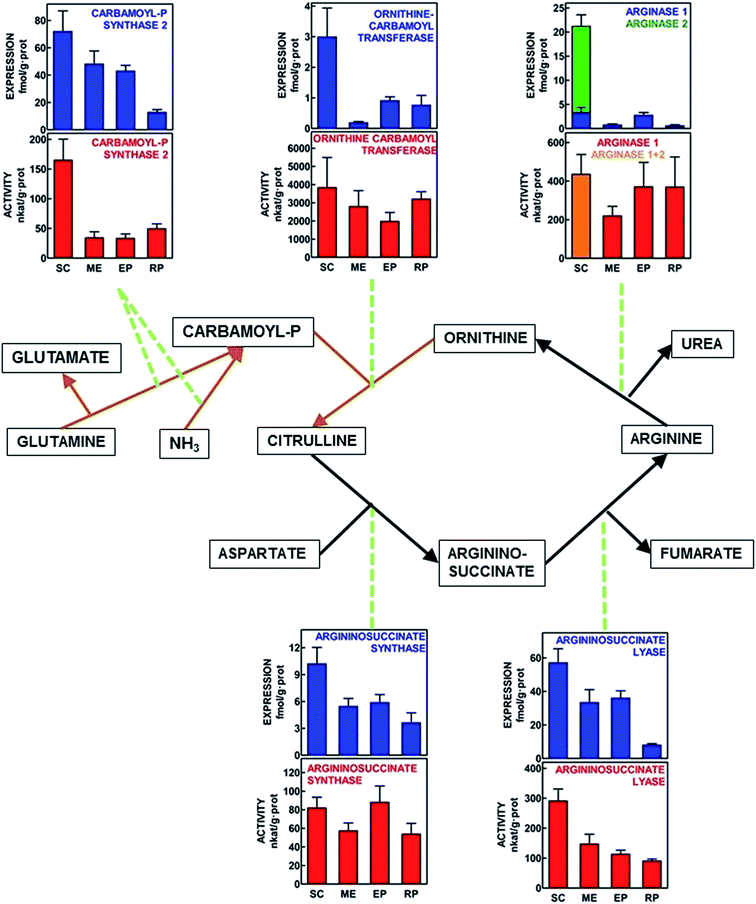
Fig. 1 shows a scheme of the urea cycle with the Vmax enzyme activities measured in WAT tissue sites, as well as the expression of the genes for the same enzymes, all referred to unit of protein weight for direct comparison between different sites. The main peculiarities of this representation were the finding of arginase 1 in all WAT sites, with a robust activity in most of them. Arginase 2 was expressed only in subcutaneous WAT, where arginase activity reflected the combined action of both isoforms. The activity data suggest that, in comparison, the probable contribution of arginase 2 was lower than that of arginase 1, despite higher expression values for arginase 2. Most enzyme activities (and gene expressions) showed the maximal values for subcutaneous WAT. The presence of measurable enzyme activities, backed by the expression of the corresponding genes in all WAT sites confirm that all enzymes of urea cycle were present and active in the four WAT sites studied. The statistical analysis of differences between sites is presented in Fig. 1.Activity/expression ratios
There were marked differences in the order of magnitude between the enzyme activities and their gene expressions, the differences being maximal for ornithine carbamoyl transferase. Thus, we calculated the quotients between enzyme activity and gene expression for each site. The data are presented in Table 3. Carbamoyl-P synthase activity/expression ratios were similar for SC and RP vs. ME and EP, with a 3–5-fold difference between both groups. The differences for ornithine carbamoyl transferase were the highest, with all sites showing similar ratios except for ME, which ratio was in the range of 3–10× that of the other sites. Arginino-succinate synthase and total arginase activity vs. arginase 1 expression ranges of ratios were rather uniform, and no significant differences between sites were observed. Arginino-succinate lyase showed similar ratios, except for RP, with values 2–3× higher. The SC ratio values for arginase were underestimated, since enzyme activity combined arginases 1 and 2.| Enzyme | Units | SC WAT | ME WAT | EP WAT | RP WAT | P |
|---|---|---|---|---|---|---|
| a The data are the quotients of enzyme activity (in μkat per nkat per g protein) and gene expression (fmol of the corresponding mRNA per g of protein). All values are the mean ± SEM of 6 different animals. Statistical significance of the differences between WAT sites was determined using a one-way ANOVA analysis; post hoc Tuckey test: in each row, different superscript letters represent different (p < 0.05) site values. | ||||||
| Carbamoyl-P synthase 2 | nkat fmol−1 | 2.65 ± 0.62A | 0.77 ± 0.21B | 0.76 ± 0.13B | 3.59 ± 0.46A | <0.0001 |
| Ornithine carbamoyl transferase | μkat fmol−1 | 1.22 ± 0.33A | 14.4 ± 4.4B | 2.28 ± 0.54A | 4.39 ± 0.90A | <0.0001 |
| Arginino-succinate synthase | nkat fmol−1 | 10.5 ± 2.8 | 11.4 ± 1.6 | 16.3 ± 3.7 | 16.7 ± 3.9 | NS |
| Arginino-succinate lyase | nkat fmol−1 | 5.95 ± 1.26A | 4.45 ± 0.70A | 3.22 ± 0.30A | 10.3 ± 1.4B | 0.0005 |
| Arginase 1 | nkat fmol−1 | 232 ± 72 | 253 ± 35 | 124 ± 35 | 378 ± 95 | NS |
The differences between enzymes were subjected to two additional analyses. First, using the data in Fig. 1, enzyme activities and gene expressions (both referred to g of tissue protein) were plotted and the correlation coefficients between the pairs of data were calculated for each enzyme. The results were carbamoyl-P synthase P = 0.0003, ornithine carbamoyl transferase P = 0.0008, arginino-succinate synthase P = 0.8844, arginino-succinate lyase P = 0.0012, and arginase 1, P = 0.0430. The only non-significant correlation between gene expression and the corresponding enzyme activity was that of arginino-succinate synthase.
The second analysis, shown in Table 4, depicts the statistical significance of the correlations between all pairs of urea cycle enzymes, both using activities (per g of tissue protein) and gene expressions (corrected by tissue RNA content). The objective was to find whether there was a uniform mechanism (shown by a high degree of correlation) linking the enzyme activities or gene expressions of the different enzymes involved in the cycle. Most data were highly correlated (in spite of the use of different WAT sites). The most patent exceptions being ornithine carbamoyl transferase and carbamoyl-P synthase (both mitochondrial) which activities were not correlated with neither of the arginino-succinate enzymes. In addition, the expression of ornithine carbamoyl transferase was not correlated with that of the lyase.
| a EA: P value for the correlation of paired enzyme activities for each animal and site, expressed in nkat per g of tissue protein; GE: P value for paired enzyme gene expressions for each animal and site (corrected by RNA tissue content), expressed in fmol of the corresponding mRNA per g of tissue RNA. | ||||
|---|---|---|---|---|
| Ornithine carbamoyl transferase | EA: P < 0.0001 | |||
| GE: P = 0.0026 | ||||
| Arginino-succinate synthase | EA: NS | EA: NS | ||
| GE: P = 0.0079 | GE: P < 0.0001 | |||
| Arginino-succinate lyase | EA: NS | EA: NS | EA: P = 0.0074 | |
| GE: P < 0.0001 | GE: NS | GE: P < 0.0001 | ||
| Arginase 1 | EA: P < 0.0001 | EA: P = 0.0008 | EA: P = 0.0252 | EA: P = 0.0270 |
| GE: P = 0.0059 | GE: P = 0.0306 | GE: P < 0.0001 | GE: P = 0.0036 | |
| Carbamoyl-P synthase | Ornithine carbamoyl transferase | Arginino-succinate synthase | Arginino-succinate lyase | |
Expressions of other amino acid metabolism enzymes
In Fig. 2 the gene expressions for enzymes complementary of the urea cycle are shown. This analysis includes the main ammonium-handling enzymes (glutamine synthetase, glutaminase, glutamate dehydrogenase and AMP deaminase, which are completed by carbamoyl-P synthase 2 shown in Fig. 1). The trend towards ammonium disposal (i.e. mitochondrial glutamate dehydrogenase, and cytoplasmic glutamine synthetase) showed higher expressions than glutaminase and AMP deaminase in all WAT sites, with the highest expression values for subcutaneous WAT and the lowest for retroperitoneal. N-Acetyl-glutamate synthase was expressed in all sites, albeit at the limit of safe expression measurement. Endothelial nitric oxide synthase was also expressed in all sites, subcutaneous WAT showing more than 2-fold expression values than the other sites, in a pattern similar to that AMP deaminase, and the glutamine enzymes.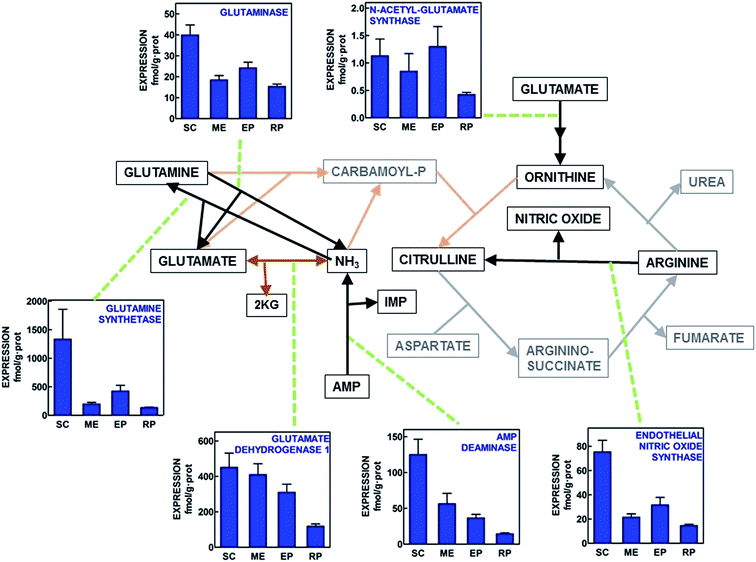 | ||
| Fig. 2 Schematic representation of the placement of additional enzyme gene expressions related to the operation of the urea cycle in four WAT sites of adult male Wistar rats. All data are presented as the mean ± SEM of 6 different animals per group. Blue columns depict the corresponding enzyme gene expression in fmol per g protein. Black lines represent cytosolic pathways analyzed; brown lines correspond to the mitochondrial pathways studied. Grey/orange lines depict the paths analyzed in Fig. 1 SC = subcutaneous WAT; ME = mesenteric WAT; EP = epididymal WAT and RP = retroperitoneal WAT. Statistical analysis of differences between groups (one-way ANOVA applied to WAT site). Glutaminase (P < 0.0001). N-Acetyl-glutamate synthase (NS). Glutamine synthetase (P = 0.0098). Glutamate dehydrogenase 1 (P = 0.0021). AMP deaminase (P < 0.0001). Endothelial nitric oxide synthase (P < 0.001). | ||
Comparison of WAT and liver urea-cycle enzyme activities
Table 5 depicts the mean values for liver urea-cycle enzyme activities, including total carbamoyl-P synthase. These values when expressed per g of protein were close in some cases to those presented in Fig. 1 for the four main WAT sites. However, arginase, and to a lesser extent carbamoyl-P synthase and arginino-succinate synthase showed higher values in liver. When the data are presented as the total content in catalytic potential (i.e. nanokatals in the whole organ or tissue site), we obtain the composite data also presented in Table 5. The direct comparison of the catalytic potential for urea cycle of liver vs. the combined (sum) of four main WAT sites showed an enormous difference for arginase (ratio liver/WAT in the range of 0.01). The differences were also marked for carbamoyl-P synthase and, to a lower extent, arginino-succinate synthase; however, the ratios were close to 1/3 to 1/2 of those of liver for arginino-succinate lyase and ornithine carbamoyl-transferase.| Weight | Protein | Carbamoyl-P synthase (nkat per g P) | Ornithine carbamoyl-transferase (μkat per g P) | Arginino-succinate synthase (nkat per g P) | Arginino-succinate lyase (nkat per g P) | Arginase (μkat per g P) | |
|---|---|---|---|---|---|---|---|
| a The liver values are presented in nkat/or μkat per g of tissue protein, and are the mean ± SEM of six different rats. The values for the four main WAT sites were calculated from the data of Table 2 and Fig. 1 and 2. They represent the total enzyme catalytic activity in the tissue as a whole (in nanokatals). | |||||||
| Liver | 509 ± 57 | 2.88 ± 0.44 | 274 ± 28 | 117 ± 15 | 227 ± 19 | ||
| Weight (g) | Protein (g) | Carbamoyl-P synthase (nkat) | Ornithine carbamoyl-transferase (nkat) | Arginino-succinate synthase (nkat) | Arginino-succinate lyase (nkat) | Arginase (nkat) | |
|---|---|---|---|---|---|---|---|
| Liver | 11.6 ± 0.5 | 2.21 ± 0.22 | 1110 ± 115 | 6183 ± 877 | 725 ± 78 | 273 ± 37 | 42![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 900 ± 3800 900 ± 3800 |
| Sum of 4 WAT sites | 30.8 ± 1.7 | 1.85 ± 0.14 | 75.0 ± 9.7 | 2107 ± 68 | 78.9 ± 7.5 | 134 ± 9 | 411 ± 61 |
| WAT/liver ratio | 2.6 | 0.84 | 0.07 | 0.34 | 0.11 | 0.50 | 0.01 |
Tissue urea and guanido amino acid levels
Table 6 shows the plasma molal concentrations of urea, arginine and citrulline, as well as the combined (molal) levels of urea + arginine + citrulline in the four WAT sites studied and liver. In all cases, the combined value for tissues was higher than their sum in plasma. The molal ratios of WAT/plasma for this combined figure of urea and urea-cycle intermediary amino acids were in the range of 3.5–5.0 for subcutaneous, mesenteric and epididymal WAT, and lower ratios (higher than 1 in any case) for retroperitoneal WAT. The ratio for liver was twice that of subcutaneous WAT.| μmol per g water | Sum of urea + arginine + citrulline (μmol per g water) | Tissue/plasma ratio | |
|---|---|---|---|
| a The data are the mean ± SEM of 6 animals. The tissue/plasma ratio is the quotient between the (measured) tissue combined urea + arginine + citrulline values expressed in molal units divided by the composite sum of plasma urea + arginine + citrulline also in molal units. The data used for composite values and ratios were homologous (i.e. obtained from the same individual rats). | |||
| Plasma urea | 4.27 ± 0.20 | 4.46 ± 0.19 | |
| Plasma arginine | 0.171 ± 0.019 | ||
| Plasma citrulline | 0.024 ± 0.002 | ||
| SC WAT | 19.7 ± 3.5 | 4.3 ± 0.8 | |
| ME WAT | 21.8 ± 4.2 | 5.0 ± 1.0 | |
| EP WAT | 16.2 ± 4.9 | 3.5 ± 0.9 | |
| RP WAT | 6.48 ± 1.78 | 1.4 ± 0.4 | |
| Liver | 39.0 ± 6.3 | 8.7 ± 1.4 | |
Unfortunately, the values obtained did not discriminate between arginine, citrulline and urea, but clearly showed that there must be a gradient of concentrations between any combination of these and plasma. These results seem to corroborate the hypothesis of an active urea cycle in WAT helping produce citrulline and arginine, but the wide differences with plasma, and the much higher relative levels of urea in plasma suggest that most WAT sites may produce significant amounts of urea.
Discussion
The main conclusion derived from the present study is the presence of all enzymes of the urea cycle (liver type) in WAT. The activities and their relationships were variable, depending on the site studied, but all enzymes were expressed in a degree sufficient to allow and justify the overall operation of the cycle. This assumption was reinforced by the presence of all enzyme activities of the urea cycle in the four WAT sites studied, which are, quantitatively, the largest discrete WAT masses in the rat.WAT nitrogen metabolism has been sparsely studied, with only a few publications dealing directly with this question;11,19 a small number of papers included WAT in enzyme distribution studies.27–29 This explains, in part, that the possible significant implication of WAT in amino acid metabolism has been seldom analyzed. Perhaps, the only clear exception is the postulated implication of WAT in the glucose–alanine cycle.10 We have recently found that WAT lactate production goes well beyond the simple limited export of 3C units under starvation.13 In addition, WAT contains several amino acid metabolism enzyme activities, such as glutamine synthetase,28 glutaminase,29 and AMP-deaminase.27 Taken together, these data hint to a functional coordinated metabolism of ammonium.
Evidently, the main problems that WAT present as an “active” metabolic organ are dual: (a) its wide dispersion and function-adapted specialization, depending on the site, and (b) the limited proportion of “active tissue” with respect to tissue mass, since most of the tissue is fat. The (a) question has been widely analyzed,30,31 but in any case there is a substantial pattern of uniformity encompassing all sites, which act coordinately as a single energy store;4 WAT has been considered to act as a disperse organ.32
With respect to the (b) question, it is true that excess tissue fat makes comparisons difficult, and needs the application of a number of methodological modifications and assumptions to allow work on it, but the non-fat and non-fiber (collagen, elastin) remaining tissue, including the non-fat part of adipocytes has a differentiated metabolic activity.33 In fact, a number of enzyme activities or expressions, such as those we present for arginino-succinate lyase and ornithine carbamoyl transferase were in the same range (per unit of protein weight) than those of the liver. When we compare the total catalytic capability of the four main WAT sites and that of liver, the hepatic preponderance is clear. The maximal differences were observed for arginase (the liver is the main site for urea production) and carbamoyl-P synthase (again liver is a main site for detoxification of ammonium), but the differences decreased considerably for the other urea cycle enzymes, reinforcing the postulated role of extra-hepatic citrulline (and arginine) provider. However, the four main WAT sites contain about half of body WAT,4 which may result, in practical terms, in doubling the ratios in Table 5 when considering WAT as a whole organ, a condition that is greatly changed in obesity. In sum, the possible capabilities of WAT urea cycle may be quantitatively important and comparable only to liver. A question that needs considerable additional work to be adequately understood.
The RNA content of a tissue is, probably, a fair (albeit indirect) indicator of overall “metabolic activity”. The data for WAT sites showed wider differences than expected. However, when these data were used to compute the gene expressions, a good degree of homogeneity between WAT sites was maintained. This is an indirect indication of shared molecular (largely genomic) regulation of urea cycle enzymes despite the specialization and diverse metabolic activity of the sites.
In spite of their expressions being correlated with those of the rest of urea cycle enzymes, the main controlling enzyme of the cycle, arginino-succinate synthase34 and the accompanying lyase, show patently lower activities than the other urea cycle enzymes. The discordance between the uniform gene regulation (highly inter-correlated expressions for all enzymes) and uncorrelated gene expressions vs. activities for arginino-succinate enzymes agree with a possible post-transcriptional modulation of this controlling section35 of the cycle. The wide differences between enzymes as to the observed ratios of activity vs. gene expression are a clear indication of a probably powerful additional mechanism of regulation, as is their turnover rates, which establish the real active life of the enzyme molecules. Higher activity vs. expression ratios, as is the case of ornithine carbamoyl transferase, suggest longer half-lives (and probably limited regulation via inactivation) compared with the critical controlling enzymes such as carbamoyl-P synthase and, especially arginino-succinate synthase. The isolated interpretation of either activity or gene expression alone may result in conflicting conclusions, not necessarily coincident with those we obtain from their combined analysis.
The sites for control of the urea cycle derived from the correlation analyses are: an overall genomic control of the cycle as a whole, affecting all sites, and two key control points, both well known: carbamoyl-P synthesis36 and arginino-succinate synthesis.34 However, the data for arginino-succinate lyase, an enzyme seldom analyzed separately, point to its activity sharing with the synthase the control of arginino-succinate, and thus the ultimate control of arginine production.37 Probably, both arginino-succinate-related enzymes are subject to additional post-transcriptional regulation that breaks up the relative uniformity of the correlation data.
The finding of a complete urea cycle in WAT represents a departure of the concept of liver as the only factual regulator of N disposal. Altered urea cycle in metabolic syndrome20 and decreased urea production38 attest a decreasing role of liver in amino acid disposal under conditions of inflammation. This fact, so far, remains unexplained in spite of the excess energy and 2-amino N that characterizes dietary-induced metabolic syndrome.
The present study opens the question of what is the role of urea cycle in WAT. The cycle we described probably does not seem, initially, focused on the conversion of ammonia to urea. Our data show that the main ammonia producing enzymes (glutaminase, AMP deaminase) are less expressed than the enzymes using it to synthesize amino-N, amido-N or guanido-N. On the ammonia-disposing side, we find glutamate dehydrogenase, probably working mainly in the direction of glutamate synthesis,39 glutamine synthetase (to produce glutamine) and carbamoyl-P synthases. The purine nucleotide cycle is probably present in adipose tissue,27 and, at least in muscle, its activity is closely related to glycolysis.40 WAT possesses a very active glycolytic capacity,13 which may help increase the up-regulation of the controlling enzyme, AMP deaminase.41 However, a main function of the purine nucleotide cycle is to decrease AMP levels; and thus, affects AMPK, and WAT metabolic regulation. We do not have enough data to establish the existence of a significant flow of amino-N to ammonia-N in WAT, but a sizeable activity28 and hormone-regulated expression42 of glutamine synthetase seems to suggest that probably the urea cycle is not, anyway, the main outlet for a significant production of ammonia in WAT under basal conditions.
The high activity of ornithine carbamoyl transferase, compared with the low arginino-succinate activity (in spite of not so different gene expressions), suggest that the main role of WAT urea cycle may not be the production of urea, but the generation of citrulline. This function seems obvious for mesenteric WAT, acting in the same way that the neighboring intestine to provide the liver (and kidney) with intermediate urea cycle substrates.43 However, this function is paralleled in all four WAT sites, which suggests a higher, possibly peripheral, demand for citrulline. This amino acid plays an important role as a controller of metabolism.44 Citrulline and arginine levels are closely related, in a way that the former is a more effective provider of arginine than arginine itself;45 this is due, probably to the strict control of arginine levels and availability in relation to nitric oxide synthesis46 and its implication in a number of other regulatory mechanisms.47 Citrulline is a floating reserve of arginine for peripheral tissues. The main known body citrulline source, intestine (including mesenteric WAT) is directly connected to the liver via portal vein, but the liver extracts only a minor fraction of portal blood citrulline for urea synthesis.48 The kidney takes up most of systemic blood citrulline (largely from liver efflux) into arginine48 under standard conditions. However, kidney arginine production from citrulline exceeds liver output.49 We postulate that WAT may justify the difference thanks to a robust ornithine carbamoyl-transferase activity sustained by the upper part of the urea cycle described here. In this respect, the difference between kidney and WAT may be the dependence of kidney of intestinal production and variable liver retention of citrulline to maintain its production of arginine. Perhaps, the immediate independence of WAT from diet as compared to the gut may justify a role for WAT to sustain the production of citrulline independently of the N-disposal function of the intestine–liver urea cycle; as well as to limit the dependence of renal-released arginine supply from this setup.
The existence of a marked gradient in WAT (and liver), for urea cycle products is not compatible with their uptake from the blood, and strongly supports that the flow of these substrates (or at least part of them) goes from WAT cells to blood. Consequently, the tissue produces them de novo. Additionally, the levels of enzyme activity in the tissues tested show a clear parallelism with the values of the molality ratios.
WAT has a significant potential for the generation of nitric oxide, largely thanks to the endothelial isozyme of nitric oxide synthase. The production of this key regulatory factor is tightly regulated, both though the modulation of the expression and activity of the enzyme,50 but also controlling the supply of arginine.51 In metabolic syndrome, the low overall production of urea13,38 has been attributed to the preservation of arginine to fuel an increased synthesis of nitric oxide,52 a consequence of inflammation. The synthesis of nitric oxide also yields citrulline as byproduct, supporting the postulated citrulline/arginine-producing role of WAT's urea cycle.
Arginase also regulates the availability of arginine to fuel nitric oxide synthesis. The widespread presence of arginases in many peripheral tissues53 has been attributed to a control role through competitive arginine disposal.54 However, in the case of WAT, we found arginase activity in all sites, but only the arginase 1 Arg1 gene was expressed; that for arginase 2, Arg2, was expressed only in subcutaneous WAT (which also presented the highest arginase activity). Arginase 1 is commonly associated with the “regular” liver urea-producing cycle, thus its presence in all WAT sites could be considered, a priori, to be more related to breakup of arginine to yield ornithine (and urea) than to control the flow of arginine towards nitric oxide. This peculiar arginase isozyme distribution also makes WAT closer to the liver in its unique configuration of urea cycle.
In general terms, the described functions were shared by the four WAT sites studied (the correlation analyses include data from all of them), but there were marked specific differences related to location. Subcutaneous WAT showed the highest activities/expressions for most enzymes, suggesting that it is the most probable WAT site to produce urea, eventually, in significant amounts. Its location, relationship with the skin and its biota may be related to its highest expression of nitric oxide synthase and the presence of arginase 2. The role of nitric oxide within the defense system and in inflammation may be a primary justification for these differences. The also higher gene expressions of both glutamine cycle enzymes hint to an active amino acid metabolism, helped by proteolysis55 and active branched-chain amino acid catabolism.56 The large mass of subcutaneous adipose tissue confers a prominent position in its possible role in amino acid metabolism to this part of the adipose organ.
The data available hint at WAT acting not only locally and peripherally, but also complementing the function of the splanchnic bed organs in the regulation of – at least—arginine metabolism. We have described a complete, (mainly) expression-regulated (and coordinated), active urea cycle in the four largest WAT sites; we hinted at its possible function in the control of arginine handling through citrulline synthesis. These results, however, do not preclude the possibility of WAT to carry out a full urea cycle, eliminating excess N via a “peripheral” urea cycle. In our opinion, the metabolic potential and control role of energy metabolism of WAT may be second only to liver; this impression should be extended to amino acid metabolism too. The large WAT mass and widespread distribution probably compensate the deceptive low proportion of “live cytosol”. The results shown in this study correspond to a “basal” state, of male rats under a standard diet. It can be speculated that the powerful urea cycle machinery installed in WAT may play a different role under other dietary and metabolic circumstances (i.e. obesity, inflammation). Our study confirms that WAT contains a full urea cycle, probably fully functional, but we are yet far from knowing its possible function in such complex organ as is WAT.
Conclusions
All these considerations bring us to suggest that the urea cycle is operative (but its functionality under basal conditions remains to be proven) in all four WAT sites. The right enzymes are expressed, and show measurable activities; there is a clear coordinated regulation on their expression (and, partly, of their activities); and the key control points: ammonium availability and arginino-succinate enzymes are the obviously best controlled bottlenecks for the urea cycle operation. The data presented suggest that WAT may produce urea but its urea cycle is geared, essentially, to provide citrulline as precursor of arginine, which is probably produced too, largely for peripheral tissues utilization. In this sense, WAT, by providing urea cycle intermediaries, as the intestine does,48 complements liver function.Conflict of interest
The authors declare that they have no conflict of interests.Author contributions
SAr and SAg did the entire animal handling and laboratory work. JAFL and SAr carried out the statistical analyses. XR, JAFL and MA designed the experiments and established the main conclusions. MA conceived the study and wrote the paper. All authors participated in the discussion of the results and in the preparation of the final text.Acknowledgements
This study was done with the partial support of grants of the Plan Nacional de Investigación en Biomedicina (SAF2012-34895) and the Plan Nacional de Ciencia y Tecnología de los Alimentos (AGL-2011-23635) of the Government of Spain, as well as of the CIBER-OBN Research web. S. Agnelli was the recipient of a Leonardo da Vinci fellowship, and S. Arriarán had a predoctoral fellowship of the Catalan Government, in both cases covering part of the time invested in this study.References
- H. Hauner, Physiol. Behav., 2004, 83, 653–658 CrossRef CAS PubMed.
- C. Deveaud, B. Beauvoit, B. Salin, J. Schaeffer and M. Rigoulet, Mol. Cell. Biochem., 2004, 267, 157–166 CrossRef CAS PubMed.
- P. A. Tataranni, D. E. Larson and E. Ravussin, J. Am. Coll. Nutr., 1994, 13, 569–574 CrossRef CAS PubMed.
- M. M. Romero, S. Roy, K. Pouillot, M. Feito, M. Esteve, M. M. Grasa, J. A. Fernández-López, M. Alemany and X. Remesar, PLoS One, 2014, 9, e90995 Search PubMed.
- J. Sanchez-Gurmaches and D. A. Guertin, Nat. Commun., 2014, 5, 4099 CrossRef CAS PubMed.
- R. Cancello, J. Tordjman, C. Poitou, G. Guilhem, J. L. Bouillot, D. Hugol, C. Coussieu, A. Basdevant, A. Bar Hen, P. Bedossa, M. Guerre-Millo and K. Clément, Diabetes, 2006, 55, 1554–1561 CrossRef CAS PubMed.
- M. Alemany, J. Clin. Endocrinol. Metab., 2011, 96, 66–68 CrossRef CAS PubMed.
- E. Fuentes, F. Fuentes, G. Vilahur, L. Badimon and I. Palomo, Mediators Inflammation, 2013, 2013, 136584 Search PubMed.
- K. N. Frayn and S. W. Coppack, Diabetologia, 1990, 33, 740 CrossRef CAS PubMed.
- K. Snell and D. A. Duff, Biochem. Biophys. Res. Commun., 1977, 77, 925–931 CrossRef CAS PubMed.
- T. J. Kowalski, G. Y. Wu and M. Watford, Am. J. Physiol., 1997, 273, E613–E622 CAS.
- K. R. Markan, M. J. Jurczak and M. J. Brady, Mol. Cell. Endocrinol., 2010, 318, 54–60 CrossRef CAS PubMed.
- S. Arriarán, S. Agnelli, D. Sabater, X. Remesar, J. A. Fernández-López and M. Alemany, PLoS One, 2015, 10, e0119572 Search PubMed.
- F. J. López-Soriano, J. A. Fernández-López, T. Mampel, F. Villarroya, R. Iglesias and M. Alemany, Biochem. J., 1988, 252, 843–849 CrossRef.
- G. Pilon, P. Penfornis and A. Marette, Horm. Metab. Res., 2000, 32, 480–484 CrossRef CAS PubMed.
- L. Millet, P. Barbe, M. Lafontan, M. Berlan and J. Galitzky, J. Appl. Physiol., 1998, 85, 181–188 CAS.
- F. Karpe, B. A. Fielding, J. L. Ardilouze, I. A. Macdonald and K. N. Frayn, J. Physiol., 2002, 540(3), 1087–1093 CrossRef CAS.
- J. Conde, M. Scotece, R. Gómez, V. López, J. J. Gómez-Reino, F. Lago and O. Gualillo, BioFactors, 2011, 37, 413–420 CrossRef CAS PubMed.
- F. J. López-Soriano and M. Alemany, Arch. Int. Physiol. Biochim., 1986, 94, 121–125 Search PubMed.
- D. Sabater, S. Agnelli, S. Arriarán, J. A. Fernández-López, M. M. Romero, M. Alemany and X. Remesar, BioMed Res. Int., 2014, 2014, 959420 Search PubMed.
- O. H. Lowry, R. W. Rosebrough, A. L. Farr and R. J. Randall, J. Biol. Chem., 1951, 193, 265–275 CAS.
- S. Arriarán, S. Agnelli, J. A. Fernández-López, X. Remesar and M. Alemany, J. Enzyme Res., 2012, 3, 29–33 Search PubMed.
- E. L. Oginsky, Methods Enzymol., 1957, 3, 639–643 Search PubMed.
- L. Arola, E. Herrera and M. Alemany, Anal. Biochem., 1977, 82, 236–239 CrossRef CAS PubMed.
- M. M. Romero, M. M. Grasa, M. Esteve, J. A. Fernández-López and M. Alemany, Nutr. Metab., 2007, 4, 26 Search PubMed.
- G. Bamias, D. Goukos, E. Laouidi, I. G. Balla, S. I. Siakavellas, G. L. Daikos and S. D. Ladas, Inflammatory Bowel Dis., 2013, 19, 2840–2847 CrossRef PubMed.
- L. Arola, A. Palou, X. Remesar and M. Alemany, Horm. Metab. Res., 1981, 13, 264–266 CrossRef CAS PubMed.
- L. Arola, A. Palou, X. Remesar and M. Alemany, Horm. Metab. Res., 1981, 13, 199–202 CrossRef CAS PubMed.
- J. M. Kowalchuk, R. Curi and E. A. Newsholme, Biochem. J., 1988, 249, 705–708 CrossRef CAS PubMed.
- S. Cinti, Dis. Models & Mech., 2012, 5, 588–594 CAS.
- T. Tchkonia, T. Thomou, Y. Zhu, I. Karagiannides, C. Pothoulakis, M. D. Jensen and J. L. Kirkland, Cell Metab., 2013, 17, 644–656 CrossRef CAS PubMed.
- S. Cinti, Prostaglandins, Leukotrienes Essent. Fatty Acids, 2005, 73, 9–15 CrossRef CAS PubMed.
- J. R. Peinado, Y. Jiménez-Gomez, M. R. Pulido, M. Ortega-Bellido, C. Díaz-López, F. J. Padillo, J. López-Miranda, R. Vázquez-Martínez and M. A. M. Malagón, Proteomics, 2010, 10, 3356–3366 CrossRef CAS PubMed.
- A. J. Meijer, C. Lof, I. C. Ramos and A. J. Verhoeven, Eur. J. Biochem., 1985, 148, 189–196 CrossRef CAS PubMed.
- G. Hao, L. J. Xie and S. S. Gross, J. Biol. Chem., 2004, 279, 36192–36200 CrossRef CAS PubMed.
- T. Saheki, T. Ohkubo and T. Katsunuma, J. Biochem., 1978, 84, 1423–1430 CAS.
- R. J. Haines, L. C. Pendleton and D. C. Eichler, Int. J. Biochem. Mol. Biol., 2011, 2, 8–23 CAS.
- T. Barber, J. R. Viña, J. Viña and J. Cabo, Biochem. J., 1985, 230, 675–681 CrossRef CAS PubMed.
- J. D. McGivan and J. B. Chappell, FEBS Lett., 1975, 52, 1–7 CrossRef CAS PubMed.
- K. Tornheim, J. Theor. Biol., 1979, 79, 491–541 CrossRef CAS PubMed.
- K. Tornheim and J. M. Lowenstein, J. Biol. Chem., 1972, 247, 162–169 CAS.
- M. J. Lee, D. W. Gong, B. F. Burkey and S. K. Fried, Am. J. Physiol., 2011, 300, E571–E580 CAS.
- G. Wu, A. G. Borbolla and D. A. Knabe, J. Nutr., 1994, 124, 2437–2444 CAS.
- C. Breuillard, L. Cynober and C. Moinard, Amino Acids, 2015, 47, 685–691 CrossRef CAS PubMed.
- E. Schwedhelm, R. Maas, R. Freese, D. Jung, Z. Lukacs, A. Jambrecina, W. Spickler, F. Schulze and K. H. Böger, Br. J. Clin. Pharmacol., 2008, 65, 51–59 CrossRef CAS PubMed.
- F. Mariotti, K. J. Petzke, D. Bonnet, I. Szezepanski, C. Bos, J. F. Huneau and H. Fouillet, Am. J. Clin. Nutr., 2013, 97, 972–979 CrossRef CAS PubMed.
- J. E. Huh, J. Y. Choi, Y. O. Shin, D. S. Park, J. W. Kang, D. Nam, D. Y. Choi and J. D. Lee, Int. J. Mol. Sci., 2014, 15, 13010–13029 CrossRef CAS PubMed.
- H. G. Windmueller and A. E. Spaeth, Am. J. Physiol., 1981, 241, E473–E480 CAS.
- Y. M. Yu, J. F. Burke, R. G. Tompkins, R. Martin and V. R. Young, Am. J. Physiol., 1996, 271, E1098–E1109 CAS.
- K. Kikuchi-Utsumi, B. Gao, H. Ohinata, M. Hashimoto, N. Yamamoto and A. Kuroshima, Am. J. Physiol., 2001, 282, R623–R626 Search PubMed.
- K. Velickovic, M. Markelic, I. Golic, V. Otasevic, A. Stancic, A. Jankovic, M. Vucetic, B. Buzadzic, B. Korac and A. Korac, Eur. J. Nutr., 2014, 53, 813–821 CrossRef CAS PubMed.
- J. G. Zhou, D. D. Kim and R. D. Peluffo, Am. J. Physiol., 2010, 299, C230–C239 CrossRef CAS PubMed.
- S. Y. Choi, C. N. Park, M. J. Ahn, J. H. Lee and T. K. Shin, Acta Histochem., 2012, 114, 487–494 CrossRef CAS PubMed.
- W. Shin, D. E. Berkovitz and S. Ryoo, Exp. Mol. Med., 2012, 44, 594–602 CrossRef CAS PubMed.
- B. W. Patterson, J. F. Horowitz, G. Wu, M. Watford, S. W. Coppack and S. Klein, Am. J. Physiol., 2002, 282, E931–E936 CAS.
- D. E. Lackey, C. J. Lynch, K. C. Olson, R. Mostaedi, M. Ali, W. H. Smith, F. Karpe, S. Humphreys, D. H. Bedinger, T. N. Dunn, A. P. Thomas, P. J. Oort, D. A. Kieffer, R. Amin, A. Bettaieb, F. G. Haj, P. Permana, T. G. Anthony and S. H. Adams, Am. J. Physiol., 2013, 304, E1175–E1187 CrossRef CAS PubMed.
| This journal is © The Royal Society of Chemistry 2015 |