Preparation of keratin/chlorhexidine complex nanoparticles for long-term and dual stimuli-responsive release†
Xuelian
Zhi
,
Yanfang
Wang
,
Pengfei
Li
,
Jiang
Yuan
* and
Jian
Shen
*
Jiangsu Key Laboratory of Biofunctional Materials, College of Chemistry and Materials Science, Nanjing Normal University, Nanjing 210023, China. E-mail: bioalchem@yahoo.com; jshen@njnu.edu.cn; Fax: +86 25 83599188; Tel: +86 25 85891536
First published on 16th September 2015
Abstract
Nanoscale polyion complex formation via the electrostatic complexation of a polyelectrolyte and a charged drug is the most convenient method for building a drug delivery system that simultaneously realizes the carrier preparation and drug embedding. Herein, we prepared keratin/chlorhexidine complex nanoparticles (KCNPs) based on a drug-induced ionic gelation technique without using a crosslinker, organic solvent or surfactant. These KCNPs exhibited good stability even after having been long-standing for 14 days. The KCNPs were characterized using FTIR, DLS, SEM and TEM. It was found that these nanoparticles had a spherical morphology with a diameter of about 180 nm, and a negatively charged surface with a zeta potential of about −39.1 mV. The cell toxicity of the KCNPs at different dosage levels was evaluated using the MTT assay method, indicating their slight cytotoxicity at lower dosages. The antibacterial activity against E. coli and S. aureus was determined using the zone of inhibition method. It seemed that the KCNPs had better antibacterial activity against S. aureus than against E. coli. Drug delivery profiles showed that the chlorhexidine (CHX) loaded nanoparticles exhibited both pH- and glutathione-responsive character. These keratin-based complex nanoparticles can be regarded as a valuable stimuli-responsive strategy for the delivery of anticancer agents.
1. Introduction
Keratin is a chief component found in hair, skin, fur, wool, horn, and feathers. It has a high content of cysteine and serine, and a large number of hydroxyl amino acids. Keratin based biomaterials have emerged as potential candidates for many biomedical and biotechnological applications due to their intrinsic biocompatibility, biodegradability, mechanical durability, and natural abundance.1 Keratin based films can be used as a wound dressing,2,3 implantable device coating4,5 and cell encapsulant,6,7 and in ocular surface reconstruction.8,9 Keratin-based hydrogels are neuroinductive and capable of facilitating regeneration in a peripheral nerve injury model in mice.10,11 Keratin hydrogels can also act as a hemostatic agent in a rabbit model for lethal liver injury,12 a carrier for rhBMP-2,13 and a platform for antibiotics.14 Keratin-based fibers have a potential use in tissue engineering scaffolds. Due to its poor mechanical property, keratin is usually blended with biodegradable polymers such as poly(3-hydroxybutyrate-co-3-hydroxyvalerate),15 poly(lactic acid),16 and poly(ε-caprolactone).17Keratin should be an ideal drug carrier due to its advantages such as good biocompatibility, biodegradability, absorbability, and non-immunogenicity. In addition, keratin is richly negatively charged, which favors the electrostatic absorption of small positively charged molecules, such as acid salt typed drugs. Furthermore, the large amount of carboxyl groups within keratin makes it a pH responsive smart drug carrier candidate. However, little work has been done on keratin-based drug carriers. The van Dyke group has described the delivery and activity of the antibiotic ciprofloxacin delivered from a keratin hydrogel.14 The Wang group has prepared a keratin hydrogel and film for controlling drug release.18,19 The Liu group has synthesized poly(ethylene glycol) (PEG) and poly(N-(2-hydroxypropyl)methacrylamide) (PHPMA) grafted keratin and used it as a dual reduction and enzyme responsive drug carrier.20,21
Various methods have been used to prepare protein-based nanoparticles, such as desolvation,22 nanoprecipitation,23 self-assembly,24 and so on. Among them, the desolvation and nanoprecipitation procedures need the use of organic solvents or additives, which may cause protein denaturation. For self-assembly procedures, protein and drug solutions are directly mixed to allow their interaction based on specific forces such as hydrogen bonding and hydrophobic interactions. For our modified keratin, a large amount of carboxylate groups (–COO−) can strongly attract the cationic drug. This electrostatic complexation leads to the formation of nanoparticles. Based on electrostatic interactions, Malek et al.25 have prepared cisplatin-incorporated nanoparticles using the drug-induced ionic gelation technique. The ionic gelation technique has been used widely to prepare chitosan nanoparticles.26–32 To the best of our knowledge, this method has not yet been reported to prepare protein nanoparticles.
Chlorhexidine (CHX) is widely used to treat and prevent skin and mucosal infections with low toxicity. In dentistry, it has been recognized as a gold standard against plaque and gingivitis for three decades.33 Due to its cationic charge, CHX is used as a drug model. The purpose of this study is to develop a facile method to fabricate stable keratin-based drug delivery systems without any chemical cross-linkers and surfactants. Herein, keratin/CHX complex nanoparticles (KCNPs) were prepared using the drug induced ionic gelation technique via electrostatic interactions (Fig. 1). Then, the size, zeta potential, and drug-loading capacity of the KCNPs were characterized using DLS, SEM, and TEM. An MTT assay was used to evaluate their cell toxicity. The pH dependent CHX delivery behavior was tested at pH 5.29, 7.4 and 9.18. The glutathione (GSH) sensitive release behavior was also tested at pH 7.4. These KCNPs have potential for long-term and controlled release of CHX for antibacterial applications.
 | ||
| Fig. 1 Schematic representation of the preparation of keratin/CHX complex nanoparticles (KCNPs) using the ionic gelation method. | ||
2. Materials and methods
2.1 Materials
Chlorhexidine acetate (C22H30N10Cl2·2C2H4O2) was supplied by Aladdin (Shanghai, China) with a purity of 98%. E. coli (ATCC 25922) and S. aureus (ATCC 25923) were provided by the Jiangsu Provincial Center for Disease Prevention and Control, China. All other chemicals were of analytical grade and were used without further purification.2.2 Extraction of keratin2,34,35
Human hair was washed with soap and 70% ethanol to remove surface oil, followed by being rinsed extensively with water and dried and then cut into short pieces. The pretreated hair was mixed with urea, sodium dodecyl sulfate (SDS), 2-mercaptoethanol and water. The mixture was kept under stirring for 48 h at 65 °C and then filtered. Subsequently, the filtrate was dialysed against deionized water for 48 h to afford a colorless solution. The dialysate was then allowed to react with iodoacetic acid for stability. Finally, this dialysate was dialysed again and lyophilized to obtain S-(carboxymethyl) keratin. The molecular weight of keratin was analyzed using SDS-polyacrylamide gel electrophoresis (PAGE).2.3 Preparation of KCNPs
KCNPs were prepared using the ionic gelation process. Keratin and CHX were each dissolved in an aqueous solution to obtain a stock solution with a final concentration of 1 mg mL−1. Then, the CHX solution was added dropwise at a speed of 1.5 mL h−1 under constant stirring to the keratin solution according to the ratios listed in Table 1, followed by stirring for 60 min and storage at room temperature overnight. The mixture was dialyzed for 1 day to remove unstable adsorbed CHX. Subsequently, the free CHX dialysate was measured using an ultraviolet spectrometer to determine the CHX loading content and encapsulation efficiency. The dialysate was lyophilized for future studies. The drug loading content (LC) and drug encapsulation efficiency (EE) were calculated according to the following equations:| Samples | EE (%) | LC (%) |
|---|---|---|
Weight ratio of keratin and CHX = 9![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 1 |
91.2 | 9.2 |
Weight ratio of keratin and CHX = 7![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3 3 |
64.1 | 21.5 |
Weight ratio of keratin and CHX = 5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 5 5 |
46.1 | 31.6 |
2.4 Characterization of the KCNPs
The infrared spectra of keratin and the KCNPs were recorded within the range of 4000–400 cm−1 using a Perkin-Elmer FT-IR spectrometer in KBr. The hydrodynamic diameter distribution and zeta potentials of the nanoparticles were determined using the dynamic light scattering (DLS) method using a Malvern Zetasizer Nano ZS90 system (Malvern Instruments Company Limited, British), in which freeze-dried KCNPs were dispersed in different pHs of phosphate buffer solutions. All analyses were triplicated and the results were the average of the three runs. The morphology of the KCNPs was observed using transmission electron microscopy (TEM, JEM-100S, JEOL, Japan) and scanning electron microscopy (SEM, Hitachi SU 3500, Japan).2.5 In vitro pH independent CHX release study
100 mg of KCNPs in 5 mL buffer solution was placed into a dialysis bag (MWCO 3500). Subsequently, the dialysis bag was dipped into a receptor compartment containing 200 mL dissolution medium, and was shaken gently at 37 ± 0.5 °C. Then, the medium was adjusted with pH 5.29 PBS, pH 7.4 PBS, and pH 9.18 sodium tetraborate buffer to test the release of CHX. The receptor compartment was closed to prevent evaporation losses from the dissolution medium. Periodically, 3 mL of the release medium was withdrawn and 3 mL of fresh PBS was added to the system. The CHX concentration in the sampled medium was determined using a UV spectrometer with absorption at the wavelength of 260 nm.2.6 In vitro GSH sensitive CHX release study
Regarding the GSH sensitive delivery, a similar procedure to that described above was followed except for the presence of GSH. The concentrations of GSH both inside and outside the dialysis bag were kept at 10 μM and in pH 7.4 PBS solution during the release experiments to mimic the redox conditions in blood plasma. At certain time intervals, the CHX concentrations of the outer side of the dialysis bag were determined using a UV spectrometer with absorption at the wavelength of 260 nm.2.7 Bacterial inhibition
The antimicrobial activity of the KCNPs was investigated against E. coli as the model for Gram-negative bacteria and S. aureus as the model for Gram-positive bacteria by the disc diffusion method. The bacteria were incubated in nutrient agar at 37 °C to reach 106 CFU mL−1. Agar plates were streaked with a sterile swab moistened with the KCNPs. Then, the bacteria were coated on the agar surface evenly and the plates were incubated overnight at 37 °C. The reaction of the microorganisms to the KCNPs was determined using the size of the inhibitory zone. When the materials have an excellent antibacterial activity, the inhibitory zones are very large.2.8 Cytotoxicity assay using MTT
The cytotoxicity of the KCNPs against NIH-3T3 cells was evaluated using an MTT assay. The cells in a DMEM medium containing 10% fetal bovine serum (FBS, Gibco) and 1% penicillin G–streptomycin were seeded into 96-well flat-bottomed plates with a density of 1 × 104 cells per well and then were incubated in CO2 atmosphere at 37 °C for 12 h. Next, the medium in each well was removed and the samples (100 μL) with various concentrations in DMEM were added to the wells. Subsequently, the cells were incubated for 48 h under the same conditions. The medium was replaced with 90 μL of fresh medium with 10 μL of the MTT solution (5 mg mL−1). After incubation for 4 h, the solution was removed, leaving the precipitate. Then, 100 μL of dimethyl sulfoxide (DMSO) was added to each well and was then oscillated for 30 min in the dark at room temperature. The cell viability was measured using a microplate reader (BioTek Synergy2) at a wavelength of 570 nm.2.9 Statistical analysis
Results are displayed as means ± standard deviation (SD). Statistical analysis was performed using one-way analysis of variance (ANOVA) with a Tukey posthoc method, and a significance level of p < 0.05 was chosen for all the tests.3. Results and discussion
3.1 Preparation of the KCNPs
Generally, keratin can be extracted using an oxidative method or a reduction method. Herein, we extracted keratin from human hair using the reduction strategy. The molecular weight of the extracted keratin was determined using SDS-PAGE analysis (Fig. 2). The patterns revealed two major, diffuse clusters of bands: 25 and 40 kDa. So, the molecular weight of keratin was concentrated on ca. 25 and 40 kDa.The ionic gelation method was used to prepare KCNPs with different ratios between keratin and CHX of 9/1, 7/3, and 5/5. The stability of the resulting KCNPs after 4 h and 14 days standing is shown in Fig. S1.† It shows that the KCNPs at the ratio of 9/1 are stable even when standing for as long as 14 days. As for the ratios of 7/3 and 5/5, the prepared nanoparticles are unstable after standing for 14 days. So, the ratio of 9/1 was fixed for the sample preparation.
The drug encapsulation efficiency (EE) and loading content (LC) of the KCNPs are listed in Table 1. Generally, a high drug concentration leads to a great drug loading content while a high keratin concentration tends to decrease drug encapsulation efficiency. Herein, the drug loading content was gradually increased while the encapsulation efficiency decreased with increasing mixed ratio between CHX and keratin. For keratin/CHX nanoparticles complexed at the ratio of 9 to 1, the EE and LC are 91.2% and 9.2%, respectively. From the above results, we can confirm that cationic drugs can be effectively entrapped in keratin molecules due to their strong electrostatic interaction.
3.2 Characterization of the KCNPs
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O stretching, is useful for the analysis of the secondary structure of the proteins. The peaks at 990 cm−1 and 614 cm−1 are the characteristic absorption peaks of the C–S and S–S bonds.36,37 In the case of KCNPs, there is no shift for the amide I peak, indicating the maintenance of the keratin conformation.
O stretching, is useful for the analysis of the secondary structure of the proteins. The peaks at 990 cm−1 and 614 cm−1 are the characteristic absorption peaks of the C–S and S–S bonds.36,37 In the case of KCNPs, there is no shift for the amide I peak, indicating the maintenance of the keratin conformation.
UV-Vis absorption measurement is a very simple method to explore the structural change and to know the complex formation. In the present study, we have recorded the UV-Vis absorption spectra of keratin and the keratin–CHX complex. In Fig. 3, the spectra showed a large blue shift in λmax from 276 nm to 263 nm, which can be related to complex formation between CHX and keratin through hydrogen bonds and electrostatic interaction.38,39
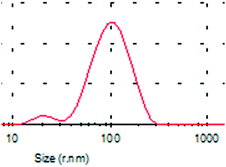
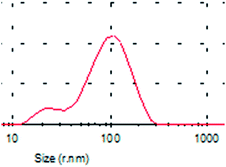
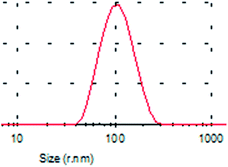
A typical TEM image of the KCNPs is shown in Fig. 4a, from which it can be seen that the KCNPs are solid spheres with a diameter of about 176 nm, which is very close to the result obtained using DLS. Generally, protein molecules exist as a form of colloid in water. As a result, there is no obvious difference between the sizes of the nanoparticles in aqueous medium and in the dry state. The morphology of the KCNPs is also observed using SEM and the results are shown in Fig. 4b. A statistical study of the particle size gives an average diameter of 170 nm.
3.3 In vitro pH independent CHX delivery profile
Keratin seems to be very appropriate to load CHX due to its rich carboxylic acid groups. Thanks to the electrostatic attraction between the cationic CHX molecules and the anionic keratin, CHX is accordingly loaded into the KCNPs with a satisfactory drug loading content of 9.2% and an encapsulation efficiency of 91.2% (Table 1). Fig. S4† shows the standard curve of CHX with a high goodness of fit as the function of concentration at 260 nm. To investigate the release characteristics of CHX from the KCNPs, the dialysis method is used. To demonstrate the pH-sensitive release, the release of CHX is studied over a period of 240 h under acidic (PBS, pH 5.29), 120 h under neutral (PBS, pH 7.4), and 120 h under basic (sodium tetraborate buffer, pH 9.18) conditions. Fig. 5 shows the whole release profiles of CHX from the KCNPs. No initial burst release occurs, indicating the strong complexation of CHX with keratin.In PBS with a pH value of 7.4, which corresponds to the physiological conditions of the blood stream, only ∼23% of CHX is delivered in the initial 100 h (Fig. 5a). The dialysate keeps clear during the whole release period. It indicates that the KCNPs are stable for a long time at physiological pH (Fig. S3†). The release of CHX is extraordinarily slow in PBS with a pH value of 9.18 (Fig. 5a). For example, only ∼8% of the loaded CHX is released within 100 h, suggesting that the CHX release from the KCNPs in basic environments is significantly suppressed. It is because the enhanced negative charges of the KCNPs adsorb CHX tightly, which inhibits CHX release.
The release of CHX is even slower at acidic pH 5.29 than at pH 7.4 in the initial 60 h (Fig. 5b). Depending on the protonation of keratin, the carboxylate groups (COO−) of the KCNPs can be converted into neutral carboxylic acid groups (COOH) by reducing the pH value; the hydrophilicity of KCNPs decreases. So, KCNPs become smaller and denser at pH 5.29 (Table 2), which suppresses the delivery. Thus, the release rate is faster than that at pH 9.18. It is due to the protonation of CHX increasing its hydrophilicity, which enhances the delivery. After releasing for the first 60 h, the release rate increases sharply. It is due to the synergetic effects of the dual protonation of CHX and keratin. The protonation of CHX increases the hydrophilicity of CHX. Meanwhile, the protonation of keratin weakens the electrostatic attraction of CHX. These two above synergetic effects both accelerate the delivery of CHX, resulting in this sharp release. It is interesting that the release profile shows a straight line, indicating a constant speed release of CHX. The pharmacokinetic release behavior follows a zero order equation. Fig. S3† shows the effect of the pH value on the stability of the KCNPs. The KCNPs tend to aggregate and deposit at acidic pH values, which are close to the iso-electric point of keratin. Such a pH-sensitive drug release behavior is desirable for tumor treatment since the ideal antitumor drug release should be slow in the neutral environment of the in vivo systemic circulation and relatively faster in the weak acidic environment of tumor tissues.40 In our study, about 39.5% of CHX is released within 240 h at pH 5.29. These results suggest that keratin-based nanocarriers are satisfactory for long term anticancer drug loading and release. The possible process of drug release could be expected. The pH-sensitive antitumor drug-loaded KCNPs would accumulate and the drug would be released at the tumor site due to the enhanced permeability and retention (EPR) effect.
The release process of CHX may be described with pseudo-first-order kinetics or pseudo-second-order kinetics equations.41 The pseudo-first-order kinetics equation can be represented in a linear form as
ln(qe − qt) = ln![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) qe − k1t qe − k1t | (1) |
The pseudo-second-order kinetics equation can be represented in a linear form as
| t/qt = 1/(k2qe2) + t/qe, | (2) |
3.4 In vitro GSH sensitive CHX release study
The release of CHX was also investigated at a GSH concentration of 10 μM corresponding to the level of GSH in blood plasma and pH 7.4 (Fig. 6a). It can be seen that about 45% of the loaded CHX can be finally released after reaching the plateau of 100 h. In comparison, only 23% of CHX can be released without GSH at the plateau of 70 h. These results could be attributed to the fact that the disulfide bonds of the KCNPs were partially cleaved by GSH through the reduction reaction, which resulted in an enhanced release of CHX. Fig. 6b shows the size of the KCNPs in the presence of 10 μM GSH at the delivery times of 0 h, 48 h, 72 h, and 120 h. The sizes of the KCNPs become larger with increasing delivery time, indicating the broken disulfide bonds of the KCNPs. The swollen KCNPs lead to a sufficient release of CHX as compared to that at pH 7.4 without GSH. Fig. 6c shows the accumulated release of CHX at pH 5.29 with and without 10 μM GSH as a function of time. The delivery is accelerated under the action of GSH as compared to that without GSH.3.5 In vitro cytotoxicity using an MTT assay
An antibacterial material requires low toxicity to normal cells at appropriate concentrations for killing bacteria efficiently. The cytotoxicity of the KCNPs towards fibroblast cells is evaluated using the MTT method. As shown in Fig. 7, at a lower concentration (2.5 μg mL−1 of KCNPs, i.e., 0.2 μg mL−1 of CHX), the cell viability of the KCNPs is 66% as compared to the FBS control, indicating their slight cytotoxicity. However, as the concentration gets even higher, they show significant cytotoxicity. Ostad et al. have used endometrial cells to assess the cytotoxicity of CHX and CHX-releasing devices. The results indicate that CHX is toxic at the concentration of 1 μg mL−1.42 It is also reported that 0.6 and 20 mg mL−1 of CHX decrease cell metabolism and viability by 60 and 70%, respectively.43 According to our drug loading content data (9.2%), 2.5 μg mL−1 of KCNPs is the equivalent of 0.23 μg mL−1 of CHX. It seems that the cytotoxicity of CHX is enhanced when CHX is complexed with keratin. | ||
| Fig. 7 Cell viability of the KCNPs at various concentrations towards NIH-3T3 cell growth determined using an MTT viability assay (* indicates a significant difference at the p < 0.05 level). | ||
3.6 Antibacterial test
CHX is used worldwide as a cationic antimicrobial agent against oral microbiota. The antibacterial activity of the KCNPs against E. coli and S. aureus is determined using the zone of inhibition method. The disks produced a clear zone of inhibition (Fig. 8). The zone of inhibition for S. aureus looks larger than that for E. coli. It seems that the KCNPs have better antibacterial activity against S. aureus than E. coli. The above results are consistent with the known conclusion that CHX is more effective against Gram-positive microbes than against Gram-negative microbes.444. Conclusion
In summary, keratin/CHX complex nanoparticles (KCNPs) are successfully prepared with a ca. 180 nm diameter. The CHX release behavior shows that these nanoparticles are both pH and GSH sensitive. Under acidic and blood plasma level GSH conditions, the KCNPs exhibit more rapid and long term drug release. Moreover, a low concentration of the KCNPs shows a slight cytotoxicity and simultaneously maintains their antibacterial property. In a word, keratin-based drug delivery systems can be regarded as a valuable pH-responsive strategy for the delivery of anticancer agents such as the doxorubicin hydrochloride salt (DOX·HCl).Acknowledgements
This work was supported by the National Natural Science Foundation of China (21274063) and PAPD of Jiangsu Higher Education Institutions.References
- J. G. Rouse and M. E. van Dyke, Materials, 2010, 3, 999–1014 CrossRef PubMed.
- J. Yuan, J. Geng, Z. Xing, K. J. Shim, I. Han, J. C. Kim, I. K. Kang and J. Shen, J. Tissue Eng. Regener. Med., 2015, 9, 1027–1035 CrossRef CAS PubMed.
- A. Vasconcelos, A. P. Pêgo, L. Henriques, M. Lamghari and A. Cavaco-Paulo, Biomacromolecules, 2010, 11, 2213–2220 CrossRef CAS PubMed.
- J. J. Martin, J. M. Cardamone, P. L. Irwin and E. M. Brown, Colloids Surf., B, 2011, 88, 354–361 CrossRef CAS PubMed.
- K. Yamauchi, M. Maniwa and T. Mori, J. Biomater. Sci., Polym. Ed., 1998, 3, 259–270 CrossRef PubMed.
- K. Yamauchi and A. Khoda, Colloids Surf., B, 1997, 9, 117–119 CrossRef CAS.
- R. Silva, B. Fabry and A. R. Boccaccini, Biomaterials, 2014, 35, 6727–6738 CrossRef CAS PubMed.
- M. Borrelli, N. Joepen, S. Reichl, D. Finis, M. Schoppe, G. Geerling and S. Schrader, Biomaterials, 2015, 42, 112–120 CrossRef CAS PubMed.
- S. Reichl, M. Borrelli and G. Geerling, Biomaterials, 2011, 32, 3375–3386 CrossRef CAS PubMed.
- L. A. Koman, A. Atala, M. V. Dyke, P. Sierpinski, J. Garrett, J. Ma, P. Apel, D. Klorig and T. Smith, Biomaterials, 2008, 29, 118–128 CrossRef PubMed.
- P. J. Apel, J. P. Garrett, P. Sierpinski, J. Ma, A. Atala, T. L. Smith, L. A. Koman and M. E. van Dyke, J. Hand Surg., 2008, 33, 1541–1547 CrossRef PubMed.
- T. Aboushwareb, D. Eberli, C. Ward, C. Broda, J. Holcomb, A. Atala and M. van-Dyke, J. Biomed. Mater. Res., Part B, 2009, 90, 45–54 Search PubMed.
- C. J. Kowalczewski, S. Tombyln, D. C. Wasnick, M. R. Hughes, M. D. Ellenburg, M. F. Callahan, T. L. Smith, M. E. van Dyke, L. R. Burnett and J. M. Saul, Biomaterials, 2014, 35, 3220–3228 CrossRef CAS PubMed.
- J. M. Saul, M. D. Ellenburg, R. C. de Guzman and M. V. Dyke, J. Biomed. Mater. Res., Part A, 2011, 98, 544–553 CrossRef PubMed.
- J. Yuan, Z. C. Xing, S. W. Park, J. Geng, J. Shen, W. Meng, K. J. Shim, I. S. Han, J. C. Kim and I. K. Kang, Macromol. Res., 2009, 17, 850–855 CrossRef CAS.
- J. Yuan, J. Shen and I. K. Kang, Polym. Int., 2008, 57, 1188–1193 CrossRef CAS PubMed.
- A. Edwards, D. Jarvis, T. Hopkins, S. Pixley and N. Bhattarai, J. Biomed. Mater. Res., Part B, 2015, 103, 21–30 CrossRef PubMed.
- J. H. Guo, S. J. Pan, X. C. Yin, Y. F. He, T. Li, R. M. Wang, J. Appl. Polym. Sci. DOI:10.1002/app.41572.
- X. C. Yin, F. Y. Li, Y. F. He, Y. Wang and R. M. Wang, Biomater. Sci., 2013, 1, 528–536 RSC.
- Q. M. Li, L. J. Zhu, R. G. Liu, D. Huang, X. Jin, N. Che, Z. Li, X. Z. Qu, H. L. Kang and Y. Huang, J. Mater. Chem., 2012, 22, 19964–19973 RSC.
- Q. M. Li, S. N. Yang, L. J. Zhu, H. L. Kang, X. Z. Qu, R. G. Liu and Y. Huang, Polym. Chem., 2015, 6, 2869–2878 RSC.
- M. Rahimnejad, N. Mokhtarian and M. Ghasemi, Afr. J. Biotechnol., 2009, 8, 4738–4743 CAS.
- S. A. Khan and M. Schneider, Macromol. Biosci., 2013, 13, 455–463 CrossRef CAS PubMed.
- S. Han, M. Li, X. Liu, H. Gao and Y. Wu, Colloids Surf., B, 2013, 102, 833–841 CrossRef CAS PubMed.
- S. J. Malek, R. Khoshchehreh, N. Goodarzi, M. R. Khoshayand, M. Amini, F. Atyabi, M. Esfandyari-manesh, S. Tehrani, R. M. Jafari, M. S. Maghazei, F. Alvandifara, M. Ebrahimi and R. Dinarvand, Colloids Surf., B, 2014, 122, 350–358 CrossRef PubMed.
- A. L. de Pinho Neves, C. C. Milioli, L. Müller, H. G. Riella, N. C. Kuhnen and H. K. Stulzer, Colloids Surf., A, 2014, 445, 34–39 CrossRef PubMed.
- J. Zhang, C. Tang and C. H. Yin, Biomaterials, 2013, 34, 3667–3677 CrossRef CAS PubMed.
- E. N. Koukaras, S. A. Papadimitriou, D. N. Bikiaris and G. E. Froudakis, Mol. Pharmaceutics, 2012, 9, 2856–2862 CrossRef CAS PubMed.
- Y. Hou, J. Hu, H. Park and M. Lee, J. Biomed. Mater. Res., Part A, 2012, 100, 939–947 CrossRef PubMed.
- W. Fan, W. Yan, Z. Xu and H. Ni, Colloids Surf., B, 2012, 90, 21–27 CrossRef CAS PubMed.
- F. M. Goycoolea, G. Lollo, C. Remuñán-López, F. Quaglia and M. J. Alonso, Biomacromolecules, 2009, 10, 1736–1743 CrossRef CAS PubMed.
- Q. Gan, T. Wang, C. Cochrane and P. McCarron, Colloids Surf., B, 2005, 44, 65–73 CrossRef CAS PubMed.
- C. G. Jones, Periodontol 2000, 1997, 15, 55–62 CrossRef CAS PubMed.
- K. Yamauchi, A. Yamauchi, T. Kusunoki, A. Kohda and Y. Konishi, J. Biomed. Mater. Res., 1996, 31, 439–444 CrossRef CAS.
- P. M. M. Schrooyen, P. J. Dijkstra, R. C. Oberthür, A. Bantjes and J. Feijen, J. Agric. Food Chem., 2000, 48, 4326–4334 CrossRef CAS PubMed.
- J. Z. Shao, J. H. Zheng, J. Q. Liu and C. M. Carr, J. Appl. Polym. Sci., 2005, 96, 1999–2004 CrossRef CAS PubMed.
- S. W. Ha, A. E. Tonelli and S. M. Hudson, Biomacromolecules, 2005, 6, 1722–1731 CrossRef CAS PubMed.
- K. Abdi, Sh. Nafisi, F. Manouchehri, M. Bonsaii and A. Khalaj, J. Photochem. Photobiol., B, 2012, 107, 20–26 CrossRef CAS PubMed.
- S. Bakkialakshmi and D. Chandrakala, Spectrochim. Acta, Part A, 2012, 88, 2–9 CrossRef CAS PubMed.
- C. J. T. Hoes, J. Grootoonk, R. Duncan, I. C. Hume, M. Bhakoo, J. M. W. Bouma and J. Feijen, J. Controlled Release, 1993, 23, 37–53 CrossRef CAS.
- L. Dong, L. Yan, W. G. Hou and S. J. Liu, J. Solid State Chem., 2010, 183, 1811–1816 CrossRef CAS PubMed.
- S. N. Ostad and P. R. Gard, J. Pharm. Pharmacol., 2000, 52, 779–784 CrossRef CAS.
- F. C. R. Lessa, A. M. F. Aranha, I. Nogueira, E. M. Giro, J. Hebling and C. A. Costa, J. Appl. Oral Sci., 2010, 18, 50–58 CrossRef CAS PubMed.
- G. J. Christensen, J. Am. Dent. Assoc., JADA, 2002, 133, 229–231 CrossRef.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c5ra16253j |
| This journal is © The Royal Society of Chemistry 2015 |