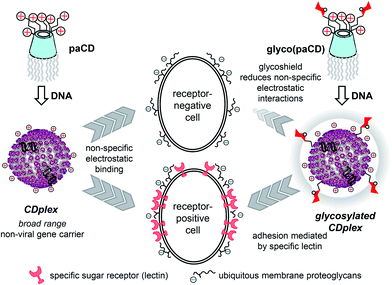
Harmonized tuning of nucleic acid and lectin binding properties with multivalent cyclodextrins for macrophage-selective gene delivery†
Alejandro Méndez-Ardoy‡
a,
Alejandro Díaz-Moscoso§
 b,
Carmen Ortiz Mellet
b,
Carmen Ortiz Mellet a,
Christophe Di Giorgio*c,
Pierre Vierlingc,
Juan M. Benito*b and
José M. García Fernándezb
a,
Christophe Di Giorgio*c,
Pierre Vierlingc,
Juan M. Benito*b and
José M. García Fernándezb
aDepartamento de Química Orgánica, Facultad de Química, Univ. Sevilla, C/Prof. García González 1, E-41012 Sevilla, Spain. E-mail: mellet@us.es
bInstituto de Investigaciones Químicas, CSIC – Univ. Sevilla, Américo Vespucio 49, E-41092 Sevilla, Spain. E-mail: jogarcia@iiq.csic.es
cInstitut de Chimie de Nice, ICN – Université de Nice Sophia Antipolis – CNRS UMR 7272, 28, Avenue de Valrose, F-06100 Nice, France. E-mail: Christophe.DI-GIORGIO@unice.fr
First published on 2nd September 2015
Abstract
Polycationic amphiphilic cyclodextrins (paCDs) have been shown to behave as efficient non-viral gene carriers paralleling the efficacy of commercial vectors towards a variety of cell lines. Their molecular framework and modular design allow the installation of saccharidic antennae to promote specific carbohydrate–protein interactions, thus potentially endowing them with selective targeting abilities. Yet, the presence of these additional functionalities onto the polycationic cluster may hamper paCD self-assembly and nucleic acid condensation. In this report we describe the influence of paCD mannosylation extent on paCD-pDNA nanocomplex stability as well as the consequences of varying glycotope density on mannose-specific lectin recognition and gene delivery capabilities. The work aims at exploring the potential of this approach to optimize both properties in order to modulate cell transfection selectivity.
1. Introduction
Gene therapy (i.e. administering nucleic acid-based therapeutics to correct a particular cellular dysfunction) represents a promising alternative to conventional drugs due to the specific and predictable mode of action of polynucleotides.1 Yet, it faces the drawback of the poor drug-likeness of nucleic acids, requiring purpose-conceived carriers, so-called vectors, to target their goal. Though their efficiency is still far from that of virus-based carriers, the advent of nanotechnology and the persisting safety concerns regarding the use of viral materials have fostered the design of a plethora of artificial gene vectors to cope with this task.2 Molecular facial amphiphiles based on macrocyclic scaffolds,3 among which polycationic amphiphilic cyclodextrins (paCDs, Fig. 1 left) are relevant representatives,4,5 have turned particularly promising in this regard. paCDs undergo nucleic acid-templated self-assembling affording supramolecular nanocomplexes (CDplexes) that promote transfection with efficiencies that rival that of formulations prepared with commercial polycationic polymers (polyplexes) or lipid vectors (lipoplexes), but with no or much milder toxicity. Selectivity is however similarly low, since cell uptake of CDplexes, as for polyplexes and lipoplexes, relies essentially in non-specific electrostatic interactions.6 Alternatively, exploiting specific ligand-receptor recognition events (e.g. between sugar epitopes and their complementary lectins) holds great promise for selectively targeting therapeutic genes.7 The concept was put forward nearly 20 years ago,8 but implementation with macromolecular systems is not that evident. Glycotargeting is presumed to arise from (i) the increase of the hydration shell of the carrier due to the hydrophilicity of the sugar motifs, which shields from non-specific (electrostatic) interactions, and (ii) specific receptor-mediated cell internalization (Fig. 1, right). Recently, we have demonstrated that installation of a homogeneous glycomultivalent display on the cationic domain of paCDs selectively promoted gene delivery to cells expressing complementary lectin receptors.9 Yet, the incorporation of glycoligands on the vector has a strong impact on the hydrophobic/hydrophilic balance and the hydrodynamic properties of the system, which may negatively affect self-assembling, nucleic acid compaction and, ultimately, transfections capabilities.10,11 Harmonizing glycotope and cationic group densities to warrant biologically useful affinities towards both the target lectin and the nucleic acid cargo is thus critical for those channels.In this paper, we report the synthesis of a series of statistically mannosylated paCDs featuring variable relative proportions of sugar and protonable amino centres and a preliminary assessment of how mannosylation extent modulates the nucleic acid condensing capacity, the dynamics and stability of the resulting glycoCDplexes, the binding affinity to mannose-specific receptors and the transfection efficiency towards macrophage mannose receptor (MMR) positive (RAW 264.7) and negative (BNL-CL2 and COS-7) cell lines.
2. Results and discussion
2.1. Selection criteria and synthesis
Compounds 1a12a and 2a12b (Scheme 1), featuring very similar structures but subtle vector performance disparities, were selected as paCD scaffolds to investigate the effect of increasing mannosylation degrees on DNA complexing abilities and MMR recognition properties. The transfection efficiency of pDNA complexes formulated with triazol-armed paCD 1a is optimal at a nitrogen/phosphorus (N/P) ratio of 10.13 Thiourea-tethered paCD 2a, on the other hand, achieves similar expression levels at a lower N/P (5), which is attributed to the privileged phosphate binding ability of the 2-aminoethylthiourea segments on the cationic cluster.12b | ||
| Scheme 1 Synthesis of statistically mannosylated paCDs from 1a and 2a and structure of the homogenously mannosylated derivative 4 used as positive control in macrophage adhesion experiments. | ||
Cyclodextrin scaffolds 1a and 2a were conjugated with 2-(α-D-mannopyranosyloxy)ethyl isothiocyanate (3)14 using thiourea-forming ligation chemistry (Scheme 1). Each individual coupling reaction results then in the replacement of a cationizable centre in the vector by a neutral biorecognizable glycotope. Reaction with increasing amounts of isothiocyanate 3 (5 to 50% relative to the paCD primary amino groups), followed by purification by size exclusion chromatography, furnished two series of paCDs with different degrees of mannosylation (1b–e and 2b–e, respectively). ESI-MS and 1H NMR spectroscopy confirmed quantitative mannoside incorporation in all cases (see ESI†).
The above multiconjugation strategy provides a very convenient method to modulate the coating sugar content of the paCD vectors while significantly reducing the synthetic effort as compared with regioselective synthetic schemes previously implemented, e.g. for the synthesis of the heptamannosylated glyco(paCD) 4 (Scheme 1).9 Although it obviously affords polydisperse mixtures of statistically glycosylates adducts, it is presumed that differences in glycocluster topology at the molecular level would be blurred when considering the glycocoating corona in the nanocomplexes after self-assembling with pDNA.7 Indeed, the scenario is very similar to that encounter for statistic glycosylation of polymeric or dendritic scaffolds, a routinely used strategy to generate multivalency.10,15,16 Glycotope density is then expected to become the main parameter influencing specific lectin recognition and gene delivery properties.
2.2. Self-assembling capabilities
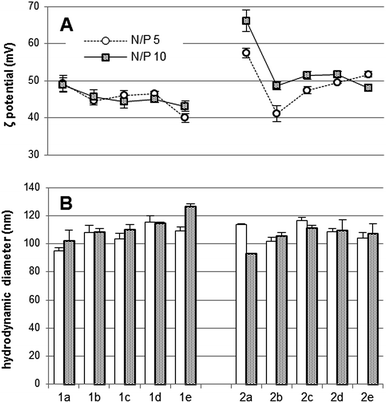
The influence of the mannosylation degree in the pDNA (luciferase-encoding pTG11236 plasmid) complexing capabilities of paCDs 1a–e and 2a–e was first screened by agarose gel electrophoresis migration experiments at different N/P ratios (1, 2, 5, and 10). No remarkable differences were noticed between the behaviour of triazol- (1a–e; Fig. 2, left column) and thiourea-tethered (2a–e; Fig. 2, right column) series. In general, mannosylation progressively decreases the ability to inhibit pDNA electrophoretic mobility of the paCDs, which is better evidenced in the thiourea-tethered series (Fig. 2, right column). Nevertheless, regardless of their mannosylation degree, all glyco(paCDs) inhibited electrophoretic mobility of pDNA at N/P ratios above 2. Comparatively, the pDNA protection against the intercalating agent ethidium bromide is more influenced by mannosylation. While pDNA is already inaccessible to ethidium bromide at N/P ≥ 2 in formulations with 1a and 2a (Fig. 2, first row), N/P 5 ratios are required for most mannosylated adducts to attain the same protection, mounting up to N/P 10 for the more heavily mannosylated triazol-linked glyco(paCD) 1e (50% mannosylation). Although electrostatic interactions with the plasmid are preserved, compaction is probably hampered as a consequence of the higher swelling capabilities imparted by the hydrophilic sugar residues.The hydrodynamic size and ζ-potential of glycoCDplexes formulated at N/P values 5 and 10 were next measured by dynamic light scattering (DLS) and mixed-mode measurement-phase analysis light scattering (M3-PALS), respectively. Dispersion of paCDs in a solution of pDNA in water furnished single populations of rather homogenous nanoparticles (polydispersity index ≈ 0.2) in all cases. (Glyco)CDplexes obtained from triazol-containing paCDs 1a–e evidenced a gradual, though subtle, increase in size and a parallel decrease in ζ-potential upon increasing the mannosylation degree, which was especially noticeable for formulations at N/P 10 (Fig. 3, left). It is worth mentioning that although an increase in sugar valency is accompanied by a decrease in the number of protonable amino groups in the glyco(paCD) conjugates, this has been compensated by normalizing at the same N/P ratio. The observed trend is probably due to the increase in the thickness of the hydration shell with heavier mannosylation, which is also expected to shield the nanoparticle surface charge to some extent. The increase in size upon glycosylation was less evident in the thiourea-tethered conjugates 2a–e (Fig. 3, right). In this case, a significant decrease in ζ-potential was already observed for the vector with the lower glycosylation rate (2b, 5% mannosylation), further increases in valency having much more modest consequences. It is hypothesized that the higher flexibility of the linker in this series allows a better accommodation of the mannosyl residues in the glycoCDplexes, minimizing the impact of glycosylation density variations.
The effect of mannosylation extent on the size and stability of the nanocomplexes formulated with the paCD derivatives 1a–e and 2a–e and pDNA in saline medium (up to 150 mM NaCl) was next investigated to further assess their behavior under physiological conditions. As a general trend, mannosylation attenuated the effect of saline stress on the colloidal stability of glycoCDplexes, as seen from the less pronounced differences in size (ESI, Fig. S11†). This result is in agreement with previous reports on glycocluster-based vectors10,17 and reinforces the notion that sugar coating prevents non-specific aggregation phenomena, probably by endowing the nanoparticles with a thicker hydration shell. The accessibility of ethidium bromide to the pDNA cargo in the most heavily mannosylated glycoCDplexes (e.g. 1e) increases under saline stress (ESI, Fig. S12†), also pointing to a non-optimal hydrophilic/hydrophobic balance at such high mannose coating densities.
2.3. Alveolar macrophage adhesion
The average mannose valencies of the paCDs prepared and evaluated in this work, taken as individual entities, are 0, 0.7, 2.1, 4.2 and 7 for 1a and 2a to 1e and 2e, respectively. According to results reported for non-amphiphilic CD-scaffolded glycoclusters,18 a significant multivalent effect would be expected only for the later derivatives. Yet, after supramolecular self-assembling in the presence of pDNA a hyper-valent display of mannosyl epitopes will be generated at the surface of the glycoCDplexes formulated with 1b–e and 2b–e. We were particularly interested at investigating the minimum mannosylation rate leading to functional DNA nanocomplexes capable of eliciting a biologically significant binding to cells expressing the macrophage mannose receptor. For such purpose, glycoCDplexes were formulated using pDNA labelled with the fluorescent probe YOYO-1 at N/P ratios 5 and 10. The ability of these particles to adhere the surface of alveolar peritoneal macrophages at 4 °C (to prevent endocytosis) was assessed by fluorescence intensity measurements (Fig. 4).19 At the lower N/P ratio, a significant increase in fluorescence was noticed even at relatively low mannosylation ratios as compared to negative controls (naked pDNA, non-glycosylated CDplexes formulated with paCDs 1a and 2a, and polyplexes formulated with the commercial vector JetPEI).20 Interestingly, fluorescence intensity plateaued at ca. 15–30% mannosylation (1c, 2c and 1d, 2d), with residual increase at higher glycan ratios. Indeed, similar macrophage adhesion levels were obtained for the positive control, the homogeneously 50% mannosylated paCD 4 (Scheme 1). At N/P 10, though the tendency is similar, the contrast is lower, due to the higher background of non-specific electrostatic interactions. As it could be expected, the higher the cationic content, the lower the contribution of specific interactions to the overall adhesion process, a phenomenon already described for a variety of cationic gene carriers.21To confirm the specificity of the interactions at play, the above experiments were run in the presence of native and mannosylated-BSA (Man-BSA) as competing agents, using the optimal N/P ratio 5. A mannose concentration-dependent inhibition of glycoCDplex adhesion to the macrophages was observed for the later, with fluorescence intensity values reduced to basal levels in the presence of 1 mg mL−1 of Man-BSA (Fig. 5). On the other hand, native BSA minimally affected adhesion. Neither BSA nor Man-BSA significantly affected the adhesion of JetPEI polyplexes and non-mannosylated CDplexes to macrophages, altogether strongly supporting the involvement of specific mannose-MMR recognition in the adhesion of the glycoCDplexes formulated with 1b–e and 2b–e to the macrophages.
2.4. Cell transfection
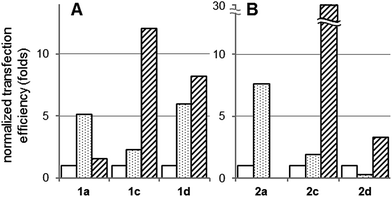
CDplexes formulated at N/P 5 with luciferase-encoding pDNA (pTG11236) and the multivalent paCD vectors having 15 and 30% of the primary amino groups replaced by mannosyl antennae (1c, 2c and 1d, 2d, respectively) were selected for cell transfection assays. According to the previously discussed results, these formulations exhibit the best balance between pDNA protection, colloidal stability and MMR-binding efficiency, which was expected to maximize receptor-mediated versus non-specific (electrostatic-mediated) cell uptake. Transfection was monitored in MMR-positive mouse leukaemic monocyte macrophages (RAW 264.7) and in MMR-devoid embryonic murine hepatocytes (BNL-CL2) and African green monkey kidney fibroblasts (COS-7). In all three cell lines and for all formulation, cell viabilities were determined to be above 80%. The corresponding luciferase expression data are plotted in Fig. 6. Naked pDNA and JetPEI polyplexes formulated at N/P 5 were used as negative and positive controls, respectively (see ESI for experimental details†).The graphs depict two different scenarios depending on the presence of triazol (1a, 1c and 1d) or thiourea linkers (2a, 2c and 2d). In the triazol-tethered series, the transfection efficiency of the non-mannosylated CDplexes formulated with paCD 1a at N/P 5 is very poor in the three cell lines, only marginally improving that of naked pDNA. Indeed, previous results have shown that triazol-tethered paCD efficiency peaks at higher N/P ratios, ca. 10.13 The mannosylated vectors 1c and 1d behaved similarly poorly in the MMR-devoid BNL-CL2 and COS-7 cell lines (Fig. 6A). Rewardingly a significant enhancement of the transfection efficiency relative to the control was observed in MMR-expressing RAW264.7 cells (29- and 12-folds for glycoCDplexes formulated with 1c and 1d, respectively). A direct comparison of the data for the three cell lines reveals a 5 to 12-fold higher protein expression in MMR-expressing as compared to MMR-devoid cells (Fig. 7A).
CDplexes formulated with the thiourea-tethered paCD 2a at N/P 5 already elicited relevant levels of luciferase activity in all three cell lines. Upon conjugation with mannose, transfection levels dropped very significantly in MMR-devoid BNL-CL2 and COS-7 cells (Fig. 6B). In contrast, transfection of RAW264.7 macrophages with the mannosylated conjugates 2c and 2d remained unaltered, probably because the detrimental effect due to attenuation of electrostatic interactions is compensated by activation of MMR-mediated cell uptake mechanisms. Even though a net enhancement in macrophage transfection efficiency is not achieved, off-target effects are very significantly reduced, imparting cell selectivity. Thus, while the parent paCD vector 2a promoted transfection of the hepatocyte and fibroblast cells with 13- and 100-fold higher efficiencies as compared with macrophages, the corresponding conjugate 2c, with a 15% mannose incorporation, transfected preferentially macrophages, with 31-fold and 16-fold higher efficiency as compared with hepatocytes and fibroblasts, respectively (Fig. 7B). This means a reversion of the cell selectivity in favour of the MMR-expressing cells by a factor of 400 to 1600.
3. Conclusions
The results presented in this work demonstrate that the installation of defined proportions of saccharidic epitopes onto molecular polycationic amphiphilic CD scaffolds can be used to modulate their DNA-templated self-assembling capabilities and the functional properties of the resulting nanocomplexes. Self-assembling of glyco(paCDs) leads to amplification of interactions with complementary lectin receptors by generating a multivalent glyocodisplay at the nanoparticles surface. Glycocoating additionally shields the positive charge of glycoCDplexes, decreasing the non-specific electrostatic interactions with proteoglycans at the cell surface. Altogether glyco(paCDs) can be exploited to enhance cell transfection selectivities by judiciously tuning the nucleic acid and lectin binding properties, as exemplified here for macrophages, after synchronized adjustment of the positive charge density and the glycosylation extent.4. Experimental
4.1. General methods
Reagents and solvents were purchased from commercial sources and used without further purification. NMR spectra were recorded at 500 MHz. 2D COSY, 1D TOCSY experiments were used to assist on NMR assignments. Gel permeation chromatography was carried out on Sephadex G-25 (GE Healthcare Life Sciences). Electrospray mass spectra (ESIMS) were obtained with a Bruker Esquire6000 instrument. Compounds 1a,12a 2a,12b 3,14 and 4![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 9 were synthesized as previously reported.
9 were synthesized as previously reported.
4.2. Synthesis
4.3. Preparation of complexes composed of paCDs and pDNA pTG11236
The plasmid pTG11236![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 22 (pCMV-SV40-luciferase-SV40 pA) employed for the preparation of the DNA complexes and for transfection assay is a plasmid of 5739 bp (base pairs). The quantities of each formulation were calculated according to the DNA concentration (0.1 mg mL−1, 303 μM phosphate), the N/P ratio, the molar weight and the number of positive charges in the paCD derivative or cationic polymer (JetPEI®).20 Experiments were performed for N/P 1, 2, 5 and 10. Concerning the preparation of the DNA complexes from paCD derivatives and JetPEI, pDNA was diluted in HEPES (20 mM, pH 7.4) to a final concentration of 303 μM (1 mg mL−1), then the desired amount of CD derivative was added from a 20 mg mL−1 stock solution (DMSO-water, 1
22 (pCMV-SV40-luciferase-SV40 pA) employed for the preparation of the DNA complexes and for transfection assay is a plasmid of 5739 bp (base pairs). The quantities of each formulation were calculated according to the DNA concentration (0.1 mg mL−1, 303 μM phosphate), the N/P ratio, the molar weight and the number of positive charges in the paCD derivative or cationic polymer (JetPEI®).20 Experiments were performed for N/P 1, 2, 5 and 10. Concerning the preparation of the DNA complexes from paCD derivatives and JetPEI, pDNA was diluted in HEPES (20 mM, pH 7.4) to a final concentration of 303 μM (1 mg mL−1), then the desired amount of CD derivative was added from a 20 mg mL−1 stock solution (DMSO-water, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2, v/v) and JetPEI was added from a 0.1 M stock solution (water). Final concentration of DMSO never exceeded 2% in these preparations. The preparation was vortexed for 2 h and used for characterization or transfection experiments.
2, v/v) and JetPEI was added from a 0.1 M stock solution (water). Final concentration of DMSO never exceeded 2% in these preparations. The preparation was vortexed for 2 h and used for characterization or transfection experiments.
4.4. Agarose gel electrophoresis
Each CD derivative/DNA sample (20 μL, 0.4 μg of plasmid) was electrophoresed for about 30 min under 150 V through a 0.8% agarose gel in TAE 1X (Tris-acetate-EDTA) buffer and stained by spreading an ethidium bromide (Sigma) solution in TAE buffer (20 μL ethidium bromide of a 10 mg mL−1 solution in 200 mL TAE). The DNA was then visualized after photographing on an UV transilluminator.4.5. Particle size and ζ potential measurements
The hydrodynamic diameters of the vesicles formed from the paCDs were measured by dynamic light scattering (DLS) at 633 nm on a Nanosizer S DLS instrument (Malvern Instruments) at 25 °C and at a detection angle of 173°. All the measurements were performed in triplicate. Data were analysed using the multimodal number distribution software included in the instrument. Results are given as volume distribution of the major population by the mean diameter with its standard deviation. Statistical analysis of the particle size was performed using a student's test (with two-sample unequal variance and two distribution tails). The ζ potential measurements were also performed in the same instrument using 'Mixed Mode Measurement' phase analysis light scattering (M3-PALS). M3 consists of both slow field reversal and fast field reversal measurements, hence the name 'Mixed Mode Measurement' that improves accuracy and resolution. PALS is a variation of laser Doppler velocimetry that uses the phase shift (i.e. frequency x time) instead of the frequency shift to determine electrophoretic mobilities and hence convert them into zeta potentials. The following specifications were applied: sampling time, automatic; number of measurements, 3 per sample; medium viscosity, 1.054 cP; medium dielectric constant, 80; temperature, 25 °C; beam mode F(Ka) 1.5 (Smoluchowsky). Experiments were run by in triplicate.4.6. Salt-induced CDplex dissociation assays
CDplexes were prepared as described above in order to obtain a final DNA concentration of 60 μM after adding a volume (representing 10% of total volume) of a NaCl solution in HEPES (in order to obtain a final NaCl concentration of 0, 50, 150 and 250 mM), and a volume (representing 2% of total volume) of a 100 μM ethidium bromide solution in DMSO. After adding, each preparation was vortexed for 15 minutes before fluorescence measurements. Each CDplex formulation was arrayed in triplicate into a white flat-bottom 96-well plate. The ethidium bromide fluorescence of each sample was measured with a luminometer (GENIOS PRO, Tecan France S.A.; excitation 485 nm, emission 590 nm). The relative ethidium bromide exclusion (% DNA excluded to ethidium bromide) was determined by the following relationship:| Relative(EtBr exclusion) = 1 − [(Fsample − Fblank)/(FDNAonly − Fblank)] |
4.7. Alveolar macrophage adhesion assays
For evaluation of the interaction of YOYO-1-labeled CDplexes with alveolar macrophages, the procedure reported by Muller and Schuber19a for mannosylated liposomes was adapted. Briefly, resident peritoneal macrophages were obtained from female Balb/c mice (6 to 8 weeks old) and maintained in Dulbecco's modified Eagle's medium (DMEM; Gibco-BRL) supplemented with 10% decomplemented fetal calf serum (FCS; Sigma) containing heparin (5 U mL−1). The cell number was adjusted to 106 cells per mL and the suspension was platted (final volume 1 mL) in multi-well plates. After 2 h in a humidified atmosphere of 5% CO2 in air (final pH 7.4), non-adherent cells were eliminated by rinsing the dishes three times with PBS. The adherent cells, 24 h after their isolation, were fed with fresh serum-less DMEM and incubated with CD-pDNA nanocomplexes, PEI polyplexes and naked DNA (control) at similar DNA concentration used in transfection assays. After the incubation time, the medium was pipetted-off and the cells washed four times with cold PBS (4 °C). The fluorescent tag associated to the cells was measured fluorimetrically (Perkin Elmer luminescence spectrometer LS50B, excitation at 491 nm and monitoring emission at 509 nm) after cell digestion in 1 mL PBS containing 0.1% of emulphogene BC-720, and scraping with a rubber policeman. Experiments were run in duplicate, and results did not differ more than 10%.4.8. In vitro transfection
Twenty-four hours before transfection, BNL-CL2, COS-7, and RAW264.7 cells were grown at a density of 2 × 104 cells per well in a 96-well plates in Dulbelco modified Eagle culture medium (DMEM; Gibco-BRL) containing 10% foetal calf serum (FCS; Sigma) in a wet (37 °C) and 5% CO2/95% air atmosphere. The complexes CD derivative/DNA or PEI/DNA polyplexes were diluted to 100 μL in DMEM in order to have 0.5 μg of DNA in the preparation. The culture medium was removed and replaced by these 100 μL of complexes in DMEM. After 4 h and 24 h, 50 and 100 μL of DMEM supplemented with 30% and 10% FCS, respectively, were added. After 48 h, the transfection was stopped, the culture medium was discarded, and the cells washed twice with 100 μL of PBS and lysed with 50 μL of lysis buffer (Promega, Charbonnières, France). The lysates were frozen at −32 °C, before the analysis of luciferase activity. This measurement was performed in a LB96P luminometer (BERTHOLD, France) in dynamic mode, for 10 s on 10 mL on the lysis mixture and using the “luciferase” determination system (Promega) in 96-well plates. The total protein concentration per well was determined by the BCA test (Pierce, MontluÇon, France). Luciferase activity was calculated as femtograms (fg) of luciferase per mg of protein. The percentage of cell viability of the nanocomplexes was calculated as the ratio of the total protein amount per well of the transfected cells relative to that measured for untreated cells x 100%.Acknowledgements
This work was supported by the Spanish Ministerio de Economía y Competitividad (MinECo; contract number SAF2013-44021-R), the Junta de Andalucía (Proyecto de Investigación de Excelencia FQM-1467), the Fondo Europeo de Desarrollo Regional (FEDER-Unión Europea), the Fondo Social Europeo (FSE) the CSIC, the CNRS, and FUSINT (CNRS project). A.M.-A. and A.D.-M. are grateful to MinECo and CSIC, respectively, for pre-doctoral fellowships.Notes and references
- V. K. Sharma, P. Rungta and A. K. Prasad, RSC Adv., 2014, 4, 16618 RSC.
- (a) M. Elsabahy, A. Nazarali and M. Foldvari, Curr. Drug Delivery, 2011, 8, 235 CrossRef CAS; (b) M. A. Mintzer and E. E. Simanek, Chem. Rev., 2009, 109, 259 CrossRef CAS PubMed; (c) C. H. Jones, C.-H. Chen, A. Ravikrishnan, S. Rane and B. A. Pfeifer, Mol. Pharmaceutics, 2013, 10, 4082 CrossRef CAS PubMed.
- (a) B. J. Ravoo and R. Darcy, Angew. Chem., Int. Ed., 2000, 39, 4324 CrossRef CAS; (b) C. Ortiz Mellet, J. M. Benito and J. M. García Fernández, Chem.–Eur. J., 2010, 16, 6728 CrossRef CAS PubMed; (c) V. Bagnacani, V. Franceschi, M. Bassi, M. Lomazzi, G. Donofrio, F. Sansone, A. Casnati and R. Ungaro, Nat. Commun., 2013, 4, 1721 CrossRef PubMed; (d) V. Bagnacani, V. F. L. Fantuzzi, F. Sansone, G. Donofrio, A. Casnati and R. Ungaro, Bioconjugate Chem., 2012, 23, 993 CrossRef CAS PubMed; (e) L. Gallego-Yerga, M. Lomazzi, V. Franceschi, F. Sansone, C. Ortiz Mellet, G. Donofrio, A. Casnati and J. M. García Fernández, Org. Biomol. Chem., 2015, 13, 1708 RSC.
- (a) B. M. D. C. Godinho, D. J. McCarthy, C. Torres-Fuentes, C. J. Beltrán, J. McCarthy, A. Quinlan, J. R. Ogier, R. Darcy, C. M. O'Driscoll and J. F. Cryan, Biomaterials, 2014, 35, 489 CrossRef CAS PubMed; (b) J. McCarthy, M. J. O'Neill, L. Bourre, D. Walsh, A. Quinlan, G. Hurley, J. Ogier, F. Shanahan, S. Melgar, R. Darcy and C. M. O'Driscoll, J. Controlled Release, 2013, 168, 28 CrossRef CAS PubMed; (c) A. M. O'Mahony, B. M. D. C. Godinho, J. Ogier, M. Devocelle, R. Darcy, J. F. Cryan and C. M. O'Driscoll, ACS Chem. Neurosci., 2012, 3, 744 CrossRef PubMed; (d) V. Villari, A. Mazzaglia, R. Darcy, C. M. O'Driscoll and N. Micalli, Biomacromolecules, 2013, 14, 811 CrossRef CAS PubMed.
- (a) N. Guilloteau, C. Bienvenu, C. Charrat, J. L. Jiménez Blanco, A. Díaz-Moscoso, C. Ortiz Mellet, J. M. García Fernández, P. Vierling and C. Di Giorgio, RSC Adv., 2015, 5, 29135 RSC; (b) C. Aranda, K. Urbiola, A. Méndez Ardoy, J. M. García Fernández, C. Ortiz Mellet and C. Tros de Ilarduya, Eur. J. Pharm. Biopharm., 2013, 85, 390 CrossRef CAS PubMed; (c) A. Martínez, C. Bienvenu, J. L. Jiménez Blanco, P. Vierling, C. Ortiz Mellet, J. M. García Fernández and C. Di Giorgio, J. Org. Chem., 2013, 78, 8143 CrossRef PubMed; (d) C. Bienvenu, A. Martínez, J. L. Jiménez Blanco, C. Di Giorgio, P. Vierling, C. Ortiz Mellet, J. Defaye and J. M. García Fernández, Org. Biomol. Chem., 2012, 10, 5570 RSC.
- (a) L. Gallego-Yerga, M. J. González-Alvarez, N. Mayordomo, F. Santoyo-González, J. M. Benito, C. Ortiz Mellet, F. Mendicuti and J. M. García Fernández, Chem.–Eur. J., 2014, 20, 6622 CrossRef CAS PubMed; (b) L. Gallego-Yerga, L. Blanco-Fernández, K. Urbiola, T. Carmona, G. Marcelo, J. M. Benito, F. Mendicuti, C. Tros de Ilarduya, C. Ortiz Mellet and J. M. García Fernández, Chem.–Eur. J., 2015, 21, 12093–12104 CrossRef CAS PubMed.
- S. Kawakami and M. Hashida, J. Controlled Release, 2014, 190, 542 CrossRef CAS PubMed.
- (a) P. Erbacher, A. C. Roche, M. Monsigny and P. Midoux, Bioconjugate Chem., 1995, 6, 401 CrossRef CAS; (b) J. S. Remy, A. Kichler, V. Mordvinov, F. Schuber and J. P. Behr, Proc. Natl. Acad. Sci. U. S. A., 1995, 92, 1744 CrossRef CAS; (c) T. Ferkol, J. C. Perales, F. Mularo and R. W. Hanson, Proc. Natl. Acad. Sci. U. S. A., 1996, 93, 101 CrossRef CAS.
- A. Díaz-Moscoso, N. Guilloteau, C. Bienvenu, A. Méndez-Ardoy, J. L. Jiménez Blanco, J. M. Benito, L. le Gourrierec, C. Di Giorgio, P. Vierling, J. Defaye, C. Ortiz Mellet and J. M. García Fernández, Biomaterials, 2011, 32, 7263 CrossRef PubMed.
- S. M. D'Addio, S. Baldassano, L. Shi, L. Cheung, D. H. Adamson, M. Bruzek, J. E. Anthony, D. L. Laskin, P. J. Sinko and R. K. Prud'homme, J. Controlled Release, 2013, 138, 41 CrossRef PubMed.
- E. M. Aguilar Moncayo, N. Guilloteau, C. Bienvenu, J. L. Jiménez Blanco, C. Di Giorgio, P. Vierling, J. M. Benito, C. Ortiz Mellet and J. M. García Fernández, New J. Chem., 2014, 38, 5215 RSC.
- (a) A. Méndez-Ardoy, M. Gómez-García, C. Ortiz-Mellet, N. Sevillano, M. D. Girón, R. Salto, F. Santoyo-González and J. M. García Fernández, Org. Biomol. Chem., 2009, 7, 2681 RSC; (b) A. Díaz-Moscoso, L. le Gourrierec, M. Gómez-García, J. M. Benito, P. Balbuena, F. Ortega-Caballero, N. Guilloteau, C. Di Giorgio, P. Vierling, J. Defaye, C. Ortiz Mellet and J. M. García Fernández, Chem.–Eur. J., 2009, 15, 12871 CrossRef PubMed.
- A. Mendez-Ardoy, N. Guilloteau, C. Di Giorgio, P. Vierling, F. Santoyo-González, C. Ortiz Mellet and J. M. García Fernández, J. Org. Chem., 2011, 76, 5882 CrossRef CAS PubMed.
- C. Grabosch, K. Kolbe and T. K. Lindhorst, ChemBioChem, 2012, 13, 1874 CrossRef CAS PubMed.
- (a) N. Symens, A. Méndez-Ardoy, A. Díaz-Moscoso, E. Sánchez-Fernández, K. Remaut, J. Demeester, J. M. García Fernández, S. C. de Smedt and J. Rejman, Bioconjugate Chem., 2012, 23, 1276 CrossRef CAS PubMed; (b) W. Yeeprae, S. Kawakami, F. Yamashita and M. Hashida, J. Controlled Release, 2006, 114, 193 CrossRef CAS PubMed; (c) L. Raviv, M. Jaron-Mendelson and A. David, Mol. Pharmaceutics, 2015, 12, 453 CrossRef CAS PubMed; (d) S. Chu, C. Tang and C. Yin, Biomaterials, 2015, 52, 229 CrossRef CAS PubMed.
- (a) H. Arima, K. Motoyama and T. Higashi, Adv. Drug Delivery Rev., 2013, 65, 1204 CrossRef CAS PubMed; (b) L. L. Kiessling and J. C. Grim, Chem. Soc. Rev., 2013, 42, 4476 RSC.
- O. Hayashida, K. Mizuki, K. Akagi, A. Matsuo, T. Kanamori, T. Nakai, S. Sando and Y. Aoyama, J. Am. Chem. Soc., 2003, 125, 594 CrossRef CAS PubMed.
- A. Martínez, C. Ortiz Mellet and J. M. García Fernández, Chem. Soc. Rev., 2013, 42, 4746 RSC.
- (a) C. D. Muller and F. Schuber, Biochim. Biophys. Acta, 1989, 986, 97 CrossRef CAS; (b) J. Rodríguez-Lavado, M. de la Mata, J. L. Jiménez-Blanco, M. I. García-Moreno, J. M. Benito, A. Díaz-Quintana, J. A. Sánchez-Alcázar, K. Higaki, E. Nanba, K. Ohno, Y. Suzuki, C. Ortiz Mellet and J. M. García Fernández, Org. Biomol. Chem., 2014, 12, 2289 RSC.
- (a) M.-H. Louis, S. Dutoit, Y. Denoux, P. Erbacher, E. Deslandes, J.-P. Behr, P. Gauduchon and L. Poulain, Cancer Gene Ther., 2006, 13, 367 CrossRef CAS PubMed; (b) O. Boussif, F. Lezoualc'h, M. A. Zanta, M. D. Mergny, D. Scherman, B. Demeneix and J.-P. Behr, Proc. Natl. Acad. Sci. U. S. A., 1995, 92, 7297 CrossRef CAS.
- (a) E. Letrou-Bonneval, R. Chèvre, O. Lambert, P. Costet, C. André, C. Tellier and B. Pitard, J. Gene Med., 2008, 10, 1198 CrossRef CAS PubMed; (b) J. S. Remy, A. Kichler, V. Mordvinov, F. Schuber and J.-P. Behr, Proc. Natl. Acad. Sci. U. S. A., 1995, 92, 1744 CrossRef CAS.
- (a) J. Gaucheron, C. Boulanger, C. Santaella, N. Sbirrazzuoli, O. Boussif and P. Vierling, Bioconjugate Chem., 2001, 12, 949 CrossRef CAS PubMed; (b) N. V. Craynest, C. Santaella, O. Boussif and P. Vierling, Bioconjugate Chem., 2002, 13, 59 CrossRef PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available: Experimental details, NMR and ESI-MS spectra, and dynamic light scattering (DLS) measurement figures. See DOI: 10.1039/c5ra16087a |
| ‡ Present address: Université de Bordeaux, Institute de Sciences Moléculaires (ISM), Nanostructures Organiques, 33405 Talence, France. |
| § Present address: Institute of Chemical Research of Catalonia (ICIQ), 43007 Tarragona, Spain. |
| This journal is © The Royal Society of Chemistry 2015 |