Toward the design of bio-solar cells: high efficiency cascade energy transfer among four donor–acceptor dyes self-assembled in a highly ordered protein–DNA matrix†
Challa V. Kumar*ab,
Marc J. Novakb,
Kyle R. Bensona,
Clive Baveghemsa,
Vindya K. Thilakarathnea,
Bobbi S. Stromera and
Filicia M. Rossa
aDepartment of Chemistry, University of Connecticut, Storrs, CT 06269-3060, USA. E-mail: challa.kumar@uconn.edu
bDepartment of Molecular and Cell Biology, University of Connecticut, Storrs, CT 06269-3125, USA
First published on 19th August 2015
Abstract
Artificial antenna complexes built via self-assembly are reported here, which indicated excellent energy transfer efficiency, macroscopic organization, unprecedented thermal stability, and ease of formation. Our system consists of four fluorescent donor–acceptor dyes, double-helical DNA and cationized bovine serum albumin, all self-assembled on cover glass slips to form functional materials. These captured radiation in the range of 330–590 nm, and excitation of any of the donor dyes resulted in efficient emission from the terminal acceptor. Excitation spectra provided unequivocal proof of energy transfer via jumper dyes, and transfer was interrupted when one of the jumper dyes was omitted, another direct evidence for cascade energy transfer. The entire assembly indicated unusually high thermal stability and continued to function efficiently even after exposure to 80 °C for >169 days, an important consideration for field applications. These unusually stable, high efficiency, multi-chromophoric, artificial antennas are the first of their kind to demonstrate self-assembled 4-dye energy cascade, converting blue photons to red photons.
Introduction
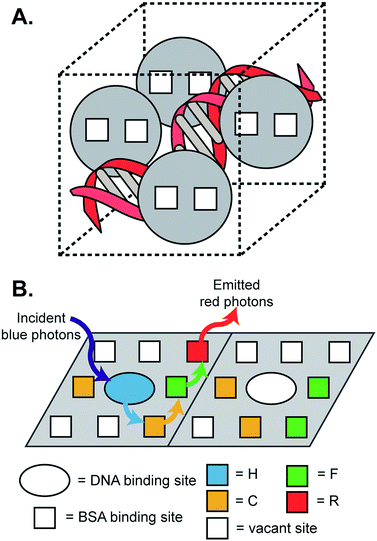
Solar energy is being touted as a long term, sustainable, green energy source.1,2 Natural photosynthetic systems use a set of pigments which are non-covalently bound to well-organized, self-assembled protein complexes and harvest light over a broad wavelength window.3 Building efficient artificial analogues of such systems continues to be a fundamental challenge, particularly when examined through the lens of solar cell applications.4 For example, the majority are solution-based systems requiring complex synthesis and extensive purification. Additionally, efficiency of energy transfer is often low. Finally, their stability over extended periods of time and their functionality in solid state has not been demonstrated, features which are important for practical solar applications. Most solar cells have poor efficiency in the UV region, and these high-energy photons are also responsible for the eventual degradation of the cell's components and failure of the cell. Conversion of UV photons to the corresponding longer wavelength photons, or downshifting, is one strategy that is expected to improve overall cell performance and longevity. Therefore, a light-harvesting device that converts UV or blue photons to red photons and operates in the solid phase would be attractive.5 Another important consideration is the environmental burden of the current solar cell components, which should be reduced for global scale production of solar energy.Inspired by nature, we designed artificial antenna complexes consisting of multiple donor–acceptor fluorescent dyes that self-assemble from readily bio-degradable components (Scheme 1). Our hypothesis is that individually-wrapped, densely packed donor–acceptor dyes could favor efficient Förster resonance energy transfer (FRET) but such dense packing must also avoid direct dye–dye contacts to suppress energy-wasting bimolecular quenching processes.6 These opposing requirements can be met by embedding the dyes in the protein–DNA matrix.
 | ||
| Scheme 1 Artificial antenna complexes constructed from donors, acceptors, cationized BSA (cBSA), and DNA. | ||
Another important consideration is finding alternatives to covalent chemistry that is often used for the construction of the antenna complexes. While there are many elegant examples of covalent coupling of dyes to form antenna complexes,7 such approaches can be challenging and expensive. Therefore, we sought out a self-assembly approach which is rugged and simple, but effective in achieving dense packing of multiple chromophores. To the best of our knowledge, efficient and highly stable energy cascade antenna films that are self-assembled from biological components have not been reported to date. Our design is rational, modular, robust, and functional in the solid state, while its synthesis can be readily scaled-up using abundant, renewable, non-toxic, environmentally benign, and inexpensive components.
The chemical novelty of the current system is that a highly organized protein–DNA matrix provides discrete, single-occupancy binding sites for jumper dyes such that the dyes are packed densely to promote FRET while inhibiting excimer/exciplex formation (Scheme 1). This strategy is in contrast to dyes packed in polymer films, covalently attached to DNA strands, or embedded in other media where the dye–dye interactions can quench excitation propagation and compete with energy transfer.8 Antennas that are obtained via covalent conjugation methods might not function well in the solid phase due to inappropriate positioning of the donor–acceptor dyes in the solid matrix, potentially allowing energy wasting excimer/exciplex formation upon film deposition. Dense population of the binding sites in the current system also assures that there would be one or more acceptors for a given donor within the Förster volume for efficient FRET while avoiding undesirable energy wasting processes.
Results and discussion
Preparation of antenna films
A set of four different dyes was chosen here, such that the emission spectrum of one donor dye had a significant overlap with the absorption spectrum of its corresponding acceptor, an important requirement for FRET. We chose Hoechst 33258 (H) as the highest energy donor, Coumarin 540A (C), which served as an acceptor for H, Fluorescein (F), which served as an acceptor for C, and Rhodamine B (R), which served as an acceptor for F (Scheme 1, ESI, Fig. S1†). The emission spectra of the donors have significant overlaps with the absorption spectra of the corresponding acceptors (Scheme 1), and these pairs are known to undergo efficient FRET.9,10DNA and bovine serum albumin (BSA) are excellent hosts for a number of dyes.11 H binds to DNA (Kb = 1 × 106 M−1) but not to BSA.12 C, F and R bind to BSA. Cationization of BSA (cBSA, ESI, Fig. S2†) promoted its binding to anionic DNA.13 The binding constants of C, F and R to cBSA in solution were determined to be 1.4 × 105, 5.1 × 104, and 3.1 × 104 M−1 respectively (ESI, Fig. S3–S5†), which were comparable to their affinities for BSA.11–14 C, F and R have poor affinities for DNA.
Upon mixing and evaporation of the solvent, these six distinct components spontaneously formed functional, ordered, self-assembled solid-state films. Aqueous buffered solutions of cBSA, R, F, C, salmon sperm DNA, and H were mixed together and drop casted onto glass substrates and air-dried. The dyes are achiral, but on binding to either cBSA or DNA the samples indicated induced circular dichroism (ICD) bands. The ICD bands of the composite films confirmed dye binding to the protein–DNA complex (ESI, Fig. S6†). Even though the dyes had only moderate affinities to their corresponding binding sites in the solution phase, dye binding in the solid state improved substantially because of the increased concentrations of both the dyes (2.5–8 mM) and their host materials (18–50 mM) in the films when compared to their corresponding solution concentrations, which were in the micromolar range. Concentrations of particular components in the film were calculated based on the number of moles of DNA, protein, and dyes used and their corresponding molecular volumes. Assuming the binding affinity was unchanged, our calculations show that the dye binding increased to nearly 99% (ESI, Table S1†) at these higher concentrations simply due to the law of mass action. Optical and electron microscopy of the DNA/BSA/dye films indicated nearly perfect grid patterns (Scheme 1; ESI, Fig. S7†), where the assemblies were highly ordered on millimeter to micrometer length scales. Neither DNA nor cBSA alone formed such ordered assemblies. A detailed description of the mechanism of the assembly is beyond the scope of this work, as we focus here on the FRET process.
Confirmation of FRET
The absorption spectra of the dyes bound to the protein–DNA assemblies in the solution phase (Fig. 1A) and the emission spectra of the individual dyes embedded in the protein–DNA films (350 nm excitation, Fig. 1B) did not indicate dye aggregation. Absorbance of DNA and cBSA did not contribute to these peaks (ESI, Fig. S8†). No excimer or exciplex emission was noted. Excitation energy cascade from H to R was examined by recording emission spectra of the assemblies by exciting at 350 nm, where H absorbs predominantly (Fig. 1A). Intense emission from R was noted at 590 nm (Fig. 1B, black line), and there was a ∼2.4-fold increase in R emission from the composite film when compared to direct excitation of R/DNA/cBSA film (Fig. 1B, purple line). Enhanced emission from R in the antenna complex was due to light absorption by H at 350 nm, followed by energy transfer via the jumper dyes to R. Energy cascade from H to R via the jumper dyes was tested by recording the emission spectra (excitation at 350 nm) of a series of samples where one particular jumper dye was omitted at a time (ESI, Fig. S9†). When R was omitted, for example, intense emission was noted from F. Omission of F led to increased emission of C, as transfer to R was interrupted by the absence of the jumper dye F. When C was omitted, emission from H increased because there was little or no excitation transfer to F or R without the jumper dye C. In every one of these cases, energy transfer from H to R was interrupted when a particular jumper dye was missing. Additionally, emission from the intermediate acceptor appeared when the sequence was broken, which was consistent with energy cascade transfer from H to C to F to R.The energy cascade was confirmed in another set of experiments where a series of excitation spectra (Fig. 1C) were recorded by monitoring R emission at 650 nm. If cascade transfer was operational, then the excitation spectrum of the 4 dye/protein/DNA film when monitored at R emission wavelengths should show excitation peaks corresponding to each dye that directly or indirectly transfers energy to R. The excitation spectrum of the composite film (Fig. 1C, black line) indicated energy transfer from each of these dyes. For comparison, the excitation spectrum of each individual dye bound to the protein/DNA complex is shown (Fig. 1C, as marked), as well as the sum of the individual dye spectra (Fig. 1C, blue dashed line). Regions where the summed spectrum is well below the observed excitation spectrum of the 4 dye-antenna (360–500 nm) indicate efficient energy transfer to R.
FRET efficiency studies
The energy transfer efficiency of the antenna was optimized by adjusting individual dye concentrations. The concentration of a given dye was varied while fixing the concentrations of cBSA, DNA, and all other dyes. Each of these films was excited at 350 nm, and the emission from R at 650 nm was monitored. The intensity of emission at 650 nm was plotted as a function of a specific dye concentration (Fig. 2A), while all other concentrations were kept constant. The overall energy transfer efficiency depended on the highest energy donor concentration, which clearly indicated donor–donor and donor–acceptor FRET. In solution, bimolecular energy transfer depends only on the acceptor concentration – the concentration of donor excited states is too small under the excitation conditions in the fluorimeter. Donor-concentration dependence on FRET was attributed to donor-to-donor energy transfer.15Calculating the efficiency (E) of energy transfer can be difficult, particularly in systems with more than one donor–acceptor pair. As such, many methods of calculation have been reported, which makes comparison of the performance of our system with the others difficult. Examples of such systems from the literature include tobacco mosaic virus coat protein,16 DNA bundles,17 DNA–lipid complexes,18 DNA/protein complexes,13 DNA origami structures,19 and DNA photonic wires.20,21 In the case of tobacco mosaic virus coat protein-3 dye system, the efficiency was reported to be ∼0.90 based on the ratio of donor absorbance to donor excitation.16 In the case of DNA-based antenna system, the efficiency was estimated as that of donor quenching (E = ∼0.90).17 The quenching efficiency of the DNA photonic wire containing four dyes and a luminescent terbium complex was reported to be 0.22 ± 0.06.21 In another study, the efficiency of a DNA–porphyrin system was reported to be 0.55 from Förster theory.18 The efficiency of the DNA origami-4 dye system was reported as the ratio of photons-in to photons-out (E = 0.36 ± 0.17).19 Finally, the efficiency of the DNA based photonic wire with 4 dyes loaded on quantum dots was reported to have an overall efficiency of 0.01–0.08.20 A summary of these works was collected in ESI, Table S2.†
The performance of our antenna system was evaluated by calculating the overall efficiency (E) using the following expression, which was derived in the ESI (S1.6†):
The overall efficiency obtained from the above analysis of our system was 0.23. To the best of our knowledge, our value is comparable to or better than that reported for other 4 dye systems (0.01–0.36).19–21 Factors that control the overall efficiency include dye excited state lifetimes, values of overlap integrals, and the quantum yield of R. Additionally, the overall efficiency of multi-step FRET processes is related to the product of the efficiencies of the individual steps, which necessarily limits the overall efficiency. In the DNA photonic wire system containing a luminescent terbium complex and four dyes, for example, individual FRET events between constituent chromophores were up to ∼80% efficient, but the step-wise nature of the overall process resulted in total efficiency of 0.22 ± 0.06.21 Individual steps could be less than 100% efficient when the excitation of the system is quenched by non-radiative pathways or by emission from an intermediary jumper dye. Emission from intermediary dyes was observed in our DNA/cBSA/four dye system, as demonstrated by the peaks at ∼420 nm and ∼540 nm (Fig. 1B, black line), which correspond to H and F, respectively. Additionally, broad emission was observed between 420 nm and 540 nm. When compared to the emission spectra of films containing DNA, cBSA, and individual dyes, this provided insight into the overall efficiency of the system.
We also examined another important parameter which is useful for applications in the solar cells. Silicon solar cells, for example, are more efficient in the red regions of the solar spectrum than the blue region, and one hypothesis is that the conversion of the blue photons to red could improve cell performance. Therefore, we calculated the conversion of the high energy blue photons (350 nm) to red photons (590 nm) by comparing the emission of the antenna with all the 4 dyes at 590 nm (Fig. 1B, black line, 350 nm excitation) with that of the film consisting of only R (2.5 mM) (Fig. 1B, purple line, 350 nm excitation). The emission intensity at 590 nm by the antenna was greater by a factor of ∼2.4 due to the energy transfer from the three additional dyes present in the film. Thus, the blue-to-red conversion efficiency is also an important characteristic of the current antenna which might be useful for solar cell applications. Similar parameters for other systems are not available for comparison but we speculate that this kind of blue-to-red conversion will be of high practical importance.
Role of matrix components
The roles of DNA and cBSA in the energy cascade mechanism were examined in a number of control experiments. When cBSA was omitted, there was no energy transfer from H to R (ESI, Fig. S11†). Similarly, films made from the four dyes without any DNA and cBSA did not produce any emission. Because the dyes were known to form aggregates at concentrations comparable to or below those studied here,22–24 films containing only DNA/four dyes or four dyes alone provided good evidence that dyes aggregated in the absence of the cBSA matrix. Energy transfer was also not observed when the 4 dyes were bound to protein/DNA in the solution phase (ESI, Fig. S12†), as the concentrations of the dyes bound to protein/DNA were not adequate to promote cascade energy transfer. The charge-conducting properties of DNA could be useful for device fabrication in future studies, where DNA could replace conducting polymers, and hence, we had to test if the antenna would function in the presence of DNA. To our surprise, FRET was noted in cBSA + 4 dye films in the absence of DNA (ESI, Fig. S13†), which demonstrated the importance of the protein scaffold in segregating the dyes and positioning them correctly for FRET, and the presence of DNA did not inhibit FRET in any manner. Additionally, the presence of DNA increased the variety of binding sites available for the self-assembly of additional chromophores, if necessary.Determination of Förster radii
To further characterize the energy transfer, the Foster radii for the donor–acceptor pairs embedded in the protein/DNA films were obtained from the Perrin equation (Φ = Φ0![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) e−VN[Q]),6 where Φ0 and Φ are the quantum yields for donor emission in the absence and presence of acceptor, respectively, N is Avogadro's number, V is the Förster volume, and [Q] is the acceptor concentration. According to this model, the donor emission is quenched only when an acceptor is present within the Förster volume. Emission spectra were collected as a function of acceptor concentration (350 nm excitation) at fixed donor concentration in the protein/DNA films (ESI, Fig. S14–16†). In the absence of R, for example, intense F emission from DNA/cBSA/H/C/F was noted. As R concentration increased, F emission was quenched and R emission was sensitized concomitantly (ESI Fig. S14†). The slope of the plot of ln(Φ0/Φ) vs. [R] gave us an estimate of the Förster radius. Thus, the Förster radii for F–R, F–C and C–H pairs were found to be 58(±1), 45(±2), and 60(±2) Å, respectively. These radii were in good agreement with literature.9,10 Taken together, the data clearly demonstrated energy cascade in the self-assembly.
e−VN[Q]),6 where Φ0 and Φ are the quantum yields for donor emission in the absence and presence of acceptor, respectively, N is Avogadro's number, V is the Förster volume, and [Q] is the acceptor concentration. According to this model, the donor emission is quenched only when an acceptor is present within the Förster volume. Emission spectra were collected as a function of acceptor concentration (350 nm excitation) at fixed donor concentration in the protein/DNA films (ESI, Fig. S14–16†). In the absence of R, for example, intense F emission from DNA/cBSA/H/C/F was noted. As R concentration increased, F emission was quenched and R emission was sensitized concomitantly (ESI Fig. S14†). The slope of the plot of ln(Φ0/Φ) vs. [R] gave us an estimate of the Förster radius. Thus, the Förster radii for F–R, F–C and C–H pairs were found to be 58(±1), 45(±2), and 60(±2) Å, respectively. These radii were in good agreement with literature.9,10 Taken together, the data clearly demonstrated energy cascade in the self-assembly.
The above observations can be readily explained in terms of the number densities of acceptors present within the Förster volume. For maximum efficiency, each Förster volume element containing a donor molecule should also have at least one acceptor. As a first approximation, the total volume of the solid film was calculated to be equal to the sum of the volumes of all the dry components; dyes, DNA, cBSA, and buffer. The occupancy number of each kind of dye molecule within the average Förster volume was calculated to be 0.71 for H, 1.8 for C, 1.4 for F, and 0.57 for R, corresponding to 1.55 mM H, 3.85 mM C, 3.1 mM F, 1.25 mM R, 49.2 mM DNA and 18.5 mM cBSA (Fig. 2B). Thus, the dye occupancies within the quenching cells were in the range of 0.57 to 1.8, with an average occupancy >1, which supported the above observations of efficient FRET in the supramolecular films. There was good probability that each of the quenching cells would have at least one primary donor (H) or one acceptor (R) dye (Scheme 2). While this does not ensure simultaneous residency of both a donor and an acceptor within the same quenching cell, it enhances the chances for such an arrangement.
Thermal stability
Since the practical utility of any solar device would require high thermal stability over a long time, we examined the stabilities of these protein/DNA/dye films and monitored their emission after heating to elevated temperatures. Using 80–85 °C as a benchmark for the thermal stability of a solar device in the field, we examined FRET as a function of heating at this temperature in ambient air. After specific time intervals, samples were cooled to room temperature for 3 h, emission/excitation spectra were collected (Fig. 2C), and the samples were returned to the oven. The emission intensities and excitation spectra showed little to no change over 0–169 days, and only small decreases were noted after 169 days. It is likely that cBSA or DNA was affected by storage at this temperature, as the films became insoluble after some time, and circular dichroism analysis of cBSA and DNA structure could not be completed. This was potentially because the elevated temperature promoted the formation of new amide bonds or disulfide exchange between neighbouring cBSA molecules, yielding an insoluble film. Regardless of the structural integrity of the biomolecules, however, the antenna complex was still functional, and this extraordinary thermal stability coupled with high efficiency of FRET is novel.Conclusions
The biomaterial based artificial antenna complexes described here were inspired by the naturally occurring photosynthetic apparatus, which is essentially edible. Inspired by the ultimate environmental compatibility of the natural system composed entirely of biological components, we attempted to mimic nature to produce naturally biodegradable components to form part of a bio-solar cell. This progress is significant, as the non-natural components such as silicon could readily accumulate in the environment or require the use of complex chemistry that produces toxic by-products. Thus, the use of the biological host materials for cell construction via self-assembly can be justified.The use of artificial protein/DNA complexes as scaffolds also allowed for a large selection of dyes that were known to bind either to DNA or to the protein at distinct, discrete, single occupancy binding sites. If the dyes were arranged too closely or aggregated, excitation would be quenched and wasted away. If the donors and acceptors were too far apart, on the other hand, there would be little to no FRET.8
Natural systems overcome this problem of adjusting the distances while preventing dye aggregation by organizing the chromophores in protein scaffolds, which enable them to increase local concentrations of the participating chromophores.25 Nature also utilizes self-assembly of the required components to form complex but functional structures which are cheap to produce but also amenable to self-repair. Similarly, the protein/DNA scaffolds developed here organized the dyes at moderately high (2.5–8 mM) concentrations via self-assembly. Since all components are self-assembled according to their respective binding affinities and bind selectivity to particular sites in the matrix, complex synthetic efforts are not required, thereby reducing the overall cost of manufacturing.
The emission and excitation spectra clearly demonstrated that low stoichiometric ratios of donors and acceptors were sufficient to achieve relatively high efficiencies. For example, for every H, one R was sufficient to produce multi-step transfer via jumper dyes. The intermediary dye compositions (C and F) were greater than 1, but considerably less than required for diffusion mediated energy transfer. Omission of specific dyes halted the excitation at particular steps, which provided direct evidence for energy cascade. The data also indicated parallel transfer pathways, and followed the well-established Perrin formalism. The ultra-high stability of the current antennas (at 80–85 °C in air, 169 days) is also important for practical applications.
When compared to other artificial antennas composed of biological materials, our system offers a number of advantages (ESI, Table S2†). First, we demonstrated efficient FRET in the solid phase, a key property for solar cell applications. Second, long term thermal stability was demonstrated, while the viability of other protein or DNA based artificial antennas under similar conditions has not been demonstrated. Third, the efficiency of FRET in the current system was comparable to or better than other four dye systems (E = 0.01–0.36), as indicated by the overall efficiency of 0.23 and the ∼2.4-fold increase in the blue to red conversion when compared to direct excitation of R in DNA and BSA. Finally, our antenna requires minimal covalent chemistry, and the protein is a waste product from the meat industry. The antenna system described here was self-assembled from inexpensive, Earth abundant and biodegradable components by drop casting onto glass. Taken together, these data represent an initial but important first step towards the design of biological solar cells.
Experimental
Materials
Bovine serum albumin (BSA) was purchased from Equitech-Bio, Inc (Texas) and salmon sperm DNA sodium salt was purchased from Amresco (Ohio). Hoechst 33258 (H) and Fluorescein (F) were purchased from Sigma (Milwaukee, Wisconsin). Coumarin 540A (C) was purchased from Exciton Chemical Co. Inc., (Ohio) and Rhodamine B (R) was purchased from Eastman Kodak Company (Rochester, NY). 1-Ethyl-3-(3-dimethylaminopropyl)carbodiimide (EDC) was purchased from TCI America (Portland, OR). Glass cover slips of 22 mm × 22 mm were purchased from Fisher Scientific (Atlanta, GA).Methods
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 MW cut off) from Millipore Inc. (Bedford, MA) until final concentration reached 1.5–2.0 mM. The chemical modification of BSA was confirmed by agarose gel electrophoresis (ESI, S1.1†).
000 MW cut off) from Millipore Inc. (Bedford, MA) until final concentration reached 1.5–2.0 mM. The chemical modification of BSA was confirmed by agarose gel electrophoresis (ESI, S1.1†).ln![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Φ0/Φ = VN[Q] Φ0/Φ = VN[Q] |
For example, the Förster radius of Fluorescein (F) and Rhodamine B (R) was determined by systematically varying R film phase concentration from 0 to ∼10 mM and recording the emission spectra. Perrin plots were constructed by plotting ln![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Φ0/Φ vs. [R]. From the slopes of the plots, quenching volumes were extracted and quenching radii determined. Other quenching radii were obtained similarly.
Φ0/Φ vs. [R]. From the slopes of the plots, quenching volumes were extracted and quenching radii determined. Other quenching radii were obtained similarly.
Acknowledgements
We acknowledge NSF EAGER grant (DMR-1441879) for financial support of this work. CVK thank the Fulbright Foundation for supporting the visit to the Indian Institute of Science Bengaluru, India and thanks Professor P. K. Das for useful discussions during the visit.Notes and references
- N. S. Lewis, Science, 2007, 315, 798–801 CrossRef CAS PubMed.
- N. S. Lewis and D. G. Nocera, Proc. Natl. Acad. Sci. U. S. A., 2006, 103, 15729–15735 CrossRef CAS PubMed.
- G. McDermott, S. Prince, A. Freer, A. M. Hawathornthwaite-Lawless, R. J. Papiz and N. W. Isaacs, Nature, 1995, 374, 517–521 CrossRef CAS PubMed.
- F. Odobel, Y. Pellegrim and J. Warnan, Energy Environ. Sci., 2013, 6, 2041–2052 CAS.
- C. Strumpel, M. McCann, G. Beaucarne, V. Arkhipov, A. Slaoui, V. Svrcek, C. D. del Canizo and I. Tobias, Sol. Energy Mater. Sol. Cells, 2007, 91, 238–249 CrossRef CAS PubMed; T. Trupke, M. A. Green and P. Wurfel, J. Appl. Phys., 2002, 92, 1668–1674 CrossRef PubMed.
- N. J. Turro, Modern Molecular Photochemistry, Benjamin/Cummings, Menlo Park, CA, 1978, p. 296 Search PubMed.
- L. O. Andersson, A. Rehnström and D. L. Eaker, Eur. J. Biochem., 1971, 20, 371–380 CrossRef CAS PubMed.
- S. Prasad, K. H. Ibnaouf, M. S. AlSalhi and V. Masilamani, Polymer, 2014, 55(3), 727–732 CrossRef CAS.
- Y. O. Posokhov, M. Merzlyakov, K. Hristova and A. S. Ladokhin, Anal. Biochem., 2008, 380, 134–136 CrossRef CAS PubMed.
- E. Johansson, E. Choi, S. Angelos, M. Liong and J. I. Zink, J. Sol-Gel Sci. Technol., 2008, 46, 313–322 CrossRef CAS.
- C. V. Kumar and M. R. Duff, Photochem. Photobiol. Sci., 2008, 7(12), 1522–1530 CAS.
- P. E. Pjura, K. Grzeskowiak and R. E. Dickerson, J. Mol. Biol., 1987, 197, 257–271 CrossRef CAS.
- C. V. Kumar and M. R. Duff, J. Am. Chem. Soc., 2009, 131, 16024–16026 CrossRef CAS PubMed.
- J. Zhu, J. J. Li and J. W. Zhao, Sens. Actuators, B, 2009, 138, 9–13 CrossRef CAS PubMed.
- C. V. Kumar, A. Chaudhari and G. L. Rosenthal, J. Am. Chem. Soc., 1994, 116, 403–404 CrossRef CAS.
- R. A. Miller, A. D. Presley and M. B. Francis, J. Am. Chem. Soc., 2007, 129, 3104–3109 CrossRef CAS PubMed.
- P. K. Dutta, R. Varghese, J. Nangreave, S. Lin, H. Yan and Y. Liu, J. Am. Chem. Soc., 2011, 133, 11985–11993 CrossRef CAS PubMed.
- J. G. Woller, J. K. Hannestad and B. Albinsson, J. Am. Chem. Soc., 2013, 135, 2759–2768 CrossRef CAS PubMed.
- I. H. Stein, C. Steinhauer and P. Tinnefeld, J. Am. Chem. Soc., 2011, 133, 4193–4195 CrossRef CAS PubMed.
- C. M. Spillmann, M. G. Ancona, S. Buckhout-White, W. R. Algar, M. H. Stewart, K. Susumu, A. L. Huston, E. R. Goldman and I. L. Medintz, ACS Nano, 2013, 7(8), 7101–7118 CrossRef CAS PubMed.
- M. Massey, M. G. Ancona, I. L. Medintz and W. R. Algar, ACS Photonics, 2015, 2, 639–652 CrossRef CAS.
- S. Das, A. P. Chattopadhyay and S. de, J. Photochem. Photobiol., A, 2008, 197, 402–414 CrossRef CAS PubMed.
- N. O. Mchedlov-Petrosyan and Y. V. Kholin, Russ. J. Appl. Chem., 2004, 77(3), 414–422 CrossRef CAS.
- P. Verma and H. Pal, J. Phys. Chem. A, 2014, 118(34), 6950–6964 CrossRef CAS PubMed.
- G. D. Scholes, G. R. Fleming, A. Olaya-Castro and R. van Grondelle, Nat. Chem., 2011, 3, 763–774 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available: Including experimental methods, characterization of modified BSA, fluorescence, circular dichroism, etc. See DOI: 10.1039/c5ra14208c |
| This journal is © The Royal Society of Chemistry 2015 |